Abstract
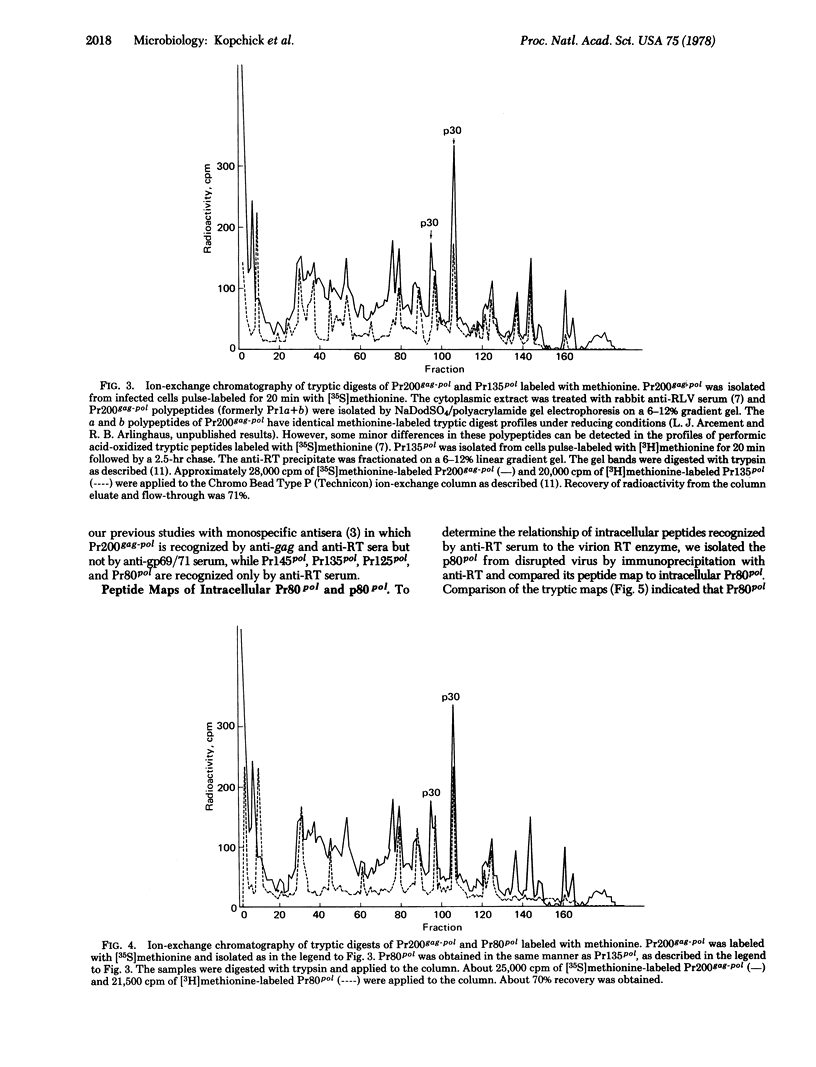
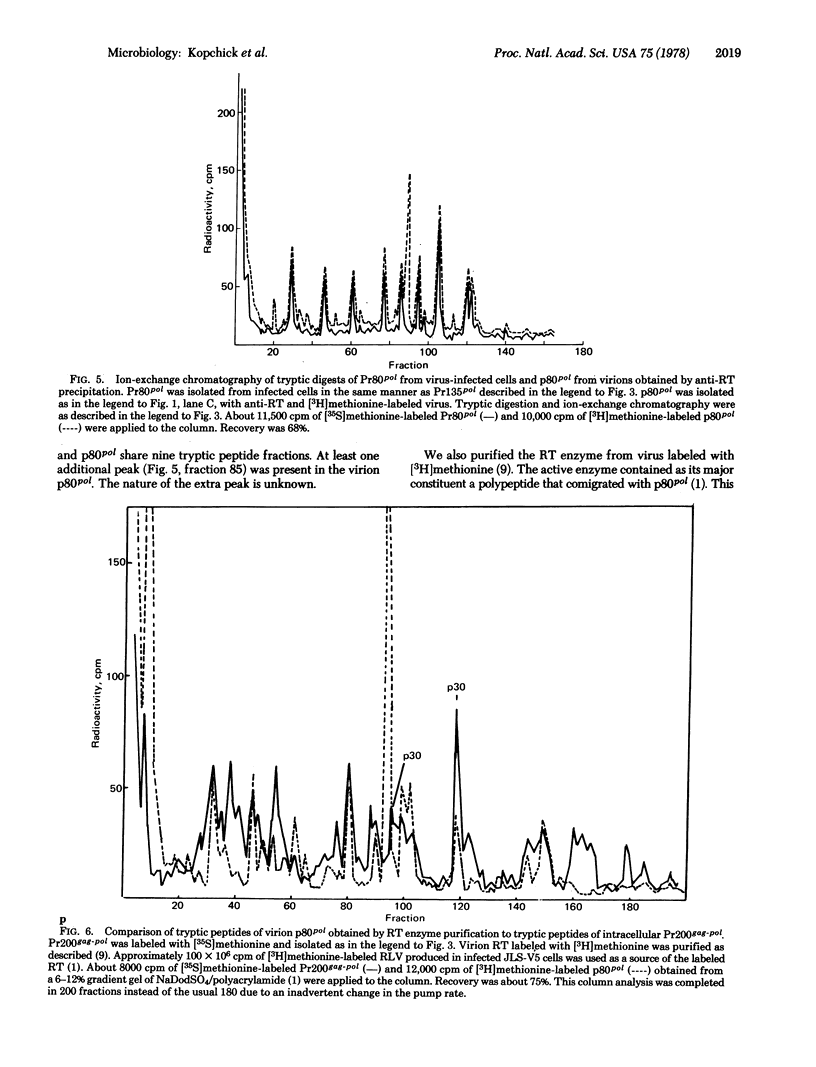
Reverse transcriptase (RT; RNA-dependent DNA nucleotidyltransferase) from Rauscher leukemia virus is synthesized in infected cells by way of a read-through poly- rotein of 200,000 molecular weight. This polyprotein (Pr200gag-pol) was precipitated by antiserum to RT; in a previous study all the monospecific antisera to gag proteins recognized Pr200gag-pol. Pr200gag-pol contains both p30 and RT peptide sequences. Intermediate RT-related precursors of 145,000 (Pr145pol), 135,000 (Pr135pol), and 125,000 (Pr125pol) molecular weights were specifically recognized by precipitation from infected cell extracts by antiserum to RT. These proteins shared methionine-containing tryptic peptide sequences with a virion polypeptide of 80,000 molecular weight (p80pol) precipitate by antiserum to RT. Purification of active RT enzyme from virions labeled with [3H]methionine showed that p80pol was the major component, based on analysis by gel electrophoresis and tryptic peptide mapping experiments. A polypeptide (Pr80pol), similar in size to mature viral p80pol, was also precipitated from infected cells by antiserum to RT. Its peptide map was nearly identical to that of virion p80pol. Pulse-chase studies showed that Pr80pol, Pr125pol, and Pr135pol were stable polypeptides, whereas Pr200gag-pol and Pr145pol were unstable precursors. Pulse-chase studies with the protein synthesis inhibitor, cycloheximide, showed that the processing of Pr200gag-pol occurred for a short time in the absence of protein synthesis.
Keywords: RNA-dependent DNA nucleotidyltransferase, processing of viral proteins, oncornaviral protein synthesis
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arcement L. J., Karshin W. L., Naso R. B., Arlinghaus R. B. "gag" polyprotein precursors of Rauscher murine leukemia virus. Virology. 1977 Sep;81(2):284–297. doi: 10.1016/0042-6822(77)90145-3. [DOI] [PubMed] [Google Scholar]
- Arcement L. J., Karshin W. L., Naso R. B., Jamjoom G., Arlinghaus R. B. Biosynthesis of Rauscher leukemia viral proteins: presence of p30 and envelope p15 sequences in precursor polypeptides. Virology. 1976 Feb;69(2):763–774. doi: 10.1016/0042-6822(76)90504-3. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Tumor viruses: 1974. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):1187–1200. doi: 10.1101/sqb.1974.039.01.137. [DOI] [PubMed] [Google Scholar]
- Jamjoom G. A., Naso R. B., Arlinghaus R. B. Further characterization of intracellular precursor polyproteins of Rauscher leukemia virus. Virology. 1977 May 1;78(1):11–34. doi: 10.1016/0042-6822(77)90075-7. [DOI] [PubMed] [Google Scholar]
- Jamjoom G. A., Naso R. B., Arlinghaus R. B. Selective decrease in the rate of cleavage of an intracellular precursor to Rauscher leukemia virus p30 by treatment of infected cells with actinomycin D. J Virol. 1976 Sep;19(3):1054–1072. doi: 10.1128/jvi.19.3.1054-1072.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacian D. L., Watson K. F., Burny A., Spiegelman S. Purification of the DNA polymerase of avian myeloblastosis virus. Biochim Biophys Acta. 1971 Sep 24;246(3):365–383. doi: 10.1016/0005-2787(71)90773-8. [DOI] [PubMed] [Google Scholar]
- Karshin W. L., Arcement L. J., Naso R. B., Arlinghaus R. B. Common precursor for Rauscher leukemia virus gp69/71, p15(E), and p12(E). J Virol. 1977 Sep;23(3):787–798. doi: 10.1128/jvi.23.3.787-798.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakower J. M., Barbacid M., Aaronson S. A. Radioimmunoassay for mammalian type C viral reverse transcriptase. J Virol. 1977 May;22(2):331–339. doi: 10.1128/jvi.22.2.331-339.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Rich A. The nature of the polypeptide chain termination signal. J Mol Biol. 1971 Jun 14;58(2):513–531. doi: 10.1016/0022-2836(71)90368-8. [DOI] [PubMed] [Google Scholar]
- Moelling K. Characterization of reverse transcriptase and RNase H from friend-murine leukemia virus. Virology. 1974 Nov;62(1):46–59. doi: 10.1016/0042-6822(74)90302-x. [DOI] [PubMed] [Google Scholar]
- Murphy E. C., Jr, Kopchick J. J., Watson K. F., Arlinghaus R. B. Cell-free synthesis of a precursor polyprotein containing both gag and pol gene products by Rauscher murine leukemia virus 35S RNA. Cell. 1978 Feb;13(2):359–369. doi: 10.1016/0092-8674(78)90204-0. [DOI] [PubMed] [Google Scholar]
- Naso R. B., Arcement L. J., Karshin W. L., Jamjoom G. A., Arlinghaus R. B. A fucose-deficient glycoprotein precursor to Rauscher leukemia virus gp69/71. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2326–2330. doi: 10.1073/pnas.73.7.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naso R. B., Vidrine J. B., Wang C. S., Arlinglaus R. B. C-type virus particles produced by normal mouse spleen-thymus cultures. Virology. 1972 Feb;47(2):521–524. doi: 10.1016/0042-6822(72)90293-0. [DOI] [PubMed] [Google Scholar]
- Panet A., Baltimore D., Hanafusa T. Quantitation of avian RNA tumor virus reverse transcriptase by radioimmunoassay. J Virol. 1975 Jul;16(1):146–152. doi: 10.1128/jvi.16.1.146-152.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M. Studies on reverse transcriptase of RNA tumor viruses III. Properties of purified Moloney murine leukemia virus DNA polymerase and associated RNase H. J Virol. 1975 Apr;15(4):843–854. doi: 10.1128/jvi.15.4.843-854.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]