Abstract
Embryonic stem cells (ESCs) are hypersensitive to many DNA damaging agents and can rapidly undergo cell death or cell differentiation following exposure. Treatment of mouse ESCs (mESCs) with etoposide (ETO), a topoisomerase II poison, followed by a recovery period resulted in massive cell death with characteristics of a programmed cell death pathway (PCD). While cell death was both caspase- and necroptosis-independent, it was partially dependent on the activity of lysosomal proteases. A role for autophagy in the cell death process was eliminated, suggesting that ETO induces a novel PCD pathway in mESCs. Inhibition of p53 either as a transcription factor by pifithrin α or in its mitochondrial role by pifithrin μ significantly reduced ESC death levels. Finally, EndoG was newly identified as a protease participating in the DNA fragmentation observed during ETO-induced PCD. We coined the term Charontosis after Charon, the ferryman of the dead in Greek mythology, to refer to the PCD signaling events induced by ETO in mESCs.
Keywords: Embryonic stem cell, ES cell, etoposide, DNA damage, Apoptosis, Autophagy, Necrosis, Necroptosis, Cell death, Programmed Cell Death, PCD, Caspase, Parp-1, RIP, cathepsin, p53, pifithrin, zVAD, ZFA, lysosome, Spautin 1, Bafilomycin A1
Introduction
Embryonic stem cells (ESCs) must maintain genomic integrity to prevent the accumulation of mutations in the cells that will give rise to an organism. Evidence supporting this proposition derives predominantly from mouse ESCs (mESCs), where mutation frequencies are significantly suppressed compared with mouse embryo fibroblasts (MEFs) (Cervantes et al., 2002). Suppression of mutation may be accomplished by the cells’ capacity to rapidly repair DNA damage by constitutively upregulating DNA repair pathways. Alternatively, cells harboring extensively damaged DNA are removed from the self-renewing stem cell population by the induction of cell differentiation or cell death (Tichy, 2011).
When cells are stressed or when they respond to a range of stimuli, such as damage to DNA, they can undergo cell death, which can be characterized as programmed cell death (PCD) or necrotic death. Necrotic death is primarily a passive process that involves rapid ATP depletion, swelling and rupture of organelles and nuclei, and random DNA breakage (Edinger et al., 2004). Under certain conditions, however, necrosis may be described as a PCD pathway, termed necroptosis, which displays many of the features of necrosis, but in which cell death is dependent on the activities of the RIP1 and/or RIP3 kinases to facilitate death (Galluzzi et al., 2011).
In addition to necroptosis, there are several other known PCD pathways, of which the most extensively described relies upon the activity of executioner caspases. These proteases cleave specific target sequences in select proteins, including Parp-1, lamins, actins, and ICAD, the inhibitor of caspase-activated DNase (Kawahara et al., 1998). Destruction of ICAD allows for DNA fragmentation by caspase activated DNase (CAD), which is one of many nucleases capable of producing DNA fragmentation during PCD.
Autophagy is typically utilized by cells to maintain energy homeostasis, particularly during times of nutrient deprivation. During autophagy, certain cellular components are recycled or damaged organelles are eliminated. As a result, autophagy can function as a pro-survival pathway, and is often activated in response to chemotherapeutic treatments. Alternatively, autophagy may also promote PCD in response to several types of stimuli (Yu et al., 2004).
The level of p53 protein, which is required for PCD under many conditions, is tightly controlled in most cell types through proteosome-mediated degradation. Following stress signals, p53 can be released from the E3 ubiquitin ligases Mdm2 or Mdm4 and becomes stabilized. It can then translocate to the nucleus to activate target gene expression, including the expression of genes involved in cell cycle inhibition and cell death. Alternatively, it can migrate to the mitochondria and promote cytochrome C release through interaction with Bcl-xL and Bcl2 (Marchenko et al., 2000; Mihara et al., 2003).
DNA fragmentation of defined size is characteristic of many PCD pathways and is typically dependent upon the activity of CAD, Apoptosis-inducing factor (AIF), or Endonuclease G (EndoG) nucleases. Under basal conditions, the inhibitor of CAD, ICAD, binds to CAD, preventing its activity. Caspase activation and activity degrades ICAD, which allows CAD to cleave DNA into nucleosomal-sized fragments. AIF localizes to the mitochondria and exhibits oxidoreductase activity (Klein et al., 2002), however, following death signal stimulus, AIF can be released from the mitochondria and translocate to the nucleus to cleave DNA into large molecular weight fragments (Joza et al., 2001; Susin et al., 1999). EndoG, like AIF, is a mitochondrial protein which functions to generate primers for mitochondrial DNA replication (Cote et al., 1993). Similar to AIF, death signals can trigger EndoG translocation to the nucleus to cleave DNA, however, DNA fragmentation is of nucleosomal size (Li et al., 2001).
The current study has investigated the cell death pathway(s) utilized by mESCs in response to etoposide (ETO), a topoisomerase II poison that indirectly induces DNA double strand breaks (DSBs) (Baldwin et al., 2005). The data show that ETO not only induces cell death with hallmarks of both PCD and necrosis, but also that this cell death is predominantly caspase- and RIP-independent. We also demonstrate a role for lysosomal cathepsins as well as p53 in this PCD pathway. Finally, we show that the EndoG nuclease contributes to DNA fragmentation in ETO-induced PCD in mESCs.
Materials and Methods
Cell Culture and Cell Treatments
C57Bl/6 ESCs (designated WD44) were used for all experiments unless otherwise noted and were maintained as previously described (Tichy et al., 2011) on feeder cell layers comprised of mitomycin C-treated mouse embryo fibroblasts. An additional C57Bl/6 ESC line, CMT1-2 was used to confirm data obtained using WD44 ESCs. MESCs deficient for p53 were a kind gift from J. Huang (NCI). Twenty-four hours prior to any experiment, ESCs were separated from the feeder cells and plated on culture dishes coated with 0.2% gelatin. Primary mouse embryo fibroblasts (MEFs) were cultured as described (Tichy et al., 2010) and used at passage 2 for experiments. Cells were never allowed to become more than 70% confluent during experimental procedures.
Additional materials and methods can be found in the supplementary methods section and a list of pharmacological inhibitors and their targets are listed in supplemental table 2.
Results
Embryonic stem cells are sensitive to ETO treatment
Embryonic stem cells of mouse (mESCs) or human (hESCs) origin are hypersensitive to DNA damaging agents and activate cell death or differentiation pathways to remove damaged cells from the stem cell pool (Aladjem et al., 1998; Tichy, 2011; Van Sloun et al., 1999). To characterize the sensitivity of mESCs to ETO, cells were treated with a low dose of drug (10µM) for 4 hours and then harvested either immediately after treatment (labeled 4/0), or after 2 (4/6), 20 (4/24), or 44 (4/48) hours of recovery prior to assaying. This concentration was chosen based on a previous report measuring DNA DSB repair kinetics in mESCs following ETO treatment (Tichy, 2011). We first examined the level of DNA DSBs produced by ETO treatment and the extent of their repair as a function of time. There was a marked increase in γH2AX foci, indicative of DSBs, at 4 hours post-treatment (Fig. 1A). During the recovery periods, γH2AX levels significantly decreased, reflective of DNA repair (Fig. 1A). The levels of γH2AX, however, did not return to baseline within the timeframe of the experiment. A consistent and substantial increase in nuclear size was also observed 20 hours post-treatment (Supplemental Fig. 1). Analysis of the cell cycle and measurement of phosphorylated histone H3, a marker of mitosis, revealed that treatment of mESCs with ETO causes these cells to accumulate in the G2 phase of the cell cycle (Fig 1B), consistent with reports for other cell types exposed to ETO (Clifford et al., 2003; Nam et al., 2010; Sleiman et al., 2000; Smith et al., 1994).
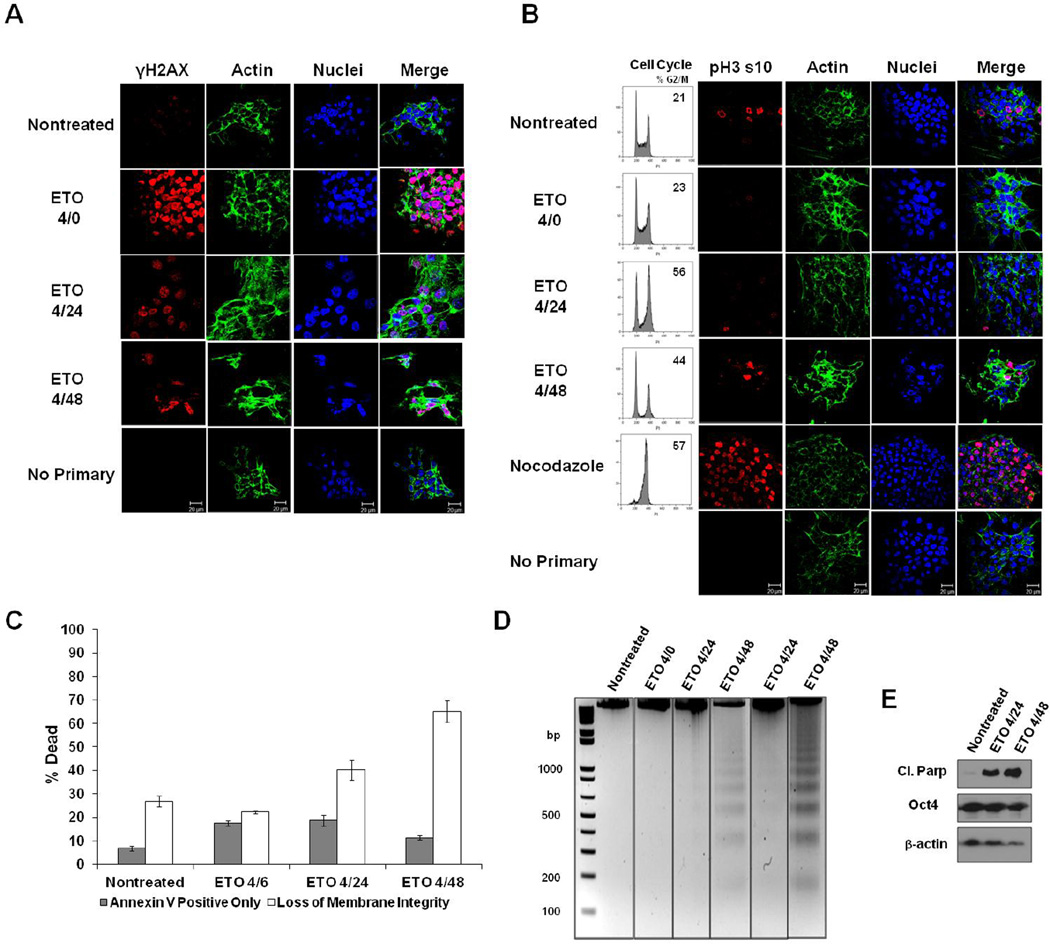
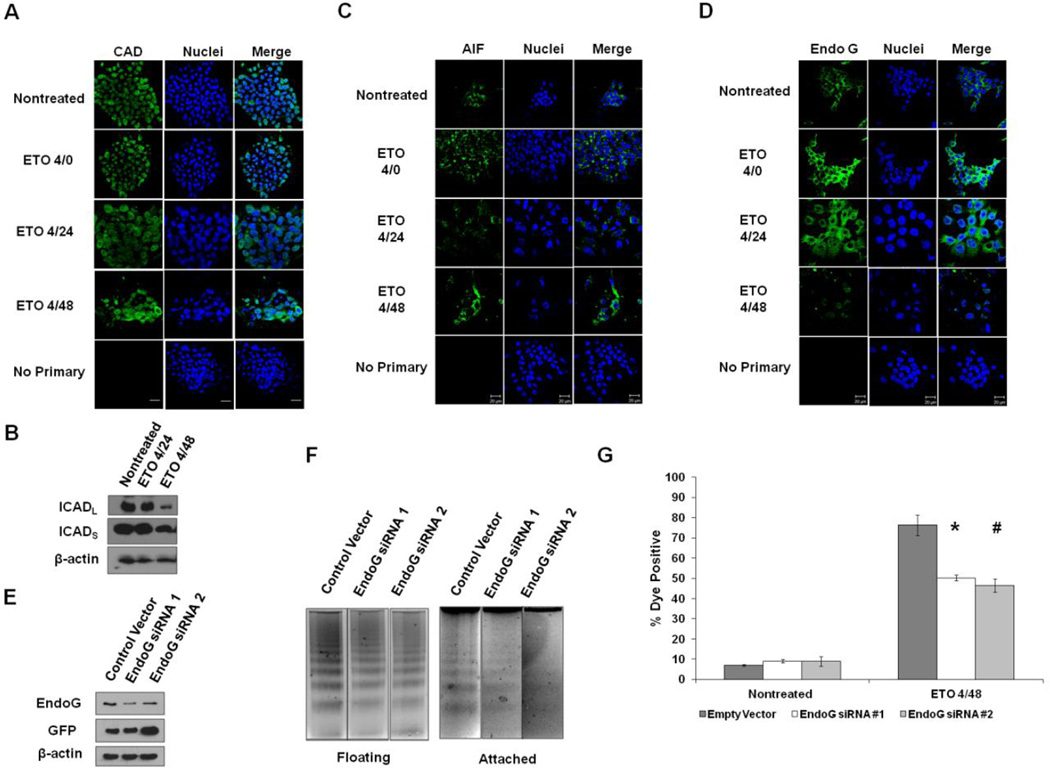
Figure 1. ESC responses to ETO.
A. ESCs were fixed immediately after 10µM etoposide (ETO) treatment (4/0-4 hour treatment; no recovery), or after 20 or 44 hours after treatment and recovery (4/24 or 4/48), or left nontreated. Cells were stained with antibody to γH2AX, and actin was stained with Alexa fluor 488-conjugated phalloidin. Nuclei were stained with Draq5. Cells stained with only secondary fluorescent antibody served as a control. B. Cells were treated as indicated and analyzed for their position in the cell cycle. To determine whether ETO induces a G2- or M-phase arrest, cells were stained with antibody to Histone H3 phospho-serine 10 (pH3 s10). Nocodazole-treated cells served as a control for mitotic arrest. C. ESCs were treated as indicated and death levels measured by flow cytometry after staining with Annexin V and a membrane impermeable dye. Data is the result of three trials. Error bars represent the standard error of the mean (SEM). D. DNA fragmentation assay on ESCs treated or left nontreated. DNA was isolated from all cells (floating and attached), and electrophoresed on 2% agarose gels. Two trials are displayed. E. Western blot using whole cell protein lysates from treated or nontreated ESCs. Expression of cleaved Parp-1 was monitored during the recovery period. Oct4 served as a marker for the level of ESC differentiation. Actin was used as a loading control.
To assess the level of cell death in ESCs treated with ETO, bivariate flow cytometry was utilized to assess Annexin V binding and plasma membrane permeability by vital dye exclusion. The numbers of Annexin V positive cells increased marginally after treatment with a short recovery period but decreased at later times of recovery (Fig.1C). There was also a significant increase in membrane permeability 20 and 44 hours post-treatment, as compared to nontreated controls. Similar trends were observed with a 129/Sv strain mESC line (Supplementary Fig. 2). Consistent with PCD signaling, DNA isolated from ETO-treated cells revealed distinct DNA laddering, characteristic of nucleosomal fragmentation, rather than a DNA smear, indicative of necrosis. The DNA laddering is first observed after four hours treatment and 20 hours recovery and became more pronounced after 44 hours of recovery (Fig. 1D), consistent with increased loss of plasma membrane integrity observed as a function of time. To confirm activation of a PCD pathway and eliminate a role for necrosis in ETO-induced death, Parp-1 cleavage was assessed and shown to increase substantially post-treatment (Fig. 1E). The level of Oct4 protein remained stable after ETO treatment, indicating that the mESCs retained pluripotency and that ETO did not induce them to differentiate (Fig. 1E).
Caspases are not significantly activated in mESCs after challenge with ETO
Our data show that caspase 3 is not activated in response to ETO in C57Bl/6 and 129/Sv mESCs (Supplemental Fig. 3A and 3B), confirming a previous study (Mantel et al., 2007). Under atypical conditions, however, a small fraction of caspase 3 was cleaved following ETO treatment, as well as following treatment of cells with UV radiation (Supplemental Fig. 3C). When the activation status of the other executioner caspases, 6 and 7, and the initiator caspases 8 and 9 was examined following ETO treatment, there was little or no increase in the cleavage of these caspases to indicate their activation (Supplemental Fig. 3A). The lack of cleavage of known caspase targets, including lamin A and C and actin (Fischer et al., 2003), confirmed the absence of caspase activity (Supplemental Fig. 3A). Intriguingly, expression of lamins A and C increased after treatment, which is consistent with the observed increase in nuclear size observed following treatment with ETO. Since mESCs treated with ETO undergo Parp-1 cleavage, caspase activity was further assessed. Treatment of mESCs with zVAD, a pan caspase inhibitor, in combination with ETO showed no reduction of Parp-1 cleavage compared with cells treated with ETO alone, indicating that caspases are not responsible for Parp-1 cleavage in these cells (Supplemental Fig. 3D). Similarly, treatment of mESCs with ETO in the presence or absence of zVAD showed no difference in the level of cell viability based on dye exclusion, confirming that caspases do not play a major role in ETO-induced PCD in mESCs (Supplemental Fig. 3E).
RIP-dependent necrosis does not significantly contribute to ETO-induced PCD
Necroptosis is a form of PCD that is dependent on the RIP1 and/or RIP3 kinases (Christofferson et al., 2010; Declercq et al., 2009; Galluzzi et al., 2008). We examined the necroptotic pathway more closely following ETO treatment of mESCs, since several features of this pathway were manifested during the process of mESC PCD. There was nuclear swelling following ETO treatment, a characteristic of necrotic cell death, as well as DNA fragmentation of defined sizes and cleavage of Parp-1, both characteristic of a PCD pathway, suggesting that necroptosis may play a role in ETO-induced PCD. Both RIP1 and RIP3 kinases are present in mESCs at a basal level and increase in abundance at later time points following exposure to ETO (Supplementary Fig. 4A), suggesting that RIP1 and RIP3 may contribute to ETO-induced PCD. To test this proposition, the protein levels of RIP1 or RIP3 were reduced with siRNAs (Supplementary Fig. 4B) and the extent of cell death was measured by flow cytometry, based on exclusion of a vital dye. Knocking down RIP3 had a significant effect on cell death at the 4/24 time point, but this effect did not persist into the 4/48 time point. The RIP1 knockdown had no effect on the level of cell death at any time point (Supplementary Fig. 4C). To independently address these findings, cells were treated with ETO and necrostatin-1, a RIP1 kinase inhibitor, either individually or in combination. Cells were treated with either ETO alone or ETO in combination with necrostatin-1 for four hours, at which time necrostatin-1 was added back for the duration of the recovery period. Consistent with the results using siRNA, necrostatin-1 did not reduce the level of ETO-induced cell death (Supplementary Fig. 4D). Thus, if necroptosis contributes to ETO-induced mESC PCD, its role must be a minor one.
Etoposide activates pro-survival autophagy in mESCs
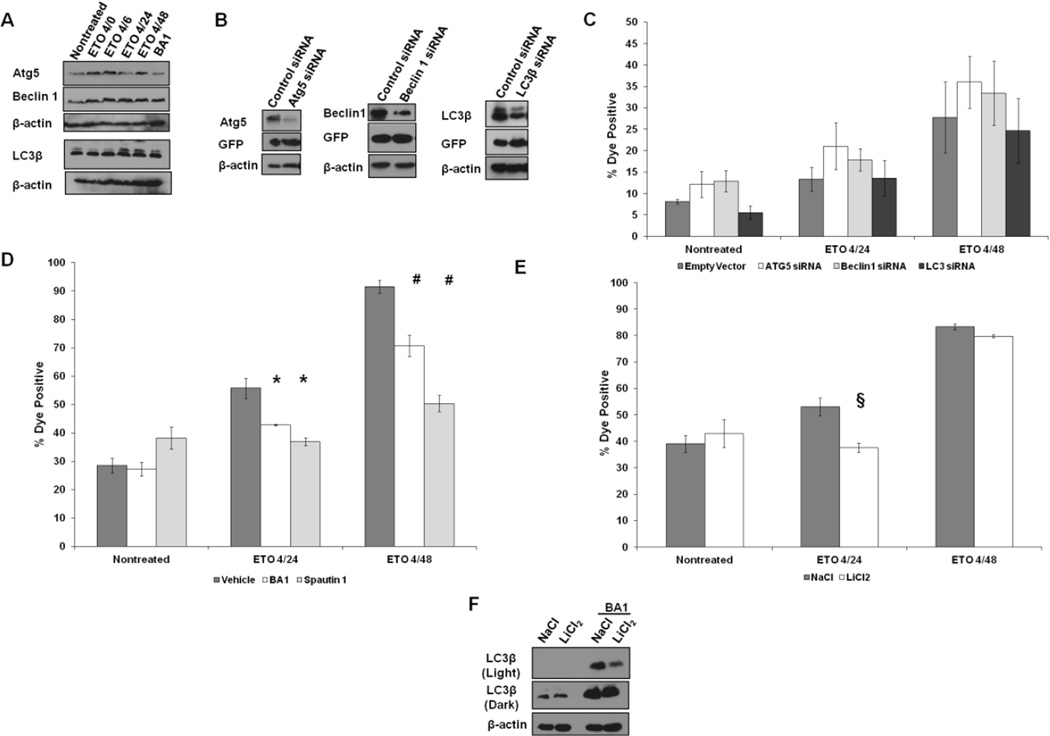
Autophagy has been implicated in the cell death of cervical cancer cells following treatment with ETO (Lee et al., 2007). To assess whether autophagy may also participate in mESC PCD after treatment with ETO, several autophagic proteins were assayed by Western blot. The levels of Atg5 and Beclin 1 proteins increased following treatment, as did the level of LC3β expression and LC3β flux (Fig. 2A), suggestive of autophagy activation. There was also a significant increase in endogenous LC3 foci, which serves as a marker of autophagosomes, based on Imagestream flow cytometry (Supplementary Fig. 5), further supporting an ETO-induced autophagic process. Suppression of Atg5, Beclin 1, or LC3β by siRNA (Fig. 2B), however, provided no evidence for involvement of autophagy in PCD following treatment of mESCs with ETO (Fig. 2C). Since expression of siRNA targeted proteins was not completely eliminated, the possibility remains that residual levels of autophagy proteins are sufficient to induce PCD. To probe this possibility, mESCs were treated with ETO in the presence or absence of the autophagy inhibitors Bafilomycin A1 (BA1) and Spautin 1. The BA1 inhibitor targets the vacuolar H+ ATPase, which is responsible for the acidification of the lysosome, and can prevent their fusion with autophagosomes (Yamamoto et al., 1998). Spautin 1 inhibits two ubiquitin-specific proteases, USP10 and USP13, both of which can prevent the ubiquitin-mediated degradation of Beclin 1 by removal of ubiquitin groups necessary for targeted degradation (Liu et al., 2011). Both of these inhibitors significantly reduced the level of ETO-induced PCD at the 24 hour time point and even more so at the 48 hour time point (Fig. 2D). Since these data are in conflict with findings obtained with the siRNA knockdown experiments, BA1 and Spautin 1 may target proteins and pathways other than the vaculolar H+ ATPase and USP 10 and 13, respectively, whose inhibition protects cells from PCD. Since LiCl2 can promote cells to undergo autophagy by inhibiting IMPase and GSK3 (Chang et al., 2011), mESCs were treated with ETO in the presence or absence of LiCl2. Activating autophagy in this manner results in more rapid degradation of LC3β-docked cargo, and can be monitored by a reduction in LC3β protein levels. If autophagy functions as a cell survival pathway rather than a cell death pathway as a result of ETO challenge, then administration of LiCl2 with ETO should reduce or show no change in the level of cell death. Consistent with this prediction, the level of cell death was reduced at 20 hours post-treatment after stimulating autophagy, with little or no effect on death levels by the 4/48 time point (Fig. 2E). The levels of LC3β decrease after the mESCs are treated with LiCl2 (Fig. 2F), which reflects degradation of LC3-docked cargo.
Figure 2. Autophagy is not involved in ETO-induced PCD.
A. Western blots characterizing the expression patterns of proteins involved in autophagy in mESCs prior to and after ETO treatment and recovery. Bafilomycin A1 (BA1) served as a control and inhibits autophagy while allowing for the accumulation of autophagic intermediates. B. Western blot showing level of knockdown of Atg5, Beclin1, or LC3β, after transient transfection of the indicated siRNAs. Cells transfected with siRNAs express GFP, which was probed for to demonstrate equal levels of transfection compared to control vector. C. ESCs were transfected with siRNAs. Twenty four hours posttransfection, cells were treated as indicated and flow cytometry was performed to measure plasma membrane integrity. Cells were first sorted for GFP positive cells prior to analysis for levels of death. Error bars represent the SEM. At least 6 trials are displayed per group. D. ESCs were treated with Spautin 1 or BA1 during ETO treatment and added back during the recovery period. Cells were subjected to flow cytometry to measure the loss of membrane permeability. Error bars represent the SEM. F. ESCs were treated with either NaCl or LiCl2 during and after ETO treatment and subjected to flow cytometry to analyze loss of plasma membrane integrity. F. Western blot of ESCs treated with NaCl or LiCl2 probed for LC3. To demonstrate autophagy is increased, BA1 was added to cells for 6 hours at a concentration of 1nM. Two exposures are provided. Actin served as a loading control. §p<0.07;*p<0.05; #p<0.01.
Cathepsin involvement in ETO-induced death
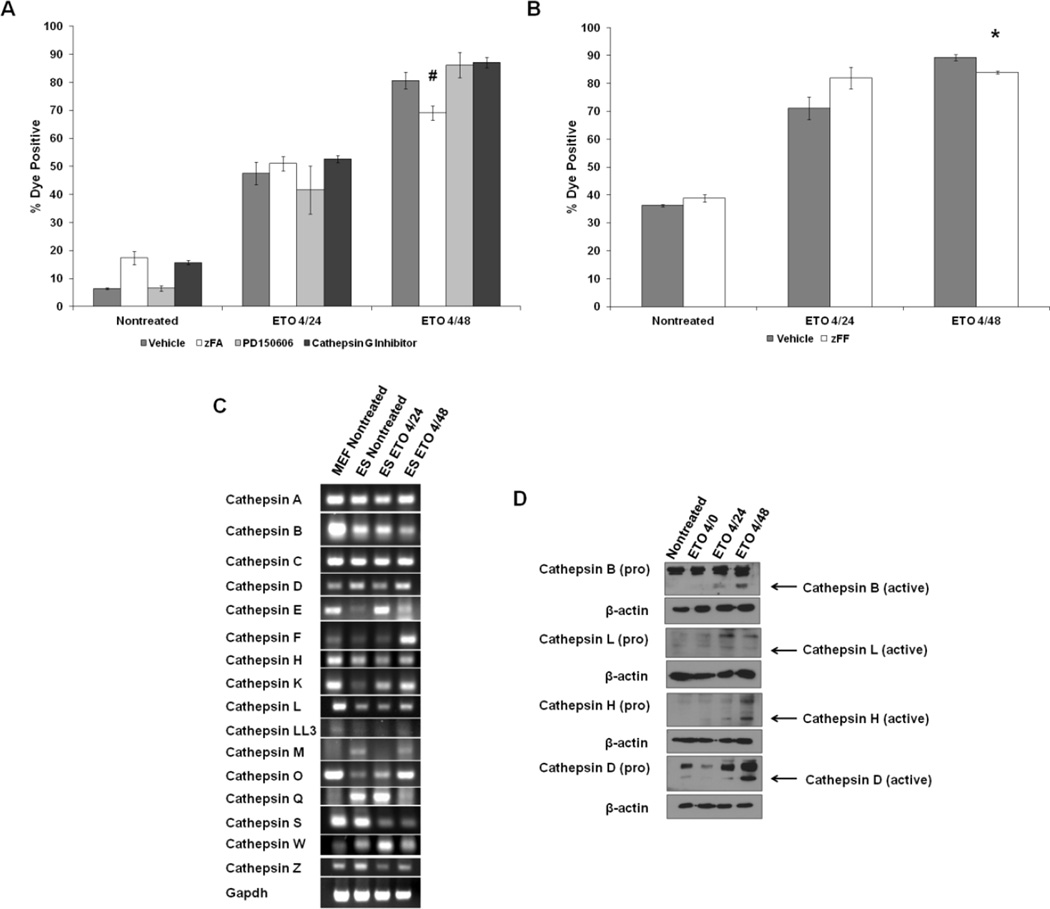
Cleavage of Parp-1, a PCD metric, is mainly affected by cathepsins and calpains, as well as caspases (Chaitanya et al., 2010; Gobeil et al., 2001). Lysosomal proteases such as the cathepsins are likely candidates as enzymes that cleave Parp-1 in mESCs, since treatment with BA1 reduced PCD levels (Fig. 2D). To test whether cathepsins and/or calpains are involved in ETO-induced PCD, ESCs were treated with ETO and treated concomitantly with a series of cathepsin and calpain inhibitors. The inhibitors included zFA, an inhibitor of cathepsins B, L, S, and H, PD150606, an inhibitor of calpains 1 and 2, and a cathepsin G-specific inhibitor. Treatment with zFA significantly reduced the level of cell death at the 4/48 time point (Fig. 3A). No effect was observed with the calpain or cathepsin G inhibitors. These data were confirmed using zFF, a cathepsin inhibitor which inhibits only cathepsins B and L. Again, there was a modest but statistically significant reduction in ETO-induced PCD by 48 hours (Fig. 3B), suggesting that a collaboration of several cathepsins is involved in ETO-induced PCD in mESCs.
Figure 3. Role of lysosomal proteases in ETO PCD.
A. ESCs were treated with ETO or left nontreated and coincubated with zFA to inhibit lysosomal proteases B, L, S, and H, PD150606 to inhibit calpains 1 and 2, or a cathepsin G inhibitor and subjected to flow cytometry to measure loss of plasma membrane integrity. A minimum of 6 experiments are displayed. Error bars represent the SEM. B. ESCs were treated with zFF and cotreated with ETO as indicated. Flow cytometry measuring violet cell stain is displayed. C. RT-PCR of murine cathepsins. RNA was isolated from nontreated MEFs, ESCs, as well as ESCs treated with ETO and harvested at the 4/24 and 4/48 time points. Complimentary DNA was generated and PCR was performed using primers designed against murine cathepsins. Gapdh served as a loading control. D. ESCs were treated as indicated and whole cell extracts were used for western blots. Levels of inactive (pro) and active cathepsins are displayed. Actin served as a loading control. *p<0.05#p<0.01.
Since none of the broad cathepsin inhibitors completely protected the mESCs from cell death, in order identify which cathepsins are induced following ETO treatment that may play also play a role in PCD, quantitative RT-PCR was used to probe the levels of all known murine cathepsin mRNAs in unchallenged MEFs and ESCs, and from ESCs treated with ETO and allowed to recover. A majority of the cathepsin genes were expressed in both cell types at varying levels; with some exceptions, the levels were generally higher in MEFs than in ESCs (Fig. 3C). Several cathepsins were undetectable in either cell type, including the placental cathepsins J, R, 3, 6, and 8, as well as cathepsins 7 and G. When nontreated ESCs were compared to ESCs treated with ETO and allowed to recover, no discernible pattern was apparent. The mRNA levels for cathepsins E, F, K, L, O, and Q, however, increased in the ESCs treated with ETO, suggesting that in addition to cathepsins B, L, S, and H, many other cathepsins may variably participate in ETO-induced PCD. To investigate the activation status of several cathepsins following ETO treatment in ESCs, western blots were conducted using commercially available antibodies. Figure 3D shows that cathepsins B, L, H, and D are all activated following ETO exposure, albeit at different levels, indicating that these cathepsins participate in ETO-induced PCD.
Role of p53 in ETO-induced death
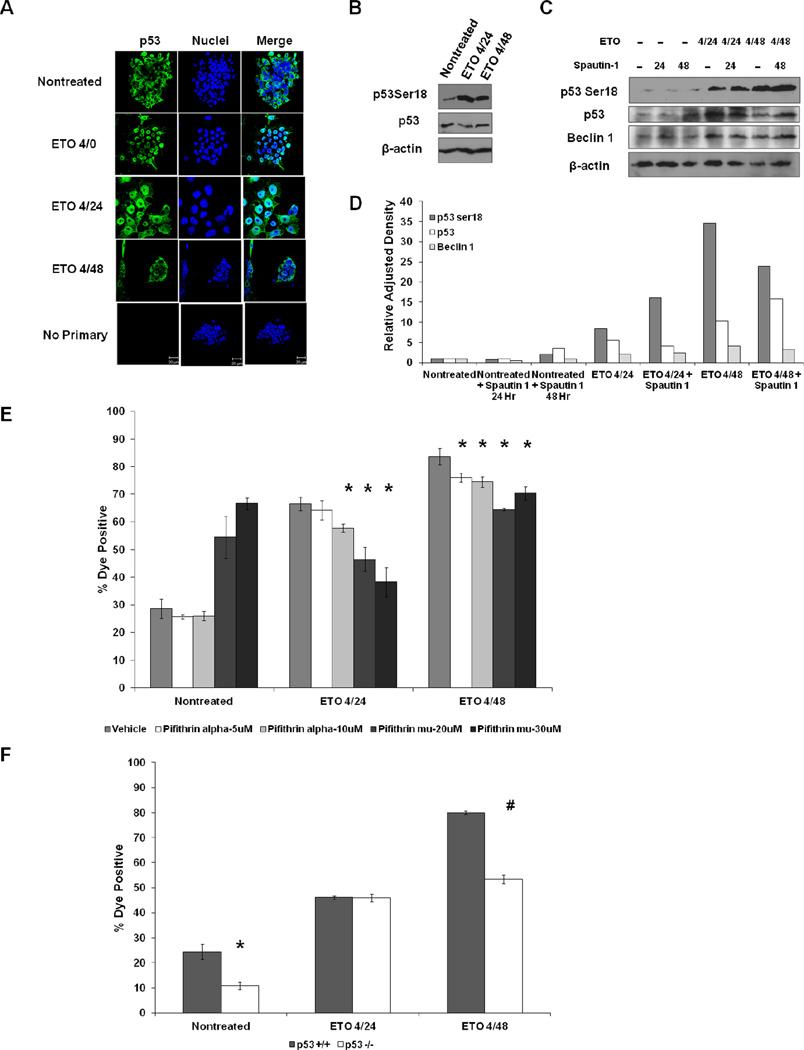
The p53 protein has been implicated in ETO-induced PCD in hESCs (Grandela et al., 2007) and other cell types (Arriola et al., 1999; Karpinich et al., 2002), and can play a role in the disruption of the lysosomal membrane, which subsequently results in cathepsin release during PCD (Johansson et al., 2010; Li et al., 2007; Yuan et al., 2002). In mESCs, p53 is stable and predominantly cytoplasmic (Bialik et al., 2004; Edinger et al., 2004; Findeisen et al., 2004), an observation we confirmed (Fig. 4A). When challenged with ETO, a large portion of p53 was translocated to the nucleus, while a fraction remained extranuclear. Phosphorylation of p53 ser-18 (p53 ser-15 in humans) by ATM is a signature of p53 stabilization and activation (Ashcroft et al., 2000; Nakagawa et al., 1999). Western blots of mESC lysates showed that phosphorylated p53 substantially increased after ETO treatment, while the total level of p53 remained stable (Fig. 4B).
Figure 4. P53 promotes ETO-induced PCD.
A. ESCs were treated with ETO or left nontreated and fixed and stained with p53 antibody. B. Western blot using ESC whole cell lysates that were treated with ETO or left nontreated and harvested at the indicated time points and probed with the indicated antibodies. Actin served as a loading control. C. Western blots displaying the levels of p53 and Beclin 1 proteins in ESCs treated with ETO or left nontreated and incubated with Spautin 1. D. Quantitation of protein levels in C using ImageJ software. Band intensities were normalized to respective actin controls. E. Flow cytometric death analysis of ESCs treated with ETO or left nontreated and incubated with either pifithrin α to prevent transactivation of p53 target genes or pifithrin μ to prevent any mitochondrial function of p53. F. Flow cytometric death analysis of p53 proficient (+/+) ESCs (WD44) or p53 deficient (−/−) ESCs.*p<0.05; #p<0.01
The level of p53 protein is regulated in multiple ways. As previously indicated, Spautin 1 inhibits proteases USP10 and USP13, which results in the stabilization of Beclin 1. USP10 also has been implicated in regulating the abundance and function of p53 (Reece et al., 2010). Since Beclin 1 does not appear to be involved in ETO-induced cell death in mESCs (Fig. 3C), we considered the possibility that p53, the other known target of Spautin 1, might contribute to ETO-mediated cell death. Treating mESCs with Spautin 1 reduced the level of p53 protein following ETO administration compared to cells treated with ETO alone, but only at the 4/24 time point (Fig. 4C and 4D). A slight reduction in Beclin 1 was also seen in the ETO 4/48 time point when cells were cotreated with Spautin 1 (Fig. 4C). However, levels of p53 were not significantly reduced in cells concomitantly with ETO and Spautin 1 at the 4/48 time point. The data suggest that Spautin 1 inhibits cell death through a novel mechanism, since knockdown of Beclin 1 had little effect on cell death levels following ETO treatment (Fig. 2C) and p53 levels are not dramatically reduced in ETO and Spautin 1 treated cells at the 4/48 time point (Fig. 4D). Spautin 1 also appears to induce a cellular stress response, as evidenced by an increase in the level of p53 phospho-ser 18 levels in the cells jointly treated with ETO and Spautin 1 compared to control cells treated only with ETO (Fig. 4C).
In many cell types, phosphorylation of ser18 on p53 results in p53 stabilization and its translocation to the nucleus, where it can induce the expression of target genes involved in cell cycle arrest and apoptosis. To address whether p53 plays a role in ETO-induced PCD in mESCs, the levels of cell death were quantified after treatment with ETO alone or together with pifithrin α to prevent the transactivation of p53. Since p53, in addition to its nuclear functions, can also play a role in PCD by translocating to the mitochondria and activating BAX (Pettersen et al., 2004), mESCs were treated with ETO and with pifithrin μ to reduce p53 association with mitochondria before assessment of cell death. Cell death was significantly reduced in a dose-dependent manner when cells were treated with either pifithrin α or pifithrin μ in addition to ETO at both the 4/24 and 4/48 time points (Fig. 4E). Although treatment with pifithrin μ by itself was slightly cytotoxic, it protected the cells from ETO-induced cell death. At lower doses (10µM), pifithrin μ was not cytotoxic but the overall reduction in ETO-induced PCD was less pronounced but still statistically significant (Supplemental Fig. 6). To confirm that p53 plays a role in ETO-induced PCD, p53 −/− and p53 +/+ mESCs were assayed for levels of cell death following ETO exposure. ETO-induced PCD was significantly lower in the p53-deficient ESCs at the 4/48 time point compared with a p53 proficient controls (Fig. 4F), indicating that p53 is a significant participant in ETO-induced PCD in mESCs.
ETO-induced DNA fragmentation
When mESCs are treated with ETO, DNA is cleaved into fragments of defined size, a hallmark of PCD. Fragment size depends on the nuclease activated in the death signaling cascade. Although CAD cleaves cellular DNA into nucleosomal-sized fragments in many cell types, it is unlikely to be the main culprit enzyme in mESC-induced PCD since caspases do not appear to be significantly activated in mESCs following ETO exposure and since caspase inhibition does not result in a reduction in cell death. Nevertheless, to test the involvement of CAD on DNA fragmentation in mESCs, the intracellular localization of CAD was determined before and after challenge with ETO. Prior to challenge, CAD was localized almost exclusively within the nucleus and remained there after treatment (Fig. 5A). This is noteworthy since in some cell types, CAD resides in the cytoplasm and translocates to the nucleus upon death (Liu et al., 1997). In other cell types CAD remains in the nucleus prior to and following DNA damaging treatments (Samejima et al., 1998). The inhibitor of CAD, ICAD, which exists as two splice variants designated ICADL and ICADS, regulates CAD activity. Caspases are among the main proteases that cleave and inactivate ICAD, but they are not alone in this activity. After treatment of mESCs with ETO and subsequent recovery, there was some ICAD degradation, particularly the ICADL isoform, suggesting that CAD could be involved in the observed DNA fragmentation (Fig. 5B). The ancillary degradation of ICAD protein during late stage cell death by nonspecific proteases, however, cannot be excluded.
Figure 5. DNA Fragmentation in ETO-induced PCD.
A. ESCs were treated as indicated, fixed, and stained with antibody to caspase-activated DNase (CAD). Nuclei were stained with Draq5. B. Western blot on ESCs treated as indicated and probed for antibody to ICAD or actin. C. ESCs were treated as indicated and probed with antibody to apoptosis-inducing factor (AIF). Draq5 was used to stain nuclei. D. Immunofluorescent staining of EndoG in ESCs treated as indicated. Note that a large portion of EndoG translocates into the nucleus after treatment. E. Western blot showing the level of knockdown of EndoG using two different siRNAs in ESCs after treatment with ETO and harvested at the 4/48 time point. Protein lysates were generated from remaining attached cells. GFP was used to show level of transfection between the transfected cells. F. DNA fragmentation assay on DNA isolated from 4/48 ETO-treated cells, either floating or attached after transfection with control or EndoG siRNAs. G. Flow cytometric analysis of levels of death in ESCs following transfection of EndoG siRNAs and ETO treatment. *p<0.05; #p<0.01
Two other enzymes that may contribute to ETO-induced DNA fragmentation in mESCs are AIF and EndoG, both of which have the capacity to fragment DNA during PCD after translocating from the mitochondria to the nucleus. When mESCs were treated with ETO and allowed to recover, AIF remained extranuclear (Fig. 5C), indicating that it does not play a role in ETO-induced DNA fragmentation. EndoG, like AIF, localizes to the mitochondria, and can relocate to the nucleus and induce nucleosomal-sized DNA fragmentation, particularly under conditions of oxidative stress (Ishihara et al., 2006). During ETO-induced PCD, much of the EndoG was found in the cytosol prior to treatment and in the nucleus after ETO treatment, suggesting that it may participate in ETO-induced DNA fragmentation (Fig. 5D). When siRNAs to EndoG were used, the level of EndoG protein was reduced (Fig. 5E), as was the level of DNA fragmentation in both attached and floating cells (Fig. 5F), indicative of EndoG involvement in PCD. When cell death levels were measured in ESCs transfected with EndoG or control siRNAs and then treated with ETO, a significant reduction in cell death was observed, confirming a role for EndoG in ETO-induced PCD in mESCs (Fig. 5G).
Discussion
Although ETO induces PCD in several cell types (Fujino et al., 2002; Mayorga et al., 2004; Mizumoto et al., 1994; Tsujimoto, 1997; Yoo et al., 2012), the pathway(s) involved are not completely understood. We now show that ETO induces a PCD pathway in mESCs that appears independent of caspase activity. Although there is a single report which argues that exposure to ETO induces caspase 3 activation (Grandela et al., 2007), it is based on hESCs, which may differ in behavior from mESCs. A separate account supports our findings that caspase 3 is not activated in mESCs (Mantel et al., 2007), arguing that cells of the same type but of different species origin may differ in their response to ETO exposure. In addition to their roles in apoptosis, the activity of caspases, particularly that of caspase 3, have been implicated in retinoic acid-induced differentiation of hESCs, and in the differentiation of both murine-derived myoblasts and neural stem cells (Fernando et al., 2005; Fujita et al., 2008; Larsen et al., 2010). Similarly, nucleoside analogs can trigger caspase-7-dependent cleavage of Oct4 to induce differentiation of embryonic carcinoma cells, which have many similarities to ESCs, suggesting that caspase activity is important during the differentiation process (Musch et al., 2010). We observe caspase 3 cleavage during retinoic acid-induced mESC differentiation (Supplemental Fig. 7), suggesting that caspase activation is stimulus-specific in mESCs. Although the data are not sufficient to argue unequivocally that caspases directly contribute to charontosis in mESCs, their participation remains a viable possibility. Survivin and XIAP, members of the inhibitor of apoptosis protein (IAP) family, can bind to and inhibit caspases, and are abundant in unchallenged mESCs (Supplemental Fig. 8A). Survivin is regulated by the Oct4 pluripotency transcription factor (Guo et al., 2008), which may explain why it is expressed at such high levels and why caspase activity appears inhibited in mESCs, at least after ETO treatment. Also, both Survivin and c-IAP2 proteins are induced following ETO treatment in mESCs (Supplemental Fig. 8B). Thus, the low caspase activity may be explained in part by the level of IAP expression in mESCs.
After a four hour exposure to ETO and 44 hours of recovery, RIP1 expression was elevated, indicative of necroptosis. This observation is similar to what is seen in cell lines treated with ETO, and is consistent with the suggestion that an increase in RIP1 may contribute to necroptosis in ETO-mediated PCD (Tenev et al., 2011). A functional role for increased RIP1 expression in ETO-induced necroptosis in mESCs, however was not seen, either by siRNA inhibition of the RIP kinases, or by necrostatin-1 inhibition of RIP1. Interestingly, necroptosis, specifically the formation of the ripoptosome, can also be inhibited by c-IAP1 and c-IAP2 (Feoktistova et al., 2011). Expression of c-IAP2 was increased following ETO treatment (Supplemental Fig. 8B), suggesting that elevated IAPs might impair necroptosis in ESCs treated with ETO.
Autophagy is frequently activated during PCD in many cell types, irrespective of how cell death is induced (Goehe et al., 2012; Hughson et al., 2012; Munoz-Gamez et al., 2009; Orlotti et al., 2012). Exposure of human cervical cancer cells to therapeutics such as ETO promotes autophagy, while inhibition of autophagy by 3-methyladenine reduces PCD (Lee et al., 2007). Similarly, ETO treatment of Bax/Bak-deficient MEFs induces cell death with autophagic features (Shimizu et al., 2004). Conflicting evidence, however, suggests that reducing autophagy in HepG2 cells with siRNA to Beclin 1 or with 3-methyladenine increases ETO-induced cell death (Xie et al., 2011). Similar results in other cancer model systems (Amaravadi et al., 2007; Apel et al., 2008; Guo et al., 2012; Han et al., 2011), have led to clinical trials in which autophagy is targeted (Amaravadi et al., 2011). Whether autophagy is a direct activator of cell death or a passive bystander activated during the process of PCD remains controversial. There are conflicting data from different cell models and a lack of specific pharmacological inhibitors of autophagy (Kroemer et al., 2008; Levine et al., 2005). The resolution to this issue may not be simple, since autophagy may be a prerequisite to some PCD pathways (Chen et al., 2011), and inhibition of autophagy may induce compensatory pathways of PCD activation. The timing at which autophagy is inhibited and the impact that this timing may have on PCD is yet another confounding factor. Inhibition of early signaling within the autophagy pathway after treatment of glioma cells with imatinib promotes cell survival, while the inhibition of autophagy at a later stage results in loss of the protective effect and increased cell death (Shingu et al., 2009). Inhibition of autophagy in mESCs at several different stages of the autophagic process using siRNAs revealed no significant reduction of PCD following ETO exposure. In fact, knockdown of the autophagy mediators Atg5 or Beclin 1 tended to produce a higher level of cell death. Although treatment of mESCs with BA1, an inhibitor of late stage autophagy, significantly reduced PCD, an unequivocal role for autophagy in PCD was not established. Indeed, the data indicated that autophagy in mESCs has a pro-survival function in mESCs after ETO treatment. This finding suggested that other pathways, including those involving the lysosome, might be active in mESC PCD.
Because cathepsins are located within lysosomes and can be targeted by BA1, and because they participate in many cell death pathways, they are attractive candidates as effectors of charontosis in mESCs (Broker et al., 2004; Droga-Mazovec et al., 2008; Hsu et al., 2009; Kagedal et al., 2001; Zuzarte-Luis et al., 2007). We have shown that a broad spectrum cathepsin inhibitor, which targets cathepsins B, L, S, and H, produces a significant reduction or delay in ETO-induced PCD. These specific cathepsins, however, do not contribute to Parp-1 cleavage (Supplemental Fig. 9). Although many other cathepsins were elevated after treatment with ETO at the RNA level, their functional involvement in Parp-1 cleavage and charontosis is only inferential. Since cathepsins exist as zymogens that are not fully active until they are cleaved, RNA expression data cannot be considered sufficient to assign functional activity. Thus, the cathepsin(s) responsible for Parp-1 cleavage, and other cathepsins that promote charontosis have not yet been identified.
Cathepsins other than B, L, S, and H can also be involved in PCD signaling. For example, the involvement of cathepsin D in PCD has been described in response to a variety of stimuli (Johnson, 2000). Cathepsin D is also released from lysosomes into the cytosol in U937 lymphoma cells and HeLa cells in response to several DNA damaging agents, including ETO (Emert-Sedlak et al., 2005). Furthermore, siRNAs that target cathepsin D in these cells can partially alleviate PCD (Emert-Sedlak et al., 2005). We have also observed cathepsin D activation following ETO exposure in mESCs (Fig. 3D), however, the lack of a specific pharmacological inhibitor to cathepsin D has made assaying its role in ETO-induced PCD difficult. In MEFs deficient for Bak and Bax, cathepsin Q plays a role in ETO-induced PCD signaling that requires p53 transcriptional activity (Tu et al., 2009). Thus, the possibility that additional cathepsins collaborate in ETO-induced charontosis remains a viable one, especially since the protective effect that zFA exerts on ETO-treated mESCs is not sufficient to completely alleviate the high level of PCD.
In hESCs, p53 transactivation of target genes is important for ETO-induced PCD (Grandela et al., 2007). Inhibition of p53 by pifithrin μ also significantly reduced levels of cell death in these cells. Consistent with these data, we show that pifithrin μ protects mESCs from cell death, suggesting that p53 plays a role in mitochondrial-dependent PCD. Additionally, treatment of mESCs with pifithrin α was also protective against ETO-induced PCD, although not to the same extent as pifithrin μ. The ability of pifithrin α to prevent p53 transactivation is somewhat controversial, particularly at doses greater than 30µM (Walton et al., 2005). However, several other studies show that pifithrin α does indeed prevent the expression of p53 target genes, particularly at doses of 30µM or lower (Bragado et al., 2007; Jiang et al., 2006; Menendez et al., 2011; Schneider-Stock et al., 2005). At a high dose (30µM), pifithrin α had no effect on ETO-induced PCD levels (Supplemental Fig. 6). At lower doses, mESCs displayed slight but statistically significant reductions in ETO-induced PCD, indicating that p53 transactivation is partially responsible for ETO-induced PCD (Fig. 4D). A comparison of ETO-induced death levels in p53 +/+ and −/− mESCs has shown that p53 is unequivocally involved in charontosis. However, since complete protection from death was not observed in p53 −/− cells, then p53-independent pathways must also be involved. We cannot rule out the contribution of the p53 family members p63 and p73, particularly since data describing the functions of these proteins in mESCs are severely lacking. Both of these proteins have been reported to induce apoptosis in a variety of cell types, dependent and independent of p53 activity in PCD signaling (Alonso et al., 2009; Codelia et al., 2010; Dohn et al., 2001; Flores et al., 2002; Melino et al., 2004; Petre-Lazar et al., 2007; Pyati et al., 2011; Rana et al., 2010).
DNA fragmentation is an essential component of PCD. Of several nucleases was tested for their involvement in PCD, EndoG emerged as a likely candidate, although a role for CAD cannot be discounted due to the degradation of ICAD observed 44 hours after ETO treatment. EndoG is activated in cells undergoing oxidative stress and translocates to the nucleus to fragment DNA (Higgins et al., 2009; Ishihara et al., 2006). ETO induces oxidative stress and reactive oxygen species (ROS) in a variety of cell types (England et al., 2004; Hirano et al., 2004; Rojas et al., 2009), an observation we have observed in mESCs after ETO treatment (Supplemental Fig. 10), adding credence to the involvement of EndoG in this process. Finally, since pifithrin μ inhibits p53 association with mitochondria and results in decreased PCD in ETO-treated mESCs, and since EndoG is released from mitochondria to promote DNA fragmentation in response to ETO treatment, p53 may contribute to EndoG.
In summary, we have shown that ETO induces massive DNA DSBs, an accumulation of cells in the G2 phase of the cell cycle, and extensive PCD in mESCs (Fig. 1; Supplemental Fig. 11). This PCD appears to be independent of caspase activity and of RIP kinases, which are active in necroptosis. Knockdown of proteins that are integral to autophagy did not reduce PCD, whereas chemical inhibitors of autophagy did significantly decrease the high levels of cell death after treatment with ETO. The possibility that autophagy itself promotes PCD was excluded, since the activation of autophagy during ETO treatment promoted cell survival and not cell death. When mESCs were exposed to a broad spectrum inhibitor of cathepsins, the level of cell death was significantly reduced suggesting that these lysosomal proteases were involved in PCD. The involvement of p53 was also queried by use of inhibitors that prevent p53 transactivation or mitochondrial translocation. Inhibitors of both p53 activities significantly protected ESCs from ETO-induced PCD, albeit at varying degrees. Lastly, we have identified a novel role for EndoG in the ETO-induced PCD pathway in mESCs, a finding that has not been described in ETO-induced PCD in any other cell type. We have coined the term charontosis to describe this PCD pathway, which is summarized in Figure 6.
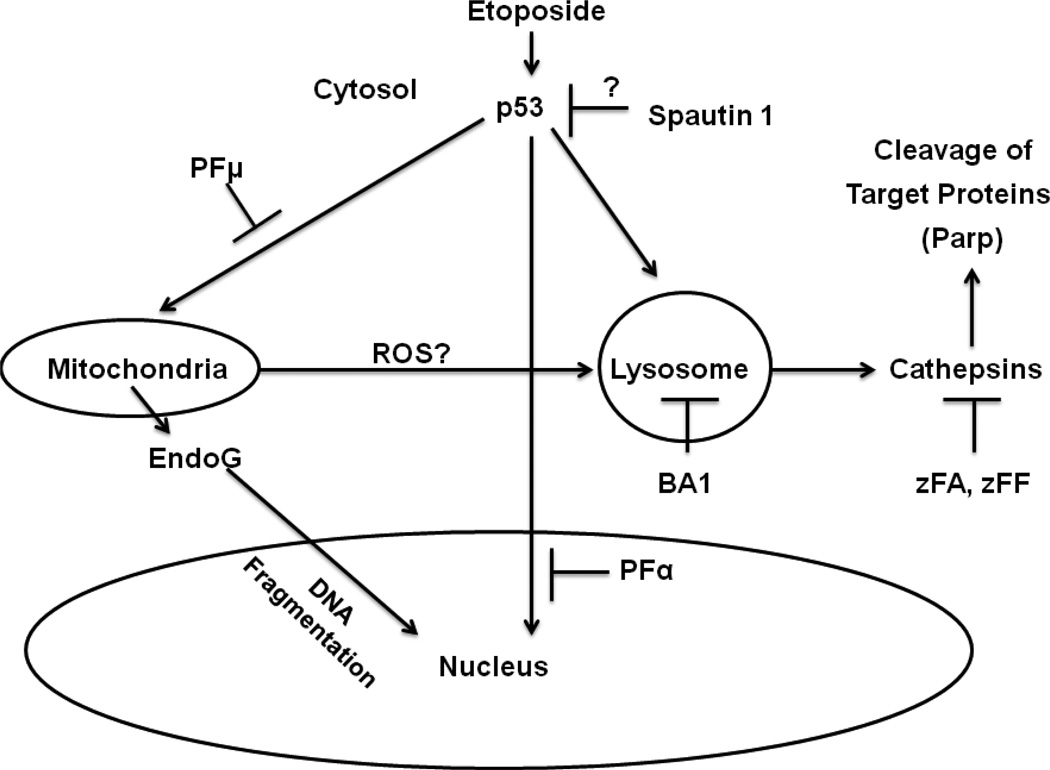
Figure 6. Model of ETO-induced PCD (Charontosis) in mESCs.
Etoposide promotes p53 translocation from the cytoplasm to the nucleus to promote transcription of target genes involved in cell death. This activity is reduced following treatment with pifithrin α (PFα). A portion of p53 remains outside the nucleus and presumably translocates to mitochondria to inactivate pro-survival Bcl-2 family members. Pifithrin μ (PFμ) can reduce p53 association with mitochondria, resulting in protection from death. The mitochondria may release ROS, which can disrupt the lysosomal membrane and cause release of cathepsins, which cleave specific target proteins. P53 may also translocate to the lysosome and result in the release of cathepsins. The activities of several cathepsins are inhibited by zFA and zFF, which reduce ETO-induced PCD. Bafilomycin A1 (BA1) inhibits lysosomal acidification, resulting in decreased cathepsin activity and reduced PCD. Spautin1 can inhibit ubiquitin-specific proteases that can regulate p53, although other targets are known to exist. Finally, EndoG is released from mitochondria to induce DNA fragmentation.
Supplementary Material
Highlights.
Mouse embryonic stem cells (mESCs) are hypersensitive to DNA damaging agents.
Etoposide induces massive cell death in mESCs.
MESC death is dependent upon cathepsins, p53 and EndoG.
We coined the term charontosis to describe this novel cell death pathway.
Acknowledgements
We thank P Hexley and G Babcock for assistance with flow cytometry, performed at Shriners Hospitals for Children - Cincinnati, supported by a grant from the Shriners of North America SSF 84070. We would also like to thank N White and the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center, supported in part by NIH AR-47363, NIH DK78392 and NIH DK90971, for assistance with Imagestream flow cytometry data acquisition and analysis. We also thank S. Mylavarapu and H. Ma for assistance with data acquisition. This work was supported in part by grants R01 ES012695 and R01 ES12695-4S1 to PJS from the National Institutes of Health and the Center for Environmental Genetics and grant P30 ES006096 from NIEHS. EDT was supported by a NIH training grant T32 ES007250.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare that there are no conflicts of interest.
References
- Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- Alonso R, Lopez-Guerra M, Upshaw R, Bantia S, Smal C, Bontemps F, Manz C, Mehrling T, Villamor N, Campo E, Montserrat E, Colomer D. Forodesine has high antitumor activity in chronic lymphocytic leukemia and activates p53-independent mitochondrial apoptosis by induction of p73 and BIM. Blood. 2009;114:1563–1575. doi: 10.1182/blood-2009-02-207654. [DOI] [PubMed] [Google Scholar]
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- Arriola EL, Lopez AR, Chresta CM. Differential regulation of p21waf-1/cip-1 and Mdm2 by etoposide: etoposide inhibits the p53-Mdm2 autoregulatory feedback loop. Oncogene. 1999;18:1081–1091. doi: 10.1038/sj.onc.1202391. [DOI] [PubMed] [Google Scholar]
- Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr Med Chem Anticancer Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- Bialik S, Kimchi A. DAP-kinase as a target for drug design in cancer and diseases associated with accelerated cell death. Semin Cancer Biol. 2004;14:283–294. doi: 10.1016/j.semcancer.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Bragado P, Armesilla A, Silva A, Porras A. Apoptosis by cisplatin requires p53 mediated p38alpha MAPK activation through ROS generation. Apoptosis. 2007;12:1733–1742. doi: 10.1007/s10495-007-0082-8. [DOI] [PubMed] [Google Scholar]
- Broker LE, Huisman C, Span SW, Rodriguez JA, Kruyt FA, Giaccone G. Cathepsin B mediates caspase-independent cell death induced by microtubule stabilizing agents in non-small cell lung cancer cells. Cancer Res. 2004;64:27–30. doi: 10.1158/0008-5472.can-03-3060. [DOI] [PubMed] [Google Scholar]
- Cervantes RB, Stringer JR, Shao C, Tischfield JA, Stambrook PJ. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc Natl Acad Sci U S A. 2002;99:3586–3590. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaitanya GV, Steven AJ, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun Signal. 2010;8:31. doi: 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JW, Choi H, Cotman SL, Jung YK. Lithium rescues the impaired autophagy process in CbCln3(Deltaex7/8/Deltaex7/8) cerebellar cells and reduces neuronal vulnerability to cell death via IMPase inhibition. J Neurochem. 2011;116:659–668. doi: 10.1111/j.1471-4159.2010.07158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Chiu LY, Maa MC, Wang JS, Chien CL, Lin WW. zVAD-induced autophagic cell death requires c-Src-dependent ERK and JNK activation and reactive oxygen species generation. Autophagy. 2011;7:217–228. doi: 10.4161/auto.7.2.14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofferson DE, Yuan J. Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol. 2010;22:263–268. doi: 10.1016/j.ceb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford B, Beljin M, Stark GR, Taylor WR. G2 arrest in response to topoisomerase II inhibitors: the role of p53. Cancer Res. 2003;63:4074–4081. [PubMed] [Google Scholar]
- Codelia VA, Cisterna M, Alvarez AR, Moreno RD. p73 participates in male germ cells apoptosis induced by etoposide. Mol Hum Reprod. 2010;16:734–742. doi: 10.1093/molehr/gaq045. [DOI] [PubMed] [Google Scholar]
- Cote J, Ruiz-Carrillo A. Primers for mitochondrial DNA replication generated by endonuclease G. Science. 1993;261:765–769. doi: 10.1126/science.7688144. [DOI] [PubMed] [Google Scholar]
- Declercq W, Vanden Berghe T, Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell. 2009;138:229–232. doi: 10.1016/j.cell.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20:3193–3205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- Droga-Mazovec G, Bojic L, Petelin A, Ivanova S, Romih R, Repnik U, Salvesen GS, Stoka V, Turk V, Turk B. Cysteine cathepsins trigger caspase-dependent cell death through cleavage of bid and antiapoptotic Bcl-2 homologues. J Biol Chem. 2008;283:19140–19150. doi: 10.1074/jbc.M802513200. [DOI] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Emert-Sedlak L, Shangary S, Rabinovitz A, Miranda MB, Delach SM, Johnson DE. Involvement of cathepsin D in chemotherapy-induced cytochrome c release, caspase activation, and cell death. Mol Cancer Ther. 2005;4:733–742. doi: 10.1158/1535-7163.MCT-04-0301. [DOI] [PubMed] [Google Scholar]
- England K, O'Driscoll C, Cotter TG. Carbonylation of glycolytic proteins is a key response to drug-induced oxidative stress and apoptosis. Cell Death Differ. 2004;11:252–260. doi: 10.1038/sj.cdd.4401338. [DOI] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Hacker G, Leverkus M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando P, Brunette S, Megeney LA. Neural stem cell differentiation is dependent upon endogenous caspase 3 activity. FASEB J . 2005;19:1671–1673. doi: 10.1096/fj.04-2981fje. [DOI] [PubMed] [Google Scholar]
- Findeisen M, Vennemann M, Brinkmann B, Ortmann C, Rose I, Kopcke W, Jorch G, Bajanowski T. German study on sudden infant death (GeSID): design, epidemiological and pathological profile. Int J Legal Med. 2004;118:163–169. doi: 10.1007/s00414-004-0433-8. [DOI] [PubMed] [Google Scholar]
- Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- Fujino M, Li XK, Kitazawa Y, Guo L, Kawasaki M, Funeshima N, Amano T, Suzuki S. Distinct pathways of apoptosis triggered by FTY720, etoposide, and anti-Fas antibody in human T-lymphoma cell line (Jurkat cells) J Pharmacol Exp Ther. 2002;300:939–945. doi: 10.1124/jpet.300.3.939. [DOI] [PubMed] [Google Scholar]
- Fujita J, Crane AM, Souza MK, Dejosez M, Kyba M, Flavell RA, Thomson JA, Zwaka TP. Caspase activity mediates the differentiation of embryonic stem cells. Cell Stem Cell. 2008;2:595–601. doi: 10.1016/j.stem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008;135:1161–1163. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vanden Berghe T, Vanlangenakker N, Buettner S, Eisenberg T, Vandenabeele P, Madeo F, Kroemer G. Programmed necrosis from molecules to health and disease. Int Rev Cell Mol Biol. 2011;289:1–35. doi: 10.1016/B978-0-12-386039-2.00001-8. [DOI] [PubMed] [Google Scholar]
- Gobeil S, Boucher CC, Nadeau D, Poirier GG. Characterization of the necrotic cleavage of poly(ADP-ribose) polymerase (PARP-1): implication of lysosomal proteases. Cell Death Differ. 2001;8:588–594. doi: 10.1038/sj.cdd.4400851. [DOI] [PubMed] [Google Scholar]
- Goehe RW, Di X, Sharma K, Bristol M, Henderson S, Valerie K, Rodier F, Davalos A, Gewirtz D. The autophagy-senescence connection in chemotherapy; must tumor cells (self) eat before they sleep? . J Pharmacol Exp Ther. 2012 doi: 10.1124/jpet.112.197590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandela C, Pera MF, Grimmond SM, Kolle G, Wolvetang EJ. p53 is required for etoposide-induced apoptosis of human embryonic stem cells. Stem Cell Res. 2007;1:116–128. doi: 10.1016/j.scr.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Guo XL, Li D, Hu F, Song JR, Zhang SS, Deng WJ, Sun K, Zhao QD, Xie XQ, Song YJ, Wu MC, Wei LX. Targeting autophagy potentiates chemotherapy-induced apoptosis and proliferation inhibition in hepatocarcinoma cells. Cancer Lett. 2012;320:171–179. doi: 10.1016/j.canlet.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Guo Y, Mantel C, Hromas RA, Broxmeyer HE. Oct-4 is critical for survival/antiapoptosis of murine embryonic stem cells subjected to stress: effects associated with Stat3/survivin. Stem Cells. 2008;26:30–34. doi: 10.1634/stemcells.2007-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Sun J, Feng L, Wang K, Li D, Pan Q, Chen Y, Jin W, Wang X, Pan H, Jin H. Autophagy inhibition enhances daunorubicin-induced apoptosis in K562 cells. PLoS One. 2011;6:e28491. doi: 10.1371/journal.pone.0028491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins GC, Beart PM, Nagley P. Oxidative stress triggers neuronal caspase-independent death: endonuclease G involvement in programmed cell death-type III. Cell Mol Life Sci. 2009;66:2773–2787. doi: 10.1007/s00018-009-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kawai K, Ootsuyama Y, Orimo H, Kasai H. Detection of a mouse OGG1 fragment during caspase-dependent apoptosis: oxidative DNA damage and apoptosis. Cancer Sci. 2004;95:634–638. doi: 10.1111/j.1349-7006.2004.tb03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KF, Wu CL, Huang SC, Wu CM, Hsiao JR, Yo YT, Chen YH, Shiau AL, Chou CY. Cathepsin L mediates resveratrol-induced autophagy and apoptotic cell death in cervical cancer cells. Autophagy. 2009;5:451–460. doi: 10.4161/auto.5.4.7666. [DOI] [PubMed] [Google Scholar]
- Hughson LR, Poon VI, Spowart JE, Lum JJ. Implications of therapy-induced selective autophagy on tumor metabolism and survival. Int J Cell Biol. 2012;2012:872091. doi: 10.1155/2012/872091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara Y, Shimamoto N. Involvement of endonuclease G in nucleosomal DNA fragmentation under sustained endogenous oxidative stress. J Biol Chem. 2006;281:6726–6733. doi: 10.1074/jbc.M510382200. [DOI] [PubMed] [Google Scholar]
- Jiang M, Wei Q, Wang J, Du Q, Yu J, Zhang L, Dong Z. Regulation of PUMA-alpha by p53 in cisplatin-induced renal cell apoptosis. Oncogene. 2006;25:4056–4066. doi: 10.1038/sj.onc.1209440. [DOI] [PubMed] [Google Scholar]
- Johansson AC, Appelqvist H, Nilsson C, Kagedal K, Roberg K, Ollinger K. Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis. 2010;15:527–540. doi: 10.1007/s10495-009-0452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE. Noncaspase proteases in apoptosis. Leukemia. 2000;14:1695–1703. doi: 10.1038/sj.leu.2401879. [DOI] [PubMed] [Google Scholar]
- Joza N, Susin SA, Daugas E, Stanford WL, Cho SK, Li CY, Sasaki T, Elia AJ, Cheng HY, Ravagnan L, Ferri KF, Zamzami N, Wakeham A, Hakem R, Yoshida H, Kong YY, Mak TW, Zuniga-Pflucker JC, Kroemer G, Penninger JM. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410:549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- Kagedal K, Johansson U, Ollinger K. The lysosomal protease cathepsin D mediates apoptosis induced by oxidative stress. FASEB J. 2001;15:1592–1594. doi: 10.1096/fj.00-0708fje. [DOI] [PubMed] [Google Scholar]
- Karpinich NO, Tafani M, Rothman RJ, Russo MA, Farber JL. The course of etoposide-induced apoptosis from damage to DNA and p53 activation to mitochondrial release of cytochrome c. J Biol Chem. 2002;277:16547–16552. doi: 10.1074/jbc.M110629200. [DOI] [PubMed] [Google Scholar]
- Kawahara A, Ohsawa Y, Matsumura H, Uchiyama Y, Nagata S. Caspase-independent cell killing by Fas-associated protein with death domain. J Cell Biol. 1998;143:1353–1360. doi: 10.1083/jcb.143.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JA, Longo-Guess CM, Rossmann MP, Seburn KL, Hurd RE, Frankel WN, Bronson RT, Ackerman SL. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419:367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci U S A. 2010;107:4230–4235. doi: 10.1073/pnas.0913089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Tong SY, Kim JJ, Um SJ, Park JS. Caspase-independent autophagic cytotoxicity in etoposide-treated CaSki cervical carcinoma cells. DNA Cell Biol. 2007;26:713–720. doi: 10.1089/dna.2007.0577. [DOI] [PubMed] [Google Scholar]
- Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Li N, Zheng Y, Chen W, Wang C, Liu X, He W, Xu H, Cao X. Adaptor protein LAPF recruits phosphorylated p53 to lysosomes and triggers lysosomal destabilization in apoptosis. Cancer Res. 2007;67:11176–11185. doi: 10.1158/0008-5472.CAN-07-2333. [DOI] [PubMed] [Google Scholar]
- Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Hao Y, Choi A, Ke H, Ma D, Yuan J. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Mantel C, Guo Y, Lee MR, Kim MK, Han MK, Shibayama H, Fukuda S, Yoder MC, Pelus LM, Kim KS, Broxmeyer HE. Checkpoint-apoptosis uncoupling in human and mouse embryonic stem cells: a source of karyotpic instability. Blood. 2007;109:4518–4527. doi: 10.1182/blood-2006-10-054247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- Mayorga M, Bahi N, Ballester M, Comella JX, Sanchis D. Bcl-2 is a key factor for cardiac fibroblast resistance to programmed cell death. J Biol Chem. 2004;279:34882–34889. doi: 10.1074/jbc.M404616200. [DOI] [PubMed] [Google Scholar]
- Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, Knight RA, Green DR, Thompson C, Vousden KH. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- Menendez D, Shatz M, Azzam K, Garantziotis S, Fessler MB, Resnick MA. The Toll-like receptor gene family is integrated into human DNA damage and p53 networks. PLoS Genet. 2011;7:e1001360. doi: 10.1371/journal.pgen.1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Rothman RJ, Farber JL. Programmed cell death (apoptosis) of mouse fibroblasts is induced by the topoisomerase II inhibitor etoposide. Mol Pharmacol. 1994;46:890–895. [PubMed] [Google Scholar]
- Munoz-Gamez JA, Rodriguez-Vargas JM, Quiles-Perez R, Aguilar-Quesada R, Martin-Oliva D, de Murcia G, Menissier de Murcia J, Almendros A, Ruiz de Almodovar M, Oliver FJ. PARP-1 is involved in autophagy induced by DNA damage. Autophagy. 2009;5:61–74. doi: 10.4161/auto.5.1.7272. [DOI] [PubMed] [Google Scholar]
- Musch T, Oz Y, Lyko F, Breiling A. Nucleoside drugs induce cellular differentiation by caspase-dependent degradation of stem cell factors. PLoS One. 2010;5:e10726. doi: 10.1371/journal.pone.0010726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K, Taya Y, Tamai K, Yamaizumi M. Requirement of ATM in phosphorylation of the human p53 protein at serine 15 following DNA double-strand breaks. Mol Cell Biol. 1999;19:2828–2834. doi: 10.1128/mcb.19.4.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam C, Doi K, Nakayama H. Etoposide induces G2/M arrest and apoptosis in neural progenitor cells via DNA damage and an ATM/p53-related pathway. Histol Histopathol. 2010;25:485–493. doi: 10.14670/HH-25.485. [DOI] [PubMed] [Google Scholar]
- Orlotti NI, Cimino-Reale G, Borghini E, Pennati M, Sissi C, Perrone F, Palumbo M, Daidone MG, Folini M, Zaffaroni N. Autophagy acts as a safeguard mechanism against G-quadruplex ligand-mediated DNA damage. Autophagy. 2012;8 doi: 10.4161/auto.20519. [DOI] [PubMed] [Google Scholar]
- Petre-Lazar B, Livera G, Moreno SG, Trautmann E, Duquenne C, Hanoux V, Habert R, Coffigny H. The role of p63 in germ cell apoptosis in the developing testis. J Cell Physiol. 2007;210:87–98. doi: 10.1002/jcp.20829. [DOI] [PubMed] [Google Scholar]
- Pettersen AA, Seljeflot I, Abdelnoor M, Arnesen H. Unstable angina, stroke, myocardial infarction and death in aspirin non-responders. A prospective, randomized trial. The ASCET (ASpirin non-responsiveness and Clopidogrel Endpoint Trial) design. Scand Cardiovasc J. 2004;38:353–356. doi: 10.1080/14017430410024324. [DOI] [PubMed] [Google Scholar]
- Pyati UJ, Gjini E, Carbonneau S, Lee JS, Guo F, Jette CA, Kelsell DP, Look AT. p63 mediates an apoptotic response to pharmacological and disease-related ER stress in the developing epidermis. Dev Cell. 2011;21:492–505. doi: 10.1016/j.devcel.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana S, Gupta K, Gomez J, Matsuyama S, Chakrabarti A, Agarwal ML, Agarwal A, Agarwal MK, Wald DN. Securinine induces p73-dependent apoptosis preferentially in p53-deficient colon cancer cells. FASEB J. 2010;24:2126–2134. doi: 10.1096/fj.09-148999. [DOI] [PubMed] [Google Scholar]
- Reece KM, Figg WD. A novel regulator (USP10) of p53: Implications for tumor suppression and therapeutic targeting. Cancer Biol Ther. 2010;9:583–584. doi: 10.4161/cbt.9.8.11690. [DOI] [PubMed] [Google Scholar]
- Rojas E, Mussali P, Tovar E, Valverde M. DNA-AP sites generation by etoposide in whole blood cells. BMC Cancer. 2009;9:398. doi: 10.1186/1471-2407-9-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima K, Earnshaw WC. ICAD/DFF regulator of apoptotic nuclease is nuclear. Exp Cell Res. 1998;243:453–459. doi: 10.1006/excr.1998.4212. [DOI] [PubMed] [Google Scholar]
- Schneider-Stock R, Diab-Assef M, Rohrbeck A, Foltzer-Jourdainne C, Boltze C, Hartig R, Schonfeld P, Roessner A, Gali-Muhtasib H. 5-Aza-cytidine is a potent inhibitor of DNA methyltransferase 3a and induces apoptosis in HCT-116 colon cancer cells via Gadd45- and p53-dependent mechanisms. J Pharmacol Exp Ther. 2005;312:525–536. doi: 10.1124/jpet.104.074195. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Shingu T, Fujiwara K, Bogler O, Akiyama Y, Moritake K, Shinojima N, Tamada Y, Yokoyama T, Kondo S. Stage-specific effect of inhibition of autophagy on chemotherapy-induced cytotoxicity. Autophagy. 2009;5:537–539. doi: 10.4161/auto.5.4.8164. [DOI] [PubMed] [Google Scholar]
- Sleiman RJ, Stewart BW. Early caspase activation in leukemic cells subject to etoposide-induced G2-M arrest: evidence of commitment to apoptosis rather than mitotic cell death. Clin Cancer Res. 2000;6:3756–3765. [PubMed] [Google Scholar]
- Smith PJ, Soues S, Gottlieb T, Falk SJ, Watson JV, Osborne RJ, Bleehen NM. Etoposide-induced cell cycle delay and arrest-dependent modulation of DNA topoisomerase II in small-cell lung cancer cells. Br J Cancer. 1994;70:914–921. doi: 10.1038/bjc.1994.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, Meier P. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Tichy ED. Mechanisms maintaining genomic integrity in embryonic stem cells and induced pluripotent stem cells. Exp Biol Med (Maywood) 2011;236:987–996. doi: 10.1258/ebm.2011.011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy ED, Liang L, Deng L, Tischfield J, Schwemberger S, Babcock G, Stambrook PJ. Mismatch and base excision repair proficiency in murine embryonic stem cells. DNA Repair (Amst) 2011;10:445–451. doi: 10.1016/j.dnarep.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy ED, Pillai R, Deng L, Liang L, Tischfield J, Schwemberger SJ, Babcock GF, Stambrook PJ. Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 2010;19:1699–1711. doi: 10.1089/scd.2010.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy ED, Pillai R, Deng L, Tischfield JA, Hexley P, Babcock GF, Stambrook PJ. The abundance of Rad51 protein in mouse embryonic stem cells is regulated at multiple levels. Stem Cell Res. 2012;9:124–134. doi: 10.1016/j.scr.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y. Apoptosis and necrosis: intracellular ATP level as a determinant for cell death modes. Cell Death Differ. 1997;4:429–434. doi: 10.1038/sj.cdd.4400262. [DOI] [PubMed] [Google Scholar]
- Tu HC, Ren D, Wang GX, Chen DY, Westergard TD, Kim H, Sasagawa S, Hsieh JJ, Cheng EH. The p53-cathepsin axis cooperates with ROS to activate programmed necrotic death upon DNA damage. Proc Natl Acad Sci U S A. 2009;106:1093–1098. doi: 10.1073/pnas.0808173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sloun PP, Jansen JG, Weeda G, Mullenders LH, van Zeeland AA, Lohman PH, Vrieling H. The role of nucleotide excision repair in protecting embryonic stem cells from genotoxic effects of UV-induced DNA damage. Nucleic Acids Res. 1999;27:3276–3282. doi: 10.1093/nar/27.16.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton MI, Wilson SC, Hardcastle IR, Mirza AR, Workman P. An evaluation of the ability of pifithrin-alpha and -beta to inhibit p53 function in two wild-type p53 human tumor cell lines. Mol Cancer Ther. 2005;4:1369–1377. doi: 10.1158/1535-7163.MCT-04-0341. [DOI] [PubMed] [Google Scholar]
- Xie BS, Zhao HC, Yao SK, Zhuo DX, Jin B, Lv DC, Wu CL, Ma DL, Gao C, Shu XM, Ai ZL. Autophagy inhibition enhances etoposide-induced cell death in human hepatoma G2 cells. Int J Mol Med. 2011;27:599–606. doi: 10.3892/ijmm.2011.607. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yoon YG, Lee JS, Song YS, Oh JS, Park BS, Kwon TK, Park C, Choi YH, Yoo YH. Etoposide induces a mixed type of programmed cell death and overcomes the resistance conferred by Bcl-2 in Hep3B hepatoma cells. Int J Oncol. 2012 doi: 10.3892/ijo.2012.1585. [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Yuan XM, Li W, Dalen H, Lotem J, Kama R, Sachs L, Brunk UT. Lysosomal destabilization in p53-induced apoptosis. Proc Natl Acad Sci U S A. 2002;99:6286–6291. doi: 10.1073/pnas.092135599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuzarte-Luis V, Montero JA, Kawakami Y, Izpisua-Belmonte JC, Hurle JM. Lysosomal cathepsins in embryonic programmed cell death. Dev Biol. 2007;301:205–217. doi: 10.1016/j.ydbio.2006.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.