Abstract
In the development of RNA interference (RNAi) therapeutics, merely selecting short, interfering RNA (siRNA) sequences that are complementary to the messenger RNA (mRNA) target does not guarantee target silencing. Current algorithms for selecting siRNAs rely on many parameters, one of which is asymmetry, often predicted through calculation of the relative thermodynamic stability of the two ends of the siRNA. However, we have previously shown that highly-active siRNA sequences are likely to have particular nucleotides at each 5’-end, independent of their thermodynamic asymmetry. Here, we describe an algorithm for predicting highly active siRNA sequences based only on these two asymmetry parameters. The algorithm uses end sequence nucleotide preferences and predicted thermodynamic stabilities, each weighted based on training data from the literature, to rank the probability that an siRNA sequence will have high or low activity. The algorithm successfully predicts weakly- and highly-active sequences for enhanced green fluorescent protein (EGFP) and protein kinase R (PKR). Use of these two parameters in combination improves the prediction of siRNA activity over current approaches for predicting asymmetry. Going forward, we anticipate that this approach to siRNA asymmetry prediction will be incorporated into the next generation of siRNA selection algorithms.
Keywords: asymmetry, siRNA, EGFP, PKR
Introduction
Therapeutic applications of RNA interference (RNAi) leverage a conserved pathway for gene expression regulation that possesses the potential for exquisite sequence specificity through the complementarity of short interfering RNAs (siRNAs) for their target [1-3]. Though the technology has yet to demonstrate its full potential in clinical applications [4, 5], there remains major interest in developing siRNA-based therapeutics [6]. Because RNAi represents a therapeutic approach that can be applied to nearly any disease [7, 8], improvements in the design and development of siRNA therapeutics have the potential for a significant impact on clinical practice.
A number of intermolecular interactions are critical to the activity of siRNAs, including those with the delivery vehicle [9-11], the target mRNA [12-15], and the pathway proteins [16-19]. While a single RNA guide strand and Argonaute 2 (Ago2) are the minimal components required for active silencing in vitro [20], the proteins Dicer and TAR RNA Binding Protein (TRBP) are important for RLC/RISC activity in vivo [21-23]. Other proteins, such as the protein activator of PKR (PACT) [24, 25] and component 3 promoter of RISC (C3PO) [26], may also have important but as yet undefined functional roles in the RNAi process. One essential process executed by the pathway proteins is the identification and loading of the siRNA guide strand into the RLC/RISC and the concomitant destruction of the passenger strand [2, 27, 28]. The likelihood of one siRNA strand becoming the guide strand relative to the other strand is termed asymmetry [27, 29].
There are currently multiple proteins thought to participate in sensing the asymmetry of siRNA duplexes [18, 30-32]. When the existence of siRNA asymmetry was first identified, it was proposed that the relative hybridization stability of the two ends of the siRNA sequence was the principal means by which asymmetry was sensed by the pathway proteins [29]. Since that time, nearly all algorithms for selecting highly-active siRNAs have used a thermodynamic calculation for asymmetry, among other parameters [29, 33-37]. More recently, evidence has begun to accumulate that the terminal nucleotides on each 5’-end of the siRNA may be valuable for predicting the activity of an siRNA [18, 30, 38], in particular when classified according to the sixteen possible combinations of nucleotides. When terminal nucleotide classification is combined with relative hybridization stability, the accuracy of predicting siRNA activity improves markedly [38].
In this work, we wanted to predict siRNA activities based only on the two asymmetry characteristics, terminal nucleotide classification and relative thermodynamic stability, and establish experimentally their relative importance in determining the activity of an siRNA. Using a logistic regression model, we successfully predicted active and inactive sequences for the exogenous protein, enhanced green fluorescent protein (EGFP), as well as the endogenous protein, protein kinase R (PKR). In addition, the combination of both end sequence and thermodynamic stability features provided improved correlation to siRNA activity when compared to either feature individually. These results demonstrate further that asymmetry may be determined by more factors than just relative stability, and algorithms for prediction of siRNA activity should also account for terminal nucleotide sequence classification in asymmetry calculations.
Results
Ranking and Selection of EGFP-targeting siRNAs
Our ranking algorithm was initially tested on siRNAs to target the EGFP mRNA. From the cDNA sequence (see Supporting Information), there were 824 possible siRNA sequences, which were ranked based on the difference between the algorithm's predicted likelihood of high and low activity. For comparison, commercial algorithms (Dharmacon and Ambion) were also used. These selection algorithms were chosen, because their predictions are based solely on the characteristics of the siRNA and not on other factors used in some selection algorithms, such as target mRNA structure, which would make it difficult to directly compare the accuracy of our asymmetry-based predictions with predictions from more detailed selection approaches. The commercial rankings only included sequences predicted to have high activity as opposed to the entire range of possible siRNA sequences. While this is adequate for those needing effective siRNA sequences, it does not provide sufficient data to compare the characteristics of high activity and low activity siRNAs. The Dharmacon algorithm ranked the recommended siRNAs, whereas for Ambion there were no distinctions among the top 35 candidate sequences. Interestingly, there was no overlap between the lists of recommended sequences provided by the commercial algorithms. Aggregating the commercial recommendations with our predictions, we chose 11 sequences to test experimentally that would allow us to preliminarily compare the relative utility of the three prediction approaches (Table 2).
Table 2.
EGFP-targeting siRNA sequences selected for this study, sorted by algorithm rank.
| 5'Target Position | siRNA Sequence | End 5' Nucleotides | Thermodynamic ΔΔG (kcal/mol) | Algorithm Rank | Testing Rationale | |
|---|---|---|---|---|---|---|
| Anti:Sense | Rank | |||||
| 415 | UGUACUCCAGCUUGUGCCC | U:G | 1 | 4.2 | 1 | Highest algorithm ranking |
| 416 | UUGUACUCCAGCUUGUGCC | U:G | 1 | 2.1 | 17 | Highest Dharm. rank (91) |
| 274 | UGCGCUCCUGGACGUAGCC | U:G | 1 | 0.8 | 38 | On Ambion list |
| 757 | UGGGCAGCGUGCCAUCAUC | U:G | 1 | −2.6 | 177 | High end, low ΔΔG |
| 306 | CUUGUAGUUGCCGUCGUCC | C:G | 7 | 2.9 | 239 | On Ambion list |
| 396 | CAGGAUGUUGCCGUCCUCC | C:G | 7 | 0.5 | 389 | On Ambion list |
| 126 | GAUGAACUUCAGGGUCAGC | G:G | 10 | 1.9 | 418 | On Ambion list |
| 711 | AUGGCUAAGCUUCUUGUAC | A:G | 2 | −3.7 | 551 | Low rank, high Dharm. (73) |
| 783 | CAUCCCGCUCUCCUGGGCA | C:U | 16 | 3.1 | 695 | Low end, high ΔΔG |
| 159 | GGGCCAGGGCACGGGCAGC | G:G | 10 | −2.3 | 700 | On Ambion list |
| 253 | CGGGCAUGGCGGACUUGAA | C:U | 16 | −5.2 | 824 | Lowest algorithm rank |
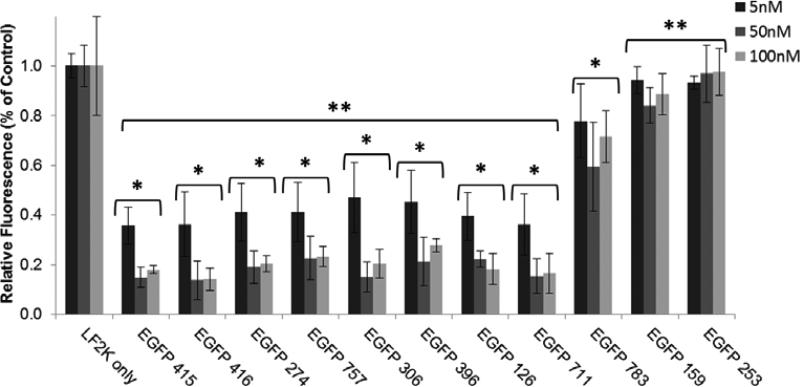
Transfection experiments were performed using H1299-EGFP cells at various siRNA concentrations (Figure 2). Surprisingly, 81% of the sequences had some silencing effect compared to control treatments, with 73% of sequences showing greater than 75% reduction in protein levels at a 50 and 100 nM siRNA concentration. One sequence (EGFP 783) showed intermediate silencing (difference from other sequences indicated by double stars), suggesting a gradient, rather than a step change, in silencing ability between active and inactive sequences. In general, sequences predicted by our algorithm to have higher activity showed increased inhibition of EGFP fluorescence. The rank order of activities was maintained at lower (5 nM) and saturating (50 and 100 nM) siRNA concentrations. The two sequences chosen based on their opposing rankings between the two features in our approach, EGFP 757 (favorable terminal sequence, unfavorable ΔΔG) and EGFP 783 (unfavorable terminal sequence, favorable ΔΔG), ultimately displayed an activity correlated with their terminal nucleotide classification rather than their thermodynamic stability.
Figure 2. Silencing of EGFP by selected siRNAs.
H1299 cells expressing EGFP were treated with 5, 50, and 100 nM of siRNA for 24 h using Lipofectamine 2000 (LF2K). Results are normalized EGFP fluorescence of LF2K-only treated control cells. siRNA treatments are ordered based on algorithm predictions (415 = highest predicted activity, 253 = lowest predicted activity). Error bars = +/− 1 standard deviation, n =4 (5 nM), 5 (50 nM), and 3 (100nM). Single stars indicate a significant difference (p < 0.05) compared to LF2K only treatments. Double stars indicate a significant difference (p < 0.05) compared to EGFP 783.
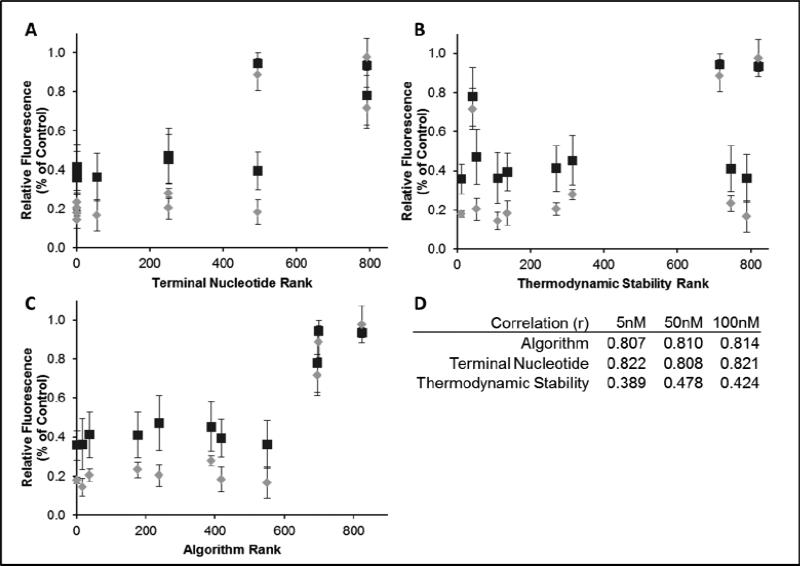
To further investigate the hypothesis that terminal sequence classification was more important than thermodynamic stability in predicting siRNA activity, silencing efficiencies were compared against algorithm rank, terminal nucleotide rank, and ΔΔG values individually (Figure 3). The correlation between activity and terminal nucleotide rank was better than for thermodynamic stability alone, with the correlation with full algorithm rank better than either alone. This agrees with our prior work showing that terminal nucleotide classification is generally a more informative predictor of siRNA activity but that inclusion of the thermodynamic calculation provides some additional complementary information [38].
Figure 3. Correlation of exogenous gene silencing with each feature of the algorithm.
EGFP silencing results graphed against terminal nucleotide rank only (A), ΔΔG values only (B) or algorithm rank (C). The squares represent 5 nM siRNA while the diamonds are 100 nM siRNA treatments. For visual clarity, the 50 nM data points were not included in the plots but follow the same trend in silencing activity. The correlation coefficients for each dataset are tabulated (D).
Ranking and Selection of PKR-targeting siRNAs
Although our algorithm successfully predicted active and inactive sequences for EGFP, we wanted to confirm similar results for an endogenous protein, the signaling pathway mediator, PKR. In addition, through systematic selection of siRNAs of high, medium, and low nucleotide ranking and siRNAs having high, medium, and low relative thermodynamic stabilities (Table 3), we aimed to further explore the relative importance of each of these characteristics in predicting silencing activity. We selected PKR as a model endogenous protein based on our prior work and expertise silencing this protein [39]. Though PKR is a double-stranded RNA responsive protein and known to be functionally connected to proteins in the RNAi pathway [40], the lack of any cytotoxicity across all of our experiments suggested that it was not initiating any generalized immune response to the siRNAs that would confound our specific silencing results.
Table 3.
PKR-targeting siRNA sequences selected for this study, sorted by end sequence ranking, followed by relative thermodynamic stability.
| 5' Target Position | siRNA Sequence | 5'-End Nucleotides | Thermodynamic ΔΔG (kcal/mol) | Algorithm Rank | Testing Rationale | |
|---|---|---|---|---|---|---|
| Anti:Sense | Rank | |||||
| 410 | UAAUGAAAUCCUUCUGGCC | U:G | 1 | 6.3 | 1 | High end, high ΔΔG |
| 193 | UCUUUGAUCUACCUUCACC | U:G | 1 | 1.9 | 61 | Previously validated |
| 816 | UUUAAAAUCCAUGCCAAAC | U:G | 1 | 0 | 230 | High. end, med. ΔΔG |
| 952 | UUGCCAAUGCUUUUACUUC | U:G | 1 | −4.2 | 948 | High end, low ΔΔG |
| 336 | AAUUCUAUUGAUAAGGCCU | A:A | 9 | 5.3 | 205 | Med. end, high ΔΔG |
| 645 | AUCUGCUGAGAAGUCACCU | A:A | 9 | 0 | 1154 | Med. end, med. ΔΔG |
| 379 | AUGCACACUGUUCAUAAUU | A:A | 9 | −5.2 | 1926 | Med end, low ΔΔG |
| 1198 | CAAAGAGUUCCAAAGCCAA | C:U | 16 | 5.5 | 1606 | Low end, high ΔΔG |
| 640 | CUGAGAAGUCACCUUCAGA | C:U | 16 | 0 | 2206 | Low end, med. ΔΔG |
| 928 | CCGCCUUCUCGUUAUUAUA | C:U | 16 | −5.2 | 2352 | Low end, low ΔΔG |
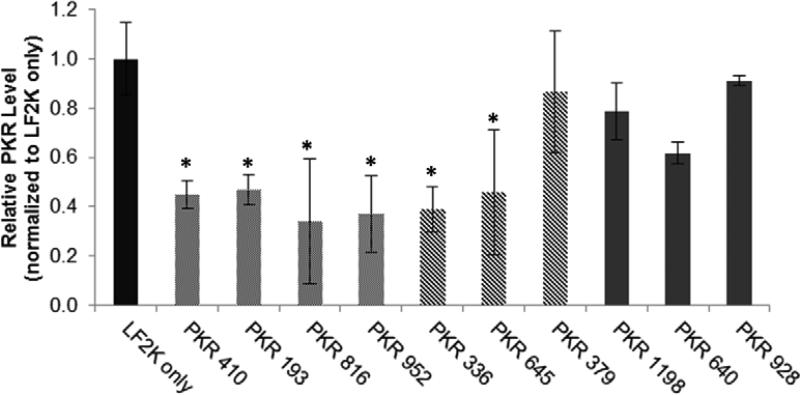
Transfection experiments were performed using HepG2 cells at 100 nM siRNA concentrations (Figure 4). In this case, 55% of sequences showed greater than 50% reduction in PKR protein levels when compared to control cells. Again, sequences predicted to have higher activity showed increased reduction in PKR protein levels. When sorted by end nucleotide classification, sequences in the UG class showed the best silencing activity, on average, regardless of their thermodynamic stability. Conversely, sequences in the low-ranking terminal nucleotide class, CU, do not show significant silencing, even when having highly positive ΔΔG values. In the intermediate category of end sequence (AA), silencing activity correlated strongly with the calculated thermodynamic stability, with a favorable value resulting in significant silencing and an unfavorable value not. Taken together with our results from the GFP experiments, these results support the argument that terminal sequence classification is a stronger predictor of siRNA activity that relative hybridization stability.
Figure 4. Silencing of PKR by selected siRNAs.
HepG2 cells were reverse-transfected with 100 nM of siRNA for 48 h using Lipofectamine 2000 (LF2K). Results are normalized to both total protein level and PKR level of LF2K-only treated control cells (black). siRNAs were grouped by end sequence rank, UG (light grey), AA (striped), and CU (med grey). Error bars = +/− 1 standard deviation, n =3. Stars indicate a significant difference (p < 0.05) when compared to LF2K only treatment.
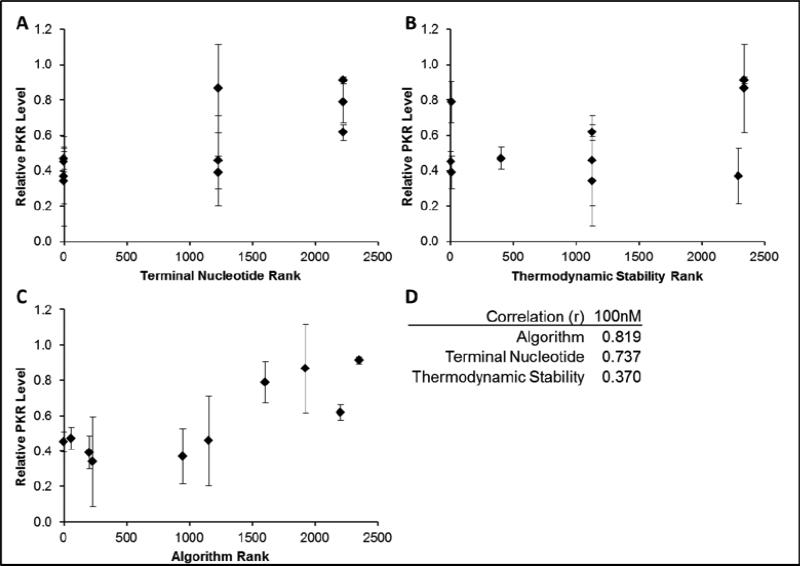
Our algorithm achieved improved correlation between rank and siRNA activity than achieved by either terminal nucleotide or thermodynamic stability independently (Figure 5). While the PKR terminal nucleotide correlation is less than that achieved from the EGFP data, the thermodynamic stability and overall algorithm rank correlation coefficients remain similar, even with a larger range of possible siRNA sequences (2352 for PKR vs. 824 for EGFP), showing the algorithm's fidelity for various sized targets. It is noteworthy that for both sequences, the top-ranked sequence, i.e., the one that would have been chosen for predicting an siRNA against a new target, was highly-active in silencing the target, further supporting our hypothesis that using only two parameters was sufficient for identifying active siRNAs against novel targets.
Figure 5. Correlation of endogenous gene silencing with each feature of the algorithm.
PKR silencing results graphed against terminal nucleotide rank only (A), ΔΔG values only (B), or algorithm rank (C). The correlation coefficients for each dataset are tabulated (D).
Discussion
The use of asymmetry is well-established as useful and important for selecting active siRNA sequences. Multi-step work flow protocols for selecting effective siRNAs have been developed [41, 42]. However the selection algorithms themselves are based in part on using relative thermodynamic stability as the sole factor in determining sequence asymmetry. Other reported algorithms lack a consensus on the best way to calculate thermodynamic asymmetry for siRNA activity prediction [36, 43-47]. When utilizing the commercially available algorithms for comparison in this study, there were no overlaps between the sequences predicted to be highly active by Dharmacon and those predicted by Ambion, further illustrating the need for a consensus approach to selecting highly active siRNAs, including which parameters are most important/useful for such predictions.
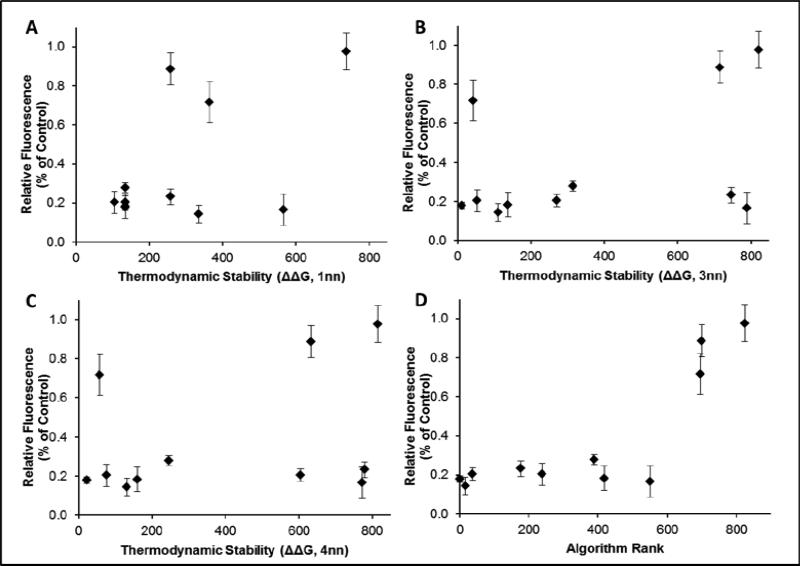
The results described here further illustrate that accounting only for thermodynamic asymmetry ignores a more important feature in asymmetry, terminal nucleotide classification. While others have identified terminal nucleotides as factors relevant to siRNA design (e.g., [35]), our approach of pairing the antisense and sense termini and weighting each pairing individually provides a unique and improved classification for sequences and their activities. Our selection technique achieves correlation coefficient values > 0.8 whereas previously reported algorithms typically achieve correlation coefficients of 0.5-0.7 between algorithm predictions and experimental results [41, 48]. As our approach is focused on the contribution of asymmetry in the siRNA to its ultimate activity, it was important to compare our approach with others means of determining asymmetry. In calculating thermodynamic asymmetry, it is common to use one, three, or four nearest neighbors at each end of the siRNA as the basis for calculation [29, 36, 48, 49]. In our prior work [38], we showed that in concert with terminal nucleotide classification, calculating thermodynamic asymmetry with three nearest neighbor parameters provided the most information while one nearest neighbor provided the most information in the absence of terminal nucleotide classification. Based on this context, we compared the correlation of our data with thermodynamic calculations performed using 1, 3, and 4 nearest neighbor parameters (Figure 6). In all cases, the correlation of our experimental data was best with rankings including terminal nucleotide classification (Table 4). This strongly supports our contention that all siRNA selection algorithms would be improved by inclusion of our asymmetry approach in place of their current asymmetry calculation.
Figure 6. Correlation of asymmetry calculations with experimental data.
100 nM EGFP silencing results graphed against rankings among all possible siRNA sequences using ΔΔG calculated using 1 nearest neighbor (A), 3 nearest neighbors (B), 4 nearest neighbors (C), or our algorithm (3 nearest neighbors and terminal nucleotide classification (D).
Table 4.
The correlation coefficients between EGFP silencing at 100 nM and the four different ranking methods in Figure 6..
| Correlation Coefficient (r) | 100nM |
|---|---|
| Algorithm | 0.814 |
| ΔΔG (Inn) | 0.539 |
| ΔΔG (3nn) | 0.424 |
| ΔΔG (4nn) | 0.327 |
Our algorithm as structured only ranks sequences based on their likelihood for silencing the intended target. It was our intent in this study to determine if the factors we tested were useful in predicting sequences of high activity. Our ranking approach does not account for potential off-target effects by the sequences. Examining the long-term design of siRNA therapeutics, it is essential that off-target effects be taken into account as well. That said, it is our belief that beginning therapeutic design with the most highly active sequence, which can then be modified, if needed, to improve its specificity, is a better approach to obtaining a useful therapeutic than beginning with the most highly specific sequence, which may then need to be modified to improve its silencing activity against the intended target.
While our approach is useful for predicting active (and inactive) siRNAs, we have not yet established the causal relationship between the terminal nucleotide classification and siRNA processing and activity. Indeed, it is well-established that the properties of the siRNA alone only provide partial information as to the likely activity of the siRNA [14, 15, 50, 51]. However, studies are increasingly reporting that more active siRNAs and miRNAs tend to contain specific nucleotides at the 5’ position of the guide strand [18, 33, 52], possibly a result of Ago2 binding specificity [30]. We expect that going forward, our ongoing work and that of others, will more firmly tie the presence of particular 5’-end nucleotides on both the guide and passenger strands with important siRNA-protein interactions that occur in the pathway to ensure proper siRNA processing.
Materials and Methods
Algorithm Design and Parameters
Using information from both terminal nucleotide classification and thermodynamic stability, we developed a 17 parameter logistic regression model based on the 16 possible end sequence combinations as well as the relative thermodynamic stability (Table 1 and Supporting Information). The relative stability is calculated by the difference between the hybridization free energy from the 5’-end of the antisense strand and the 5’-end of the sense strand, termed ΔΔG, based on the three terminal nearest neighbor pairs [38, 53] (Figure 1). ΔΔG calculations were based on 21 nt siRNAs with equivalent UU overhangs on the end of each strand. This calculation technique, when coupled with terminal sequence information, was shown to have the best predictive accuracy when tested on existing siRNA activity databases [38]. The weighting factors for each of the 17 parameters were based on fitting the model to the same siRNA databases [43, 44]. From a cDNA sequence input, the algorithm predicts the probability the given sequence will have high, medium, or low activity. Using the difference between the high and low probabilities, each siRNA sequence for a given target was ranked from the highest to lowest difference. The cDNA sequences used for EGFP and PKR are included in the Supporting Information with siRNA target regions highlighted. For comparison with other asymmetry approaches (Figure 6), asymmetry calculations were also performed using one and four nearest neighbor parameters. To ensure the most accurate comparisons across ranking approaches, sequences with equivalent values were all given the best possible ranking.
Table 1.
Values of the coefficients used as weighting factors for predicting the probability of siRNAs at high (top third among all possible sequences) and low (bottom third among all possible sequences) activity. Positive values indicate features that are positively correlated with high siRNA activity. Applying these weights in our algorithm, siRNAs were ranked by the magnitude of the difference between the probability of having high and low activity. The “Intercept” is applied to all sequences and arises from the regression model (see Supporting Information).
| Classifier | P(High) | P(Low) |
|---|---|---|
| ΔΔG | 0.2973 | −0.1661 |
| U:G | 1.8767 | −0.9852 |
| A:G | 0.3737 | −0.0193 |
| U:C | 1.8744 | −1.3088 |
| U:A | 1.7387 | −1.0301 |
| A:C | 0.4764 | −0.406 |
| U:U | 1.5084 | −1.0877 |
| C:G | −0.2463 | 0.4025 |
| G:C | −0.6404 | 0.3337 |
| A:A | 0.2381 | 0.0810 |
| G:G | −0.7873 | 0.2945 |
| A:U | −0.5707 | 0.3471 |
| C:C | −0.3552 | −0.0987 |
| G:A | −1.1939 | 0.6077 |
| G:U | −2.2329 | 1.3262 |
| C:A | −1.1755 | 0.9055 |
| C:U | −2.2846 | 1.2957 |
| Intercept | −0.0498 | −0.2145 |
Figure 1. Algorithm features for designing highly active siRNAs.
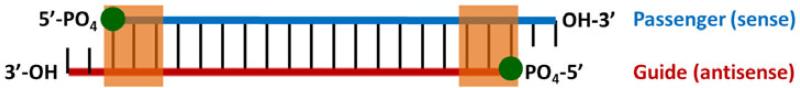
From the selected mRNA target, all potential siRNAs were classified according to their terminal sequence and thermodynamic stability. Terminal sequence (circles) uses the 5’ terminal nucleotides, antisense:sense strand and thermodynamic stability (shaded rectangles) was calculated based on ΔΔG values using the 3 terminal nearest neighbors. All sequences we synthesized to contain UU overhangs on the 3’ ends.
Dharmacon ranking information was obtained using: http://dharmacon.com/designcenter/DesignCenterPage (now available through: http://www.thermoscientificbio.com/design-center/). Potential sequences were determined by entering the nucleotide sequence directly, removing any GC restriction requirements, and choosing the “No BLAST” option to provide a direct comparison to the algorithm. Ambion ranking information was obtained through: http://ambion.com/techlib/misc/siRNA_finder.html (no longer active after acquisition of Ambion by Life Technologies). A list of potential siRNAs was generated by entering the nucleotide sequence; no other selection parameters were required.
Materials
Lipofectamine 2000 (LF2K) was purchased from Invitrogen. All EGFP and PKR siRNA sequences were 21 nt (19 bp plus UU overhangs) and were purchased from Dharmacon. Opti-Mem (GIBCO) was used for preparation of all transfection solutions. Monoclonal anti-PKR (Y117) primary antibody was purchased from Novus Biologicals. Monoclonal anti-β-actin primary antibody was purchased from Sigma. Secondary antibodies (anti-rabbit and anti-mouse) were purchased from ThermoScientific.
Cell Culture
Human lung carcinoma cells (H1299) constitutively expressing a form of EGFP, modified to contain a 2hr half-life, were generously provided by Dr. Jørgen Kjems, University of Aarhus, Denmark. Human hepatocellular carcinoma (HepG2) cells were purchased from American Type Culture Collection (ATCC). Cell culture media was prepared with Dulbecco's Modified Eagle's Medium High Glucose (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, GIBCO) and 1% penicillin-streptomycin (GIBCO). For the H1299 cells, 1% Geneticin (GIBCO) was added to maintain EGFP expression. Cells were incubated at 37°C, 5% CO2, 100% relative humidity and subcultured every 4-5 days by trypsinization.
EGFP Silencing and Fluorescence Analysis
H1299-EGFP cells were seeded in 96-well black side, clear bottom plates (Fisher Scientific) at a density of 20,000 cells/well in 0.1 mL of complete media without antibiotics. After 24 h, 50 μL solutions of varied siRNAs and LF2K were prepared in Opti-Mem and allowed to mix for 30 minutes prior to their addition to cells at final concentrations of 5-100 nM siRNA and 2.3 μg/mL LF2K. Cells were incubated in the transfection solutions at 37°C, 5% CO2, 100% humidity. After 24h, cells were washed 2 times with DPBS (Gibco), and EGFP fluorescence was quantified using a Gemini EM fluorescent plate reader (Molecular Devices) at 480 nm excitation/525 nm emission. Fluorescence intensity was normalized to control wells that were treated with transfection reagent but no siRNA. Cytotoxicity was assessed by microscopy and was not seen in any of the treatments (see Supporting Information).
PKR Silencing and Western Blotting
For siRNA transfections targeting human PKR in HepG2 cells, reverse transfection was performed. Briefly, 250 μL solutions of varied siRNAs and LF2K were prepared in Opti-Mem and allowed to mix for 30 minutes prior to their addition to standard six-well tissue culture plates. Freshly trypsinized HepG2 cells suspended in antibiotic-free media were added to the six-well culture plates at a density of 1.5 x 106 cells per well to achieve final concentrations of 100 nM siRNA and 2.3 μg/mL LF2K. The cells were then incubated at 37°C, 5% CO2, 100% humidity for 48 h and collected. PKR levels were measured by western blot analysis. The cells were washed twice with cold PBS and lysed in 200 μl/well of CelLytic M cell lysis buffer (Sigma) supplemented with protease inhibitor cocktail (Sigma). The cell lysate was clarified by centrifugation at 13,000 rpm for 10 min, and the supernatant was collected. Total protein levels were quantified by Quick Start Bradford protein assay (BioRad). Approximately 20 μg of total protein was resolved using 8% resolving, 5% stacking SDS-PAGE gels. Proteins were then transferred to nitrocellulose membranes and probed with primary and secondary antibodies. Biotinylated protein ladders (Cell Signaling Tech.) were loaded to one well of each SDS-PAGE gel, and anti-biotin antibody was used to detect the protein ladders on the western blots. Antibody detection was performed using the SuperSignal West Femto Chemiluminescence substrate kit (ThermoScientific) and imaged on the Molecular Imager ChemiDoc XRS System (Bio-Rad). Band intensity was first normalized to actin to control for protein isolation and loading and then the ratio was normalized to the ratio for control cells that received transfection reagent but no siRNA. Cytotoxicity was assessed by microscopy and CellTiter-Blue assay (Promega) (see Supporting Information) and was not seen in any of the treatments.
Statistical Analyses
Multiple comparisons between protein levels across different siRNA treatments conditions were performed using one-way (PKR data) or two-way (EGFP data) ANOVA followed by Tukey's HSD post-hoc analysis with p-value cut-off set at 0.05. Analyses were performed using either Microsoft Excel or Minitab.
Supplementary Material
Acknowledgments
Financial support for this work was provided in part by Michigan State University (MSU Foundation, Center for Systems Biology, and MSU Graduate School), the National Institutes of Health (GM079688, GM089866, RR024439, DK081768, DK088251), and the National Science Foundation (CBET 0941055).
We thank all the members of the Cellular and Biomolecular Laboratory for their advice and support. We also thank Dr Jørgen Kjems (University of Aarhus, Denmark) for providing us with the EGFP cells.
Abbreviations
- Ago2
argonaute 2
- C3PO
component 3 promoter of RISC
- EGFP
enhanced green fluorescent protein
- H1299
human lung carcinoma
- HepG2
hepatocellular carcinoma
- LF2K
lipofectamine 2000
- mRNA
messenger RNA
- nn
nearest neighbor
- PACT
protein activator of PKR
- PKR
dsRNA-dependent protein kinase R
- RISC
RNA induced silencing complex
- RLC
RISC loading complex
- RNAi
RNA interference
- siRNA
short interfering RNA
- TRBP
TAR RNA binding protein
Footnotes
Supporting Information:
See Malefyt_Supporting_Information.docx
References
- 1.Fire A, Xu S, Montgomery M, Kostas S, Driver S, Mello C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 4.Pollack A. Drugmakers’ fever for the power of RNA interference has cooled. In New York Times. 2011 [Google Scholar]
- 5.Krieg AM. Is RNAi Dead? Molecular Therapy. 2011;19:1001–1002. doi: 10.1038/mt.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gitig D. Use of siRNA in Therapeutic Arena on the Upswing. In Genetic Engineering & Biotechnology News. 2012:24–26. [Google Scholar]
- 7.Davidson BL, McCray PB. Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angart P, Vocelle D, Chan C, Walton SP. Design of siRNA Therapeutics from the Molecular Scale. Pharmaceuticals. 2013;6:440–468. doi: 10.3390/ph6040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Portis AM, Carballo G, Baker GL, Chan C, Walton SP. Confocal Microscopy for the Analysis of siRNA Delivery by Polymeric Nanoparticles. Microsc Res Tech. 2010;73:878–885. doi: 10.1002/jemt.20861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu JJ, Langer R, Chen JZ. A Novel Mechanism Is Involved in Cationic Lipid-Mediated Functional siRNA Delivery. Molecular Pharmaceutics. 2009;6:763–771. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegwart DJ, Whitehead KA, Nuhn L, Sahay G, Cheng H, Jiang S, Ma M, Lytton-Jean A, Vegas A, Fenton P, et al. Combinatorial synthesis of chemically diverse core-shell nanoparticles for intracellular delivery. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12996–13001. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 13.Brown KM, Chu C-Y, Rana TM. Target accessibility dictates the potency of human RISC. Nature structural & molecular biology. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- 14.Gredell JA, Berger AK, Walton SP. Impact of target mRNA structure on siRNA silencing efficiency: A large-scale study. Biotechnology and Bioengineering. 2008;100:744–755. doi: 10.1002/bit.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiryu H, Terai G, Imamura O, Yoneyama H, Suzuki K, Asai K. A detailed investigation of accessibilities around target sites of siRNAs and miRNAs. Bioinformatics. 2011;27:1788–1797. doi: 10.1093/bioinformatics/btr276. [DOI] [PubMed] [Google Scholar]
- 16.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proceedings of the National Academy of Sciences of the United States of America. 2007;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & Development. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gredell JA, Dittmer MJ, Wu M, Chan C, Walton SP. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry. 2010;49:3148–3155. doi: 10.1021/bi902189s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kini HK, Walton SP. Effect of siRNA terminal mismatches on TRBP and Dicer binding and silencing efficacy. FEBS Journal. 2009;276:6576–6585. doi: 10.1111/j.1742-4658.2009.07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu JD, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Structural & Molecular Biology. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 21.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO reports. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lima WF, Murray H, Nichols JG, Wu H, Sun H, Prakash TP, Berdeja AR, Gaus HJ, Crooke ST. Human Dicer binds short single-strand and double-strand RNA with high affinity and interacts with different regions of the nucleic acids. Journal of biological Chemistry. 2009;284:2535–2548. doi: 10.1074/jbc.M803748200. [DOI] [PubMed] [Google Scholar]
- 24.Kok KH, Ng MHJ, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. Journal of Biological Chemistry. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO journal. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H. Structure of C3PO and mechanism of human RISC activation. Nature structural & molecular biology. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 28.Leuschner PJF, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO reports. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz D, Hutvagner G, Du T, Xu Z, Aronin N, Zamore P. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 30.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 31.Noland CL, Ma E, Doudna JA. siRNA Repositioning for Guide Strand Selection by Human Dicer Complexes. Molecular cell. 2011;43:110–121. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betancur JG, Tomari Y. Dicer is dispensable for asymmetric RISC loading in mammals. RNA. 2012;18:24–30. doi: 10.1261/rna.029785.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nature Biotechnology. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 34.Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochemical and Biophysical Research Communications. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 35.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Research. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu ZJ, Mathews DH. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Research. 2008;36:640–647. doi: 10.1093/nar/gkm920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichihara M, Murakumo Y, Masuda A, Matsuura T, Asai N, Jijiwa M, Shinmi J, Yatsuya H, Qiao S, Takahashi M, et al. Thermodynamic instability of siRNA duplex is a prerequisite for dependable prediction of siRNA activites. Nucleic Acids Research. 2007;35:e123. doi: 10.1093/nar/gkm699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walton SP, Wu M, Gredell JA, Chan C. Designing highly active siRNAs for therapeutic applications. The FEBS journal. 2010;277:4806–4813. doi: 10.1111/j.1742-4658.2010.07903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Chan C. Repression of PKR mediates palmitate-induced apoptosis in HepG2 cells through regulation of Bcl-2. Cell Res. 2009;19:469–486. doi: 10.1038/cr.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laraki G, Clerzius G, Daher A, Melendez-Pena C, Daniels S, Gatignol A. Interactions between the double-stranded RNA-binding proteins TRBP and PACT define the Medipal domain that mediates protein-protein interactions. RNA Biol. 2008;5:92–103. doi: 10.4161/rna.5.2.6069. [DOI] [PubMed] [Google Scholar]
- 41.Birmingham A, Anderson E, Sullivan K, Reynolds A, Boese Q, Leake D, Karpilow J, Khvorova A. A protocol for designing siRNAs with high functionality and specificity. Nature Protocols. 2007;2:2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 42.Mysara M, Garibaldi JM, ElHefnawi M. MysiRNA-Designer: A Workflow for Efficient siRNA Design. Plos One. 2011;6:e25642. doi: 10.1371/journal.pone.0025642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nature Biotechnology. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 44.Shabalina SA, Spiridonov AN, Ogurtsov AY. Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinformatics. 2006;7:65. doi: 10.1186/1471-2105-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. siRNA selection server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Research. 2004;32:W130–W134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takasaki S. Selecting effective siRNA target sequences by using Bayes’ theorem. Computational Biology and Chemistry. 2009;33:368–372. doi: 10.1016/j.compbiolchem.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 47.Ladunga I. More complete gene silencing by fewer siRNAs: transparent optimized design and biophysical signature. Nucleic Acids Research. 2006;35:433–440. doi: 10.1093/nar/gkl1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matveeva OV, Nechipurenko YD, Rossi L, Moore B, Saetrom P, Ogurtsov AY, Atkins JF, Shabalina SA. Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Research. 2007;35:e63. doi: 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs Exhibit Strand Bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 50.Overhoff M, Alken M, Far RK- K, Lemaitre M, Lebleu B, Sczakiel G, Robbins I. Local RNA target structure influences siRNA efficacy: a systematic global analysis. Journal of molecular biology. 2005;348:871–881. doi: 10.1016/j.jmb.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 51.Shao Y, Chan CY, Maliyekkel A, Lawrence CE, Roninson IB, Ding Y. Effect of target secondary structure on RNAi efficiency. RNA. 2007;13:1631–1640. doi: 10.1261/rna.546207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seitz H, Tushir J, Zamore P. A 5′-uridine amplifies miRNA/miRNA* asymmetry in Drosophila by promoting RNA-induced silencing complex formation. Silence. 2011 doi: 10.1186/1758-907X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with watson-crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.