Introduction
Systemic autoimmune diseases result from interactions between genes and environmental triggers that build up overtime until clinical symptoms appear. A complex interplay between innate and adaptive immunity lies at the core of most of these diseases. This interplay is not static, as initial inflammatory cascades might change as organ damage accumulates. Furthermore, these diseases can be heterogeneous regarding the type of organs involved, clinical course and response to treatment.
The breakthrough discovery of TNF as a major player in rheumatoid arthritis (RA) [1, 2] has fueled the development of novel treatments for systemic autoimmune diseases, including monoclonal antibodies and fusion proteins targeting cytokines, as well as small molecules targeting downstream inflammatory pathways. While efficacious therapies are becoming available for many of these diseases, their underlying pathogenesis is not yet fully understood. Thus, 30–40% of RA, psoriasis and inflammatory bowel disease (IBD) patients do not respond to TNF blockade, even though the clinical presentation of non-responders is identical to that of patients achieving complete remission with this therapy. Altogether, there is a need to characterize disease pathogenesis at the individual level to predict the best treatment strategies. As many of these diseases follow a remitting and relapsing course, reliable biomarkers to predict outcome and flares also have to be identified.
Systems biology approaches enable the measurement of thousands of parameters at the genetic, transcriptional, epigenetic, protein and metabolite levels in accessible tissues such as blood, urine, synovial fluid, saliva and biopsy specimens. Previous studies in blood leukocytes and tissues have demonstrated the applicability of genome-wide microarrays to characterize molecular networks involved in cancer [3, 4], infection [5, 6], autoimmunity [7] and response to vaccination [8, 9]. Herein, we review recent developments, challenges and promising avenues in the use of systems approaches to characterize human systemic autoimmune and autoinflammatory diseases.
Prevalent and emerging systems approaches
Systems biology uses a combination of high-throughput and targeted approaches to measure the organization and dynamics of a system at the DNA, RNA and protein levels (Table 1).
Table 1.
Current and upcoming technologies
| Technology/Method | Assay Type | Target | Overview |
|---|---|---|---|
| SNP Arrays | High-throughput | DNA | DNA microarrays used to identify single nucleotide polymorphisms in populations; Widely used in GWAS to identify disease susceptibility loci |
| Sanger sequencing | Targeted | DNA | DNA sequencing method that relies on incorporation of dideoxynucleotides by DNA polymerase during DNA replication |
| "Next-gen" DNA sequencing | High-throughput | DNA | Sequences small DNA fragments in parallel, producing millions of overlapping reads that can be aligned computationally to assemble complete genomic sequences |
| DNA Microarrays | High-throughput | mRNA | Relatively quantifies mRNA levels through hybridization to cDNA probes |
| RNA-seq | High-throughput | RNAs | Quantifies RNA levels, including non-coding RNAs; Identifies splicing variants |
| Global Run-On sequencing (GRO-seq) | High-throughput | RNA polymerase | Maps the position, quantity and orientation of transcriptionally engaged RNA polymerase across the genome |
| Native Elongating Transcript sequencing (NET-seq) | High-throughput | nascent RNA | Monitors transcription at nucleotide resolution |
| Ribo-seq | High-throughput | ribosome-bound mRNA | Quantifies ribosome-bound mRNA |
| Chromatin Immuno-Precipitation sequencing (ChIP-seq) | High-throughput | protein/DNA interactions sequences | Combines chromatin immuno-precipitation with massively parallel sequencing; Used to identify regions of DNA/protein interactions, transcription regulators and chromatin modifications |
| Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE-seq) | High-throughput | chromatin/DNA | Sequencing of open regions of chromatin sensitive to folmaldehyde. Identifies DNA sequence enriched for DNAse binding sites, transcription initiation sites and active promoters |
| DNAse-seq | High-throughput | DNAse sensitive DNA regions | Sequencing of DNA regions sensitive to cleavage by DNAse I |
| Nanostring nCounter | Targeted | RNAs | Quantifies RNA and DNA levels using a barcoding approach for up to 800 different molecules at a time, without amplification steps |
| High-throughput real-time PCR platforms (OpenArray®, Fluidigm Biomark™…) | Targeted | RNAs | Real-time PCR platforms for up to 10,000 transcripts at at time; Can measure both mRNA and small non-coding RNAs |
| Protein modification arrays | targeted | Proteins | Include methylation and phosphorylation arrays; Enable the study of protein dynamics in signaling cascades and transcription regulation |
| Microsphere-based protein quantification (Luminex®) | Targeted | Proteins | Multiplexed color-codes microspheres for detection of cytokines and signal transduction proteins |
| Cytometry by Time-Of-Flight (CyTOF) | Targeted | Proteins | Cytometry using transition element isotope-labeled antibodies and quantified by mass spectrometry |
| Shotgun proteomics | High-throughput | Proteins | Combines high performance liquid chromatography with mass spectrometry for unbiased protein identification |
Over the last decade, genome-wide microarrays have been extensively used to identify transcriptional alterations in peripheral blood mononuclear cells (PBMC), whole blood or peripheral tissues from patients with systemic and organ-specific autoimmunity [7]. The advent of high-throughput sequencers is now revolutionizing the genomics field. An individual genome can now be cost-effectively sequenced at the exome level or in its entirety. As reviewed below, this is resulting in the identification of novel genes/pathways driving human inflammatory diseases. RNA-seq is quickly replacing DNA microarrays to measure transcriptional profiles, as it provides a more quantitative measure of messenger and non-coding RNAs and can detect splicing variants. Sequencing has enabled a more refined understanding of the dynamics of transcription through GRO-seq, NET-seq and Ribo-seq. It has also been applied to characterize DNA-protein interaction sites and histone modifications, using ChIP-seq, FAIRE-seq and DNAse-seq [10]. More recently, targeted sequencing assays have been developed to characterize the variable CDR3 regions of T and B cell receptor genes, which helps monitoring minimal residual disease in cancer [11, 12].
In addition, several technologies for targeted cost-effective assays are now routinely used to quantify mRNA. These include Nanostring®, OpenArray®, and Fluidigm Biomark™ [13]. Increased sensitivity and coverage of the transcriptome, combined with reduction in cost and processing time, make these targeted assays suitable for clinical applications.
Genome-wide association studies (GWAS), which use single-nucleotide polymorphism (SNP) arrays to identify genetic variants associated with disease traits in large patient populations, have been conducted for many autoimmune diseases, [14–19]. To further probe into the functional role of susceptibility loci, the results from GWAS have been combined with transcriptional profiling and/or functional assays at the protein level. Expression quantitative trait loci (eQTLs and reQTLs) take advantage of high-throughput exome and RNA sequencing technologies to identify genomic loci that regulate the expression of mRNA or proteins during homeostasis or in response to stimuli [20]. GWAS have also been combined with co-expression network-based association studies and were recently reviewed [21].
mRNA expression regulation both pre and post-transcriptionally involves epigenetic mechanisms, including DNA methylation [22], histone modifications [23], non-coding regulatory RNAs such as microRNAs [24] and recently discovered circular non-coding RNAs [25, 26]. High-throughput technologies such as methylation arrays, microRNA arrays, and high-throughput PCR platforms have helped characterize mRNA regulation mechanisms, especially in cancer [27]. In addition, new platforms for RNA knockdown in vitro have been developed and applied to primary cell populations [28], leading to increased understanding of transcriptional networks regulation.
At the protein and cellular levels, significant advances were made in both targeted and hypothesis-free proteomics assays. Flow cytometry by time-of-flight (CyTOF®), which combines traditional cytometry with proteomic readouts, can query more than 35 cell markers at once [29–31]. High-throughput phosphoproteomics platforms have enabled the characterization of signaling cascades in response to pathogens [32] and pathway inhibitors [33]. Soluble components of the immune response are routinely analyzed using Luminex xMAP® technology. Continuous advances in protein and peptide microarrays allow for large-scale interrogation of human tissues and biological fluids for biomarkers, autoantigens and drug targets [34, 35]. Finally, shotgun proteomics, which do not require a priori knowledge of the protein composition of a sample, represent a promising approach for unbiased system-level protein analysis [36]. These advances in proteomics have enabled the development of metabolomics, the systems-level analysis of metabolites, which connect biochemistry and cellular phenotype [37].
We next review recent applications of some of these approaches to the study of human systemic autoimmune and autoinflammatory diseases.
GWAS and Beyond
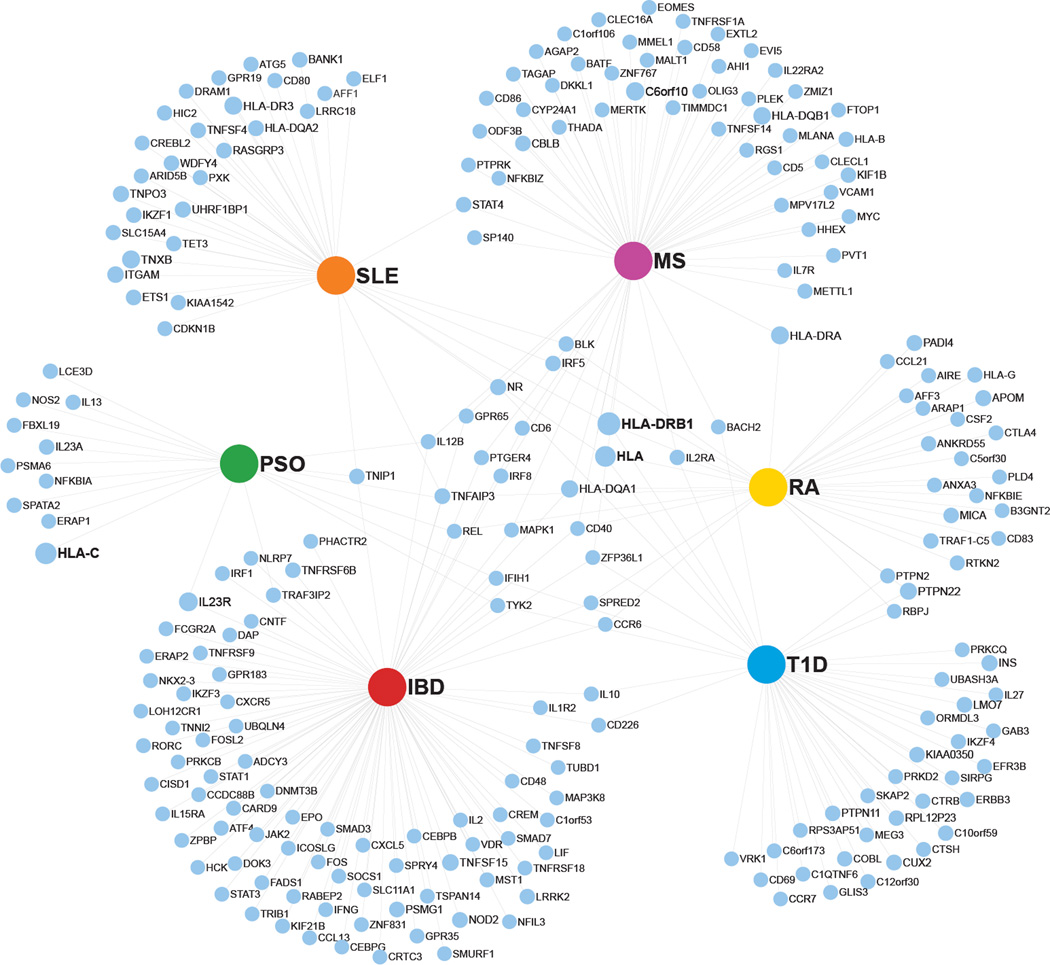
Over the last decade, GWAS have identified many alleles associated with systemic autoimmune diseases [38, 39]), including ≥50 in SLE [40] and RA [41], and 36 in psoriasis [42] (Figure 1). Interestingly, HLA variants remain at the top of the susceptibility list for all these diseases. A group of non-HLA alleles encoding molecules involved in well-defined immune activation and regulatory pathways, such as IL12B, IL2R and CD40 confer predisposition to several of these diseases as well (Figure 1).
Figure 1. Genes with risk-associated loci identified by GWAS in 6 autoimmune diseases.
Network displaying odd ratios of autoimmune disease occurence given single nucleotide polymorphisms (SNPs) in specific genes. The major hubs represent 6 autoimmune diseases (IBD: Inflammatory Bowel Disease; MS: Multiple Sclerosis; PSO: psoriasis; RA: Rheumatoid Arthritis; SLE: Systemic Lupus Erythematosus; T1D: Type I Diabetes). Each associated gene is represented by a blue dot of diameter proportional to the odds ratio (OR). The data presented herein is publicly available at http://www.genome.gov/gwastudies/index.cfm?pageid=26525384 - searchForm. Risk bearing loci without associated OR and with a p > 5×10−8 were filtered out.
Alterations in TLR and type I interferon (IFN) pathways were first noticed in diseases such as SLE, Sjögren’s Syndrome (SS) and inflammatory myopathies through blood transcriptional profiling [7]. Common variants in genes within these pathways have now been confirmed through GWAS (i.e. IRF5, IRF7, IRAK1, TNFAIP3, TNIP1, IFIH1 and TYK2) and seem to predispose to both systemic and organ-specific autoimmunity [43]. GWAS in combination with functional studies identified a variant of TNFRSF1A in MS patients [44] that might explain why TNF blockers promote MS onset and/or exacerbations.
GWAS have identified allelic variants related to autophagy, a key mechanism in degradation and recycling of large cellular components, in patients with Crohn’s Disease (CD). A variant in the autophagy-related ATG16L1 gene results in impaired regulation of the inflammasome molecules CASP1, IL-1β and IL-18 [45, 46]. Autophagy is also involved in the presentation of citrullinated self-peptides by antigen-presenting cells (APC) [47], which may contribute to autoimmunity in RA. Finally, variants in ATG5, which participates in IFN production in plasmacytoid dendritic cells (pDCs) [48], have been associated with SLE [49], although their pathogenic role remains to be demonstrated.
Deep targeted exome resequencing of susceptibility loci identified by GWAS has helped refine the identification of putative disease-relevant variants, as supported by the discovery of new NOD2-risk conferring and IL23R-protective alleles in patients with IBD [50, 51].
Monogenic diseases shed light on polygenic diseases
GWAS studies require large cohorts of patients, and are therefore not well suited for the study of rare disorders. Studies of monogenic diseases have relied on linkage association studies, SNP mapping and more recently exome or whole-genome sequencing. Monogenic disease-causing variants were long identified within the complement system in familial cases of SLE, within the apoptotic pathway in autoimmune lymphoproliferative syndrome (ALPS) (FAS, FASL, CASP10) and within key regulators of immune tolerance in organ-specific autoimmune diseases (e.g. AIRE mutation in APS1, FOXP3 mutation in IPEX syndrome) [52].
More recently, mutations in genes involved in IL-1β regulation and nucleic acid degradation have been identified as the cause of monogenic “autoinflammatory diseases” [53] and “interferonopathies” [54], respectively. Aicardi-Goutieres syndrome (AGS) is a monogenic interferonopathy manifesting as an encephalopathy that resembles congenital infection. AGS patients have extraneurological features overlapping with lupus and elevated cerebrospinal fluid IFNα levels at birth. Genes mutated in AGS are involved in cytosolic nucleic acid processing or removal. Among them, TREX1 encodes a cytoplasmic exonuclease that when deleted in mice results in cell-intrinsic production of type I IFN and autoimmunity [55]. SAMHD1 participates in retroviral control through hydrolysis of the cellular dNTPs required for viral reverse transcriptase activity [56, 57]. ADAR1 is involved in post-transcriptional dsRNA editing and miRNA-mediated gene silencing when complexed with Dicer [58, 59]. Patients with Spondyloenchondrodysplasia with Immune Dysregulation (SPENCD) have mutations in ACP5, which encodes the tartrate-resistant acid phosphatase (TRAP). They display skeletal dysplasia and SLE manifestations together with elevated blood IFNα levels [60, 61]. The information gathered through the study of these diseases is leading to novel therapeutic strategies. For example, Trex1−/− mice develop inflammatory myocarditis that responds to a cocktail of reverse transcriptase inhibitors, opening the door for this type of approach in AGS patients [62].
Exome sequencing has successfully been applied to the study of monogenic disorders combining autoinflammation and immunodeficiency. For example, a mutation in HOIL1, results in defects in the linear ubiquitination chain assembly complex (LUBAC), leading to aberrant IL-1β responses. Paradoxical coexistence of inflammation and immunodeficiency was further explained through functional assays with transcriptome read-outs, where patient fibroblasts displayed reduced response to IL-1β combined with hyper-inflammatory phenotype [63]. Deletions in PLCG2, which encodes the phospholipase Cγ2, a molecule involved in signaling within B cells, NK cells and mast cells [64], also lead to autoinflammation and immunodeficiency. Cells from patients displayed decreased signaling at 37°C, but increased signaling at sub-physiological temperatures. A PLCG2 missense mutation also leads to an inflammatory phenotype, but increased leukocyte phospholipase Cγ2 activity is present at physiological temperatures, autoimmune manifestations are absent and organ involvement different [65]. The latter finding highlights the complexity of gene structural/functional correlates even within the context of monogenic diseases.
The relevance of rare mutants to improve our understanding of more common autoimmune and inflammatory disease pathogenesis is striking. As 50% of AGS patients develop lupus symptoms, TREX1 variants were sought after and found in up to 2% of patients with non-familial SLE, revealing the importance of the cytosolic nucleic acid sensing pathway in this disease [66].
A new frontier in profiling the autoimmune transcriptome
Pioneer microarray studies of patient PBMCs and whole blood were instrumental in revealing dysregulated IFN pathways in SLE, SS, inflammatory myopathies and a subset of Systemic Sclerosis (SSc) patients. They also led to the discovery of IL-1 dysregulation and response to IL-1 blockade in systemic-onset Juvenile Idiopathic Arthritis (sJIA) [7]. Microarray-based studies in MS revealed transcriptional responses to IFNβ1α [67, 68] and enabled the stratification of patients according to disease activity [69].
RNA-seq is bringing our understanding of the transcriptional perturbations involved in autoimmunity to a new level. Two thirds of genes associated with T1D in GWAS were found expressed in human pancreatic islets using this technique [70]. RNA-seq of lesional and nonlesional psoriatic skin identified a number of differentially expressed transcripts previously undetected by microarray. A subset of them is synergistically upregulated in keratinocytes by the combination of IL-17 and TNFα, supporting the relevance of these cytokines in psoriasis pathogenesis [71].
A major contribution of RNA-seq is the characterization and quantification of disease-specific splice variants. Novel IRF5 gene isoforms, for example, have been identified using this technology as uniquely transcribed in SLE [72]. These variants can be now used to better stratify SLE patients according to risk haplotypes. Similarly, RNA-seq analysis of synovial fibroblasts identified novel genes and especially new isoforms previously not associated with RA and not detected by microarray [73]. Although larger studies need to be performed, these reports highlight the potential wealth of information that can be obtained with sequencing technologies.
Epigenetic control of autoimmunity
Protein synthesis is controlled pre-transcriptionally through epigenetic mechanisms, and post-transcriptionally by various families of non-coding RNAs. Genome-wide DNA methylation patterns have been characterized by high-throughput methylation arrays in monozygotic twins displaying disease discordance [74] for SLE [75], psoriasis [76], T1D [77] and RA [78]. These epigenetic marks could serve as additional biomarkers of disease risk. Histone modifications have been studied in the context of autoimmune inflammatory programs, with the identification of di-methylation of histone H3 at lysine 9 as a suppressor of the IFN response [79]. This suggests that epigenetic-modifying drugs could be used in the treatment of IFN-associated disorders. In type 2 diabetes, FAIRE-Seq mapping of open chromatin in human pancreatic islets linked chromatin structure to the disease-associated variant TCF7L2 [80]. This approach might help understand epigenetic factors involved in autoimmune diseases in the future.
miRNAs are small non-coding RNAs that repress translation of target mRNAs and may be major regulators of inflammatory pathways driving autoimmunity. Among the many miRNAs found modulated in autoimmune disorders, decreased expression of miR-146a might explain the increased type I interferon response observed in SLE [81]. miR-146a deletion in mice, on the contrary, results in increased TNFα and IL-6 production [82]. Interestingly, miR-146 is over-expressed in the synovial tissue [83, 84] and PBMCs [85] from RA patients, potentially as negative feedback to excess TNF signaling. miR-23b, which is down-regulated in inflammatory lesions of SLE and RA patients, acts as a regulator of inflammation by suppressing IL-17, TNFα and IL-1β induced NF-κB activation [86]. Conversely, miR-155 acts as a major inducer of inflammation. Its over-expression in the synovial fluids of RA patients was associated with the down-regulation of its target SHIP-1, an inhibitor of TNFα production [87]. miRNAs can also be used as biomarkers in various tissues/fluids. For example, their detection in the plasma and urine of patients with SLE [88] and T1D nephropathies [89] respectively provide disease- and stage-specific diagnostic signatures. Our understanding of the role of miRNA in the context of autoimmunity will benefit from integrative approaches that simultaneously quantify mRNA and genome-wide protein output by SILAC-based proteomics after miRNA introduction or knockout in cultured cells [90, 91].
Challenges and concluding remarks
Systems biology is rapidly generating large quantities of novel information in human autoimmune diseases at the genetic, transcriptional, protein and metabolic levels. Besides the raw computing capacity required for data storage and processing, which calls upon the dynamic and dematerialized capacities of the cloud, there is also a need for data management solutions [92], analysis software and novel statistical methods that will enable data interpretation on a true “systems” level.
Studies of complex diseases also need to address heterogeneity in the outbred human population, interaction with the environment, disease stages, and effects of treatments, prompting for careful experimental designs. When studying rare diseases, collaborations and consortia should be fostered to provide access to well-annotated samples, shared databases, and standardized protocols for relevant assays.
Systems biology is leading to novel discoveries in biology, medicine and pharmacotherapy. It is doing so by combining high throughput assays that generate functionally testable hypothesis in vitro and in animal models. This multi-disciplinary approach is promoting a change in culture from individual data accumulation to data and knowledge sharing across the scientific community [93, 94], leading to accelerated understanding of disease, biomarker identification and drug development.
Acknowledgements
We acknowledge support from NAID Autoimmunity Centers of Excellence (U19 AUIO82715) and the Human Immunology Project Consortium (5U19AI089987). We thank Alexander Parfenov for help with network representation of GWAS data.
References
- 1.Maini RN, et al. TNF-alpha in rheumatoid arthritis and prospects of anti-TNF therapy. Clin Exp Rheumatol. 1993;11(Suppl 8):S173–S175. [PubMed] [Google Scholar]
- 2.Elliott MJ, et al. Treatment of rheumatoid arthritis with chimeric monoclonal antibodies to tumor necrosis factor alpha. Arthritis Rheum. 1993;36(12):1681–1690. doi: 10.1002/art.1780361206. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banchereau R, et al. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One. 2012;7(4):e34390. doi: 10.1371/journal.pone.0034390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascual V, Chaussabel D, Banchereau J. A genomic approach to human autoimmune diseases. Annu Rev Immunol. 2010;28:535–571. doi: 10.1146/annurev-immunol-030409-101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentebibel SE, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med. 2013;5(176):176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10(1):116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soon WW, Hariharan M, Snyder MP. High-throughput sequencing for biology and medicine. Mol Syst Biol. 2013;9:640. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grupp SA, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter DL, et al. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin H, Marshall D. Microfluidics for single cell analysis. Curr Opin Biotechnol. 2012;23(1):110–119. doi: 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke A, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL) Nat Genet. 2010;42(4):292–294. doi: 10.1038/ng.553. [DOI] [PubMed] [Google Scholar]
- 16.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patsopoulos NA, et al. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol. 2011;70(6):897–912. doi: 10.1002/ana.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gat-Viks I, et al. Deciphering molecular circuits from genetic variation underlying transcriptional responsiveness to stimuli. Nat Biotechnol. 2013;31(4):342–349. doi: 10.1038/nbt.2519. Identification of responsiveness quantitative trait loci involved in DC response to microbial components in inbred mice
- 21.Califano A, et al. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44(8):841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 23.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20(3):259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 24.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 25.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 26. Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. Identification of circular RNAs as regulators of microRNA expression
- 27.Volinia S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20(5):589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shalek AK, et al. Nanowire-mediated delivery enables functional interrogation of primary immune cells: application to the analysis of chronic lymphocytic leukemia. Nano Lett. 2012;12(12):6498–6504. doi: 10.1021/nl3042917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maecker HT, et al. New tools for classification and monitoring of autoimmune diseases. Nat Rev Rheumatol. 2012;8(6):317–328. doi: 10.1038/nrrheum.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. Application of flow cytometry by mass spectrometry to dissect cell marker expression in hematopoietic cells at rest and in response to stimuli
- 31.Newell EW, et al. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chevrier N, et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 2011;147(4):853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bodenmiller B, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30(9):858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price JV, et al. On silico peptide microarrays for high-resolution mapping of antibody epitopes and diverse protein-protein interactions. Nat Med. 2012;18(9):1434–1440. doi: 10.1038/nm.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabakman SM, et al. Plasmonic substrates for multiplexed protein microarrays with femtomolar sensitivity and broad dynamic range. Nat Commun. 2011;2:466. doi: 10.1038/ncomms1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabido E, Selevsek N, Aebersold R. Mass spectrometry-based proteomics for systems biology. Curr Opin Biotechnol. 2012;23(4):591–597. doi: 10.1016/j.copbio.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baranzini SE. The genetics of autoimmune diseases: a networked perspective. Curr Opin Immunol. 2009;21(6):596–605. doi: 10.1016/j.coi.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sui W, et al. The applied basic research of systemic lupus erythematosus based on the biological omics. Genes Immun. 2013;14(3):133–146. doi: 10.1038/gene.2013.3. [DOI] [PubMed] [Google Scholar]
- 41.Viatte S, Plant D, Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nat Rev Rheumatol. 2013;9(3):141–153. doi: 10.1038/nrrheum.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44(12):1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bronson PG, et al. The genetics of type I interferon in systemic lupus erythematosus. Curr Opin Immunol. 2012;24(5):530–537. doi: 10.1016/j.coi.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 44. Gregory AP, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488(7412):508–511. doi: 10.1038/nature11307. Identification of a risk allele for TNFR1 in multiple sclerosis, leading to a truncated form of the TNF receptor that acts as a soluble TNF antagonist
- 45.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456(7219):264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 46.Plantinga TS, et al. Crohn's disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011;60(9):1229–1235. doi: 10.1136/gut.2010.228908. [DOI] [PubMed] [Google Scholar]
- 47.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med. 2011;208(13):2625–2632. doi: 10.1084/jem.20110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HK, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32(2):227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.International Consortium for Systemic Lupus Erythematosus, G et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40(2):204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43(11):1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Momozawa Y, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet. 2011;43(1):43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 52.Cheng MH, Anderson MS. Monogenic autoimmunity. Annu Rev Immunol. 2012;30:393–427. doi: 10.1146/annurev-immunol-020711-074953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aksentijevich I, Kastner DL. Genetics of monogenic autoinflammatory diseases: past successes, future challenges. Nat Rev Rheumatol. 2011;7(8):469–478. doi: 10.1038/nrrheum.2011.94. [DOI] [PubMed] [Google Scholar]
- 54.Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci. 2011;1238:91–98. doi: 10.1111/j.1749-6632.2011.06220.x. [DOI] [PubMed] [Google Scholar]
- 55. Stetson DB, et al. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. Identification of Trex1 as a regulator of IFNα highlighted by increased IFNα production and autoimmunity in Trex1−/− mice and patients with Aicardi-Goutieres syndrome
- 56.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 57.Lahouassa H, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13(3):223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ota H, et al. ADAR1 Forms a Complex with Dicer to Promote MicroRNA Processing and RNA-Induced Gene Silencing. Cell. 2013;153(3):575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol. 2012;13(12):252. doi: 10.1186/gb-2012-13-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Briggs TA, et al. Tartrate-resistant acid phosphatase deficiency causes a bone dysplasia with autoimmunity and a type I interferon expression signature. Nat Genet. 2011;43(2):127–131. doi: 10.1038/ng.748. Identification of ACP5 mutations in SPENCD patients leading to the loss of the TRAP protein, elevated serum IFNα and type I IFN signature comparable to the one in SLE patients
- 61.Lausch E, et al. Genetic deficiency of tartrate-resistant acid phosphatase associated with skeletal dysplasia, cerebral calcifications and autoimmunity. Nat Genet. 2011;43(2):132–137. doi: 10.1038/ng.749. [DOI] [PubMed] [Google Scholar]
- 62.Beck-Engeser GB, Eilat D, Wabl M. An autoimmune disease prevented by anti-retroviral drugs. Retrovirology. 2011;8:91. doi: 10.1186/1742-4690-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boisson B, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol. 2012;13(12):1178–1186. doi: 10.1038/ni.2457. Patients with HOIL1 mutations display increased responses to IL-1β in leukocytes but diminished in fibroblasts, which could explain paradoxical clinical phenotype of autoinflammation and immunodeficiency
- 64. Ombrello MJ, et al. Cold urticaria, immunodeficiency, and autoimmunity related to PLCG2 deletions. N Engl J Med. 2012;366(4):330–338. doi: 10.1056/NEJMoa1102140. Decreased signaling at physiological temperatures, but increased signaling at subphysiological temperatures in cells expressing mutated PLCG2 gene shed light on coexistence of atopy, autoimmunity and immunodeficiency in cold urticaria patients
- 65.Zhou Q, et al. A hypermorphic missense mutation in PLCG2, encoding phospholipase Cgamma2, causes a dominantly inherited autoinflammatory disease with immunodeficiency. Am J Hum Genet. 2012;91(4):713–720. doi: 10.1016/j.ajhg.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3'–5' DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39(9):1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 67.Weinstock-Guttman B, et al. Genomic effects of once-weekly, intramuscular interferon-beta1a treatment after the first dose and on chronic dosing: Relationships to 5-year clinical outcomes in multiple sclerosis patients. J Neuroimmunol. 2008;205(1–2):113–125. doi: 10.1016/j.jneuroim.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 68.Serrano-Fernandez P, et al. Time course transcriptomics of IFNB1b drug therapy in multiple sclerosis. Autoimmunity. 2010;43(2):172–178. doi: 10.3109/08916930903219040. [DOI] [PubMed] [Google Scholar]
- 69. Ottoboni L, et al. An RNA profile identifies two subsets of multiple sclerosis patients differing in disease activity. Sci Transl Med. 2012;4(153):153ra131. doi: 10.1126/scitranslmed.3004186. Stratification of 141 multiple sclerosis patients into 2 major disease severity groups using PBMC transcriptional profiles
- 70.Eizirik DL, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8(3):e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jabbari A, et al. Transcriptional profiling of psoriasis using RNA-seq reveals previously unidentified differentially expressed genes. J Invest Dermatol. 2012;132(1):246–249. doi: 10.1038/jid.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stone RC, et al. RNA-Seq for enrichment and analysis of IRF5 transcript expression in SLE. PLoS One. 2013;8(1):e54487. doi: 10.1371/journal.pone.0054487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heruth DP, et al. RNA-seq analysis of synovial fibroblasts brings new insights into rheumatoid arthritis. Cell Biosci. 2012;2(1):43. doi: 10.1186/2045-3701-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet. 2011;27(3):116–125. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Javierre BM, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20(2):170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gervin K, et al. DNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: identification of epigenetically dysregulated genes. PLoS Genet. 2012;8(1):e1002454. doi: 10.1371/journal.pgen.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rakyan VK, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7(9):e1002300. doi: 10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liu Y, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2):142–147. doi: 10.1038/nbt.2487. Genome-wide characterization of methylation patterns in 354 RA patients and 337 controls identifies 2 clusters within the MHC region with methylation patterns associated with disease risk
- 79.Fang TC, et al. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J Exp Med. 2012;209(4):661–669. doi: 10.1084/jem.20112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42(3):255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60(4):1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 82.Boldin MP, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208(6):1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakasa T, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58(5):1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stanczyk J, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 85.Pauley KM, et al. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10(4):R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhu S, et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-alpha. Nat Med. 2012;18(7):1077–1086. doi: 10.1038/nm.2815. Identification of the role of miR-23b as a blocker of IL-17-, TNFα- and IL1β-mediated NF-κB activation in SLE, RA and MS, and reciprocal inhibition of miR-23b by IL-17
- 87.Kurowska-Stolarska M, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci U S A. 2011;108(27):11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carlsen AL, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013;65(5):1324–1334. doi: 10.1002/art.37890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Argyropoulos C, et al. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS One. 2013;8(1):e54662. doi: 10.1371/journal.pone.0054662. A 27 microRNA signature in the urine of patients with T1D differentiates between stages of diabetic nephropathy
- 90.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 92.Chaussabel D, et al. Data management: it starts at the bench. Nat Immunol. 2009;10(12):1225–1227. doi: 10.1038/ni1209-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaye J, et al. Data sharing in genomics--re-shaping scientific practice. Nat Rev Genet. 2009;10(5):331–335. doi: 10.1038/nrg2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Piwowar HA, Day RS, Fridsma DB. Sharing detailed research data is associated with increased citation rate. PLoS One. 2007;2(3):e308. doi: 10.1371/journal.pone.0000308. [DOI] [PMC free article] [PubMed] [Google Scholar]