SUMMARY
Small-molecule ligands of nuclear hormone receptors (NHRs) govern the transcriptional regulation of metazoan development, cell differentiation, and metabolism. However, the physiological ligands of many NHRs remain poorly characterized primarily due to lack of robust analytical techniques. Using comparative metabolomics, we identified endogenous steroids that act as ligands of the C. elegans NHR, DAF-12, a vitamin-D and liver-X receptor homolog regulating larval development, fat metabolism, and lifespan. The identified molecules feature unexpected chemical modifications and include only one of two DAF-12 ligands reported earlier, necessitating a revision of previously proposed ligand biosynthetic pathways. We further show that ligand profiles are regulated by a complex enzymatic network including the Rieske oxygenase DAF-36, the short-chain dehydrogenase DHS-16, and the hydroxysteroid dehydrogenase, HSD-1. Our results demonstrate the advantages of comparative metabolomics over traditional candidate-based approaches and provide a blueprint for the identification of ligands for other C. elegans and mammalian NHRs.
INTRODUCTION
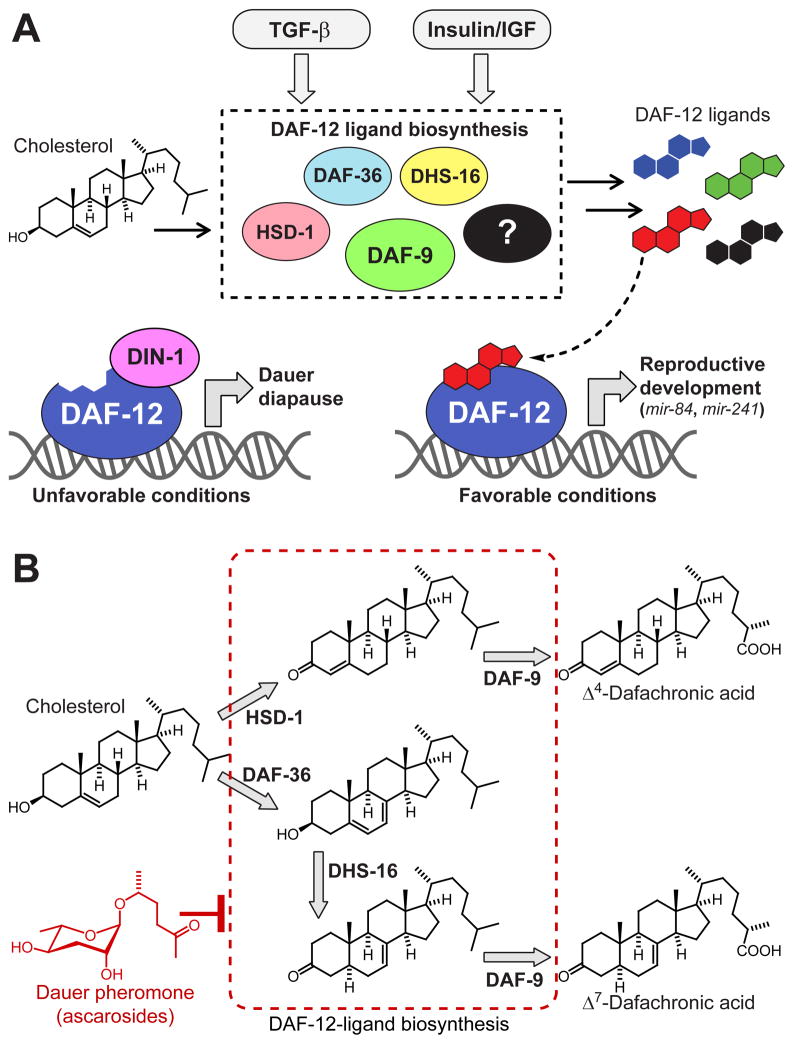
Small-molecule ligands of nuclear hormone receptors (NHRs), a conserved family of ligand-activated transcription factors, control diverse aspects of metabolism, cell differentiation, development, and aging. Precise knowledge of ligand structures and biosynthetic pathways is essential for understanding NHR function (Mangelsdorf et al., 1995; Wollam and Antebi, 2011), because even small differences in ligand structures may result in dramatic changes of transcriptional activity and specificity (Brown and Slatopolsky, 2008; Singarapu et al., 2011). However, the endogenous ligands of many NHRs remain poorly characterized, in part because ligands constitute very minor components of highly complex animal metabolomes (Schupp and Lazar, 2010). The free living nematode C. elegans has 284 NHRs, allows facile genetic manipulation, and can be grown in large quantities, providing an opportunity to investigate structures, biosynthesis, and functions of NHR ligands in a relatively simple model system (Taubert et al., 2010). Although many of the 284 C. elegans NHRs appear to be derived from extensive duplication and diversification of an ancestral gene related to mammalian HNF4 receptors and may not be ligand-regulated, several C. elegans NHRs represent orthologs of hormone-regulated NHRs in other metazoans (Antebi, 2006; Palanker et al., 2009; Taubert et al., 2010). The most prominent C. elegans NHR, DAF-12, a homolog of vertebrate vitamin D (VDR) and liver X receptors (LXR), functions as a ligand-dependent switch that regulates both adult lifespan and larval development (Antebi et al., 2000; Fielenbach and Antebi, 2008; Kenyon, 2010; Riddle et al., 1981; Riddle and Albert, 1997; Shen et al., 2012). The biosynthesis of the steroidal ligands of DAF-12 is controlled by a complex endocrine signaling network, of which many components appear to be conserved between C. elegans and mammals (Fielenbach and Antebi, 2008). Perception of environmental stimuli by chemosensory neurons modulates conserved insulin/IGF and TGF-β signaling pathways, which converge on genes implicated in DAF-12-ligand biosynthesis (Figure 1A). Under unfavorable conditions such as overcrowding or scarcity of food, ligand biosynthesis is suppressed, and unliganded DAF-12 interacts with its co-repressor DIN-1 (Ludewig et al., 2004). The resulting repression of DAF-12 target genes causes developmental arrest and entry into a highly stress-resistant larval stage called the dauer diapause (Gems et al., 1998; Hu, 2007; Larsen et al., 1995; Schaedel et al., 2012). In contrast, favorable conditions trigger upregulation of DAF-12 ligand biosynthesis. DAF-12 ligand binding then results in dissociation of the corepressor DIN-1 to allow expression of DAF-12 target genes, promoting rapid development from larvae to reproductive adults (Fielenbach and Antebi, 2008; Ludewig et al., 2004). Additionally, ligand-dependent activation of the DAF-12 target genes mir-84 and mir-241, two microRNAs of the conserved let-7 family (Bethke et al., 2009; Hammell et al., 2009), is required for lifespan regulation in response to signals from reproductive tissues (Shen et al., 2012; Yamawaki et al., 2010). These findings indicate that metazoan lifespan is coupled to the gonad via NHR signaling.
Figure 1. Steroidal ligands control C. elegans development and lifespan via the nuclear hormone receptor DAF-12.
(A) Under favorable conditions, insulin/IGF and TGF-β signaling drive biosynthesis of steroidal DAF-12-ligands. Liganded DAF-12 promotes development, in part via transcription of the let-7-family microRNA’s mir-84 and mir-241. Under unfavorable conditions, ligand biosynthesis is inhibited, resulting in interaction of unliganded DAF-12 with its co-repressor DIN-1.
(B) Previously described DAF-12-ligands and proposed biosynthetic pathways. DAF-12-ligand biosynthesis is downregulated in response to dauer pheromone, a blend of ascarosides, e.g. the shown ascr#2 (red).
Based on extensive biochemical studies, two bile acid-like steroids named Δ4- and Δ7-dafachronic acid (DA) were proposed as endogenous ligands of DAF-12 (Figure 1B) (Motola et al., 2006). Central to identification of the DAs as DAF-12 ligand candidates were precursor studies in which a variety of 3-keto sterols were identified as substrates for the cytochrome P450, DAF-9, which had been shown to act upstream of DAF-12 in DAF-12-ligand biosynthesis (Gerisch et al., 2001; Jia et al., 2002; Motola et al., 2006). DAF-9 was further shown to act on the sidechain in these steroids introducing a terminal carboxyl group (Motola et al., 2006). In a separate study, a DAF-12-activating isomer of 3β-hydroxy cholest-5-enoic acid was detected in C. elegans metabolite extracts (Held et al., 2006). However, given the very low concentrations of the putative DAF-12-ligands in C. elegans, isolation and full spectroscopic characterization of these compounds was not pursued.
Although none of the structures of the proposed DAF-12-ligands have been confirmed based on conclusive spectroscopic analysis of C. elegans-derived samples, a biosynthesis model has been developed (Figure 1B), and the biochemical roles of genes proposed to function upstream of DAF-9 in DAF-12-ligand biosynthesis have been studied extensively (Dumas et al., 2010; Gerisch et al., 2007; Patel et al., 2008; Rottiers et al., 2006; Williams et al., 2010; Wollam et al., 2011; Yoshiyama-Yanagawa et al., 2011). More recent work has shown that the proposed DAF-12 ligands do not explain all DAF-12 associated functions and suggested the possibility that other DAs may exist (Patel et al., 2008; Williams et al., 2010; Wollam et al., 2012). In this study, we identify the endogenous ligands of DAF-12 using an unbiased comparative metabolomics approach (Forseth and Schroeder, 2011), which revealed new ligand structures and differential regulation of DAF-12-ligand biosynthesis.
RESULTS
Customizing a comparative metabolomics approach
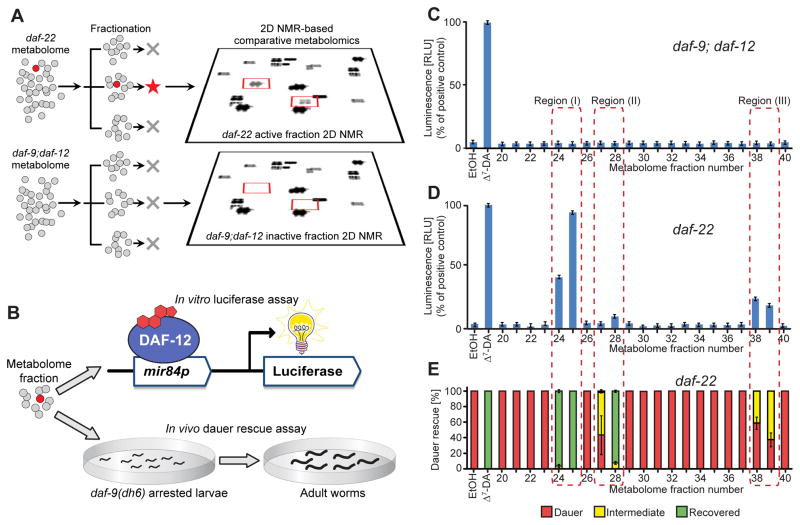
We aimed to identify endogenous DAF-12 ligands from direct spectroscopic evidence, in contrast to earlier work that relied on classical genetics and biochemical experiments. For this purpose we combined activity-guided fractionation and NMR-based comparative metabolomics via DANS (Differential Analyses by 2D NMR Spectroscopy) (Figure 2A). Two-dimensional (2D) NMR spectroscopy can provide a largely unbiased overview of metabolome composition, and comparing 2D NMR spectra of different mutant backgrounds via DANS often permits detection and partial identification of minor metabolites such as signaling molecules (Forseth and Schroeder, 2011; Pungaliya et al., 2009). DANS relies on correlating genetic changes with metabolomic changes for compound identification thereby reducing the need for extensive fractionation, which can result in activity loss or the introduction of artifacts. We envisioned that this strategy could be applied to identify DAF-12-ligands if one compared a mutant metabolome lacking DAF-12-ligands with the metabolome of worms that produces DAF-12-ligands abundantly.
Figure 2. Detection of DAF-12-ligands in C. elegans mutant metabolomes.
(A) Fractionation of active, ligand-rich daf-22 and inactive daf-9;daf-12 metabolomes is followed by 2D NMR-based comparative metabolomics of active fractions.
(B) Assessment of DAF-12-ligand content using (1) an in vitro luciferase assay in HEK-293T cells transfected with full-length DAF-12 and a mir84p-luciferase reporter vector and (2) in vivo daf-9(dh6) dauer rescue assays.
(C) daf-9;daf-12 metabolome fractions are inactive in the luciferase assay. 100 nM Δ7-DA is used as a positive control (error bars, ±SD).
(D) Luciferase assays of daf-22 metabolome fractions reveal three active regions (error bars, ±SD).
(E) daf-9(dh6) dauer rescue assays of daf-22 metabolome fractions show activity in the same three regions (error bars, ±SD). For worm images of scored phenotypes see Figure S1C.
See also Figure S1
For DANS-based ligand identification, we chose daf-9;daf-12 double mutants as the ligand-deficient strain and daf-22 mutants as a putatively ligand-rich reference strain. daf-9;daf-12 double mutants do not produce DAF-12-ligands, but nonetheless bypass the dauer stage, because lack of DAF-12 prevents the execution of genetic programs required for dauer formation (Gerisch and Antebi, 2004). Therefore, these animals can be grown in quantities large enough for NMR spectroscopic analyses. daf-22 mutant worms develop normally to adulthood, but are defective in the biosynthesis of the dauer-inducing ascarosides (Butcher et al., 2009; von Reuss et al., 2012), which we hypothesized may adversely affect DAF-12-ligand production in wild-type (WT) liquid cultures (Figure 1B). Downregulation of DAF-12-ligand biosynthesis by ascarosides is also suggested by the finding that exposure to high concentrations of dauer pheromone abolished expression of DAF-9, one of the key enzymes in the proposed biosynthetic pathway of DAF-12-ligands (Schaedel et al., 2012). Furthermore, given that the mammalian ortholog of DAF-22, SCPx, is involved in oxidative breakdown of steroid side chains in bile acid biosynthesis, it seemed possible that DAF-22 contributes to degradation of steroidal DAF-12 ligands in C. elegans.
To prepare for DANS analysis, metabolome extracts of daf-9;daf-12, WT, and daf-22 mixed-stage liquid cultures were fractionated using an automated, highly reproducible chromatography system (Figure 2A and Supplemental Experimental Procedures). The resulting parallel sets of metabolome fractions were assessed for DAF-12-ligand content using in vivo and in vitro bioassays (Figure 2B). The in vivo assay used daf-9(dh6) worms, which are defective in DAF-12-ligand production. In the absence of exogenously added DAF-12-ligand or a suitable precursor, developing daf-9(dh6) worms arrest as dauer larvae, because unliganded DAF-12 constitutively interacts with its corepressor DIN-1 (Gerisch et al., 2007; Ludewig et al., 2004). The assay scored the ability of added fractions to rescue the arrested dauer larvae and promote development to adulthood, providing a measure for the presence of a DAF-12 ligand that would dissociate the DAF-12/DIN-1 complex. The in vitro assay measured transcriptional activation by DAF-12 of a luciferase reporter in HEK-293T cells that were co-transfected with full-length DAF-12 and the reporter construct (Bethke et al., 2009). This assay provided a measure for ligand-dependent interaction of DAF-12 with mammalian coactivator(s) in the cells. Both of these assays consistently showed activity for three groups of daf-22 and WT fractions (regions I–III in Figures 2C–2E; for WT assay data, see Figure S1), indicating the presence of DAF-12-ligands or precursors, whereas all daf-9;daf-12 fractions were inactive in both assays, in accordance with previous work (Gerisch and Antebi, 2004; Gerisch et al., 2007; Motola et al., 2006). As anticipated, daf-22 fractions were significantly more active in the daf-9(dh6) dauer rescue assay than the corresponding WT fractions, suggesting higher production of DAF-12-ligands in daf-22 mutants (Figure S1).
DANS reveals steroids with unexpected structural features
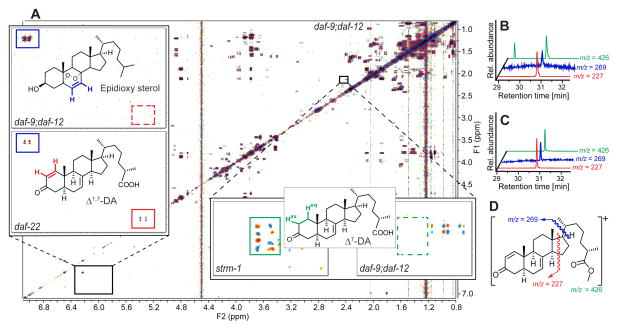
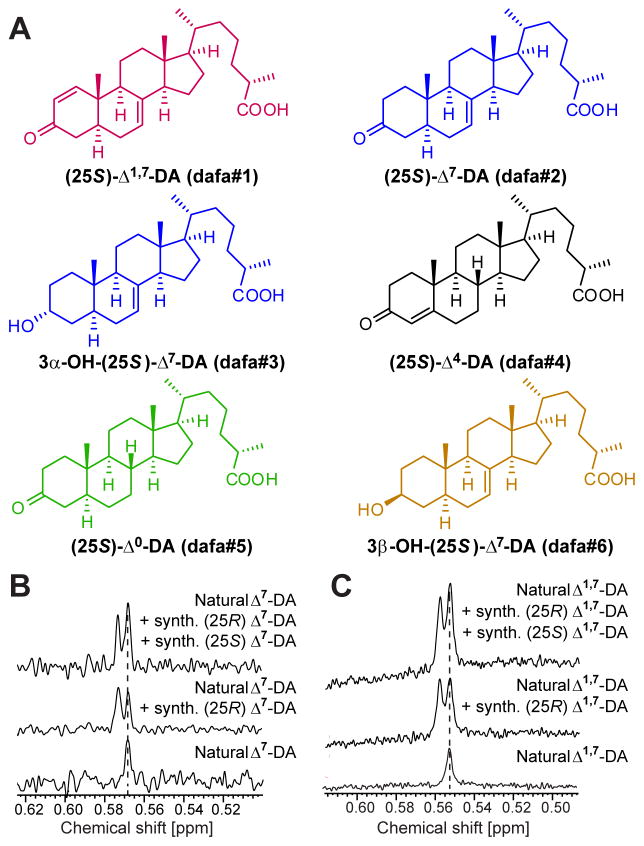
2D NMR spectra of the most active group of daf-22 fractions, active region I, revealed long-chained ascarosides (Pungaliya et al., 2009), in addition to a complex mixture of fatty acids, glycerides, other lipids, and epidioxy sterol derivatives (Figures 3A and S2A), all of which were also present in similar concentrations in corresponding daf-9;daf-12 fractions. Closer inspection of region I 2D NMR spectra revealed several sets of signals that were consistently absent in daf-9;daf-12, and thus appeared to be daf-9-dependent (Figure 3A). Further analysis of these differential signals suggested that they represent steroidal structures. Because of their very low concentrations, identification of the putative daf-9-dependent steroids required additional fractionation via HPLC, which resulted in two active samples each containing 1–2% of daf-9-dependent components (Figures S2B and S2C). NMR-spectroscopic analysis of the most active fractions showed a distinct set of daf-9-dependent signals at 5.9 and 7.0 ppm with a coupling constant of 10 Hz, which indicated the presence of an unusual Δ1-unsaturated 3-keto steroid (Figure 3A). Comparison with literature data suggested 3-oxocholesta-1,7-dienoic acid (“Δ1,7-DA”), which had not previously been described from worms, in addition to smaller amounts of the known Δ7-DA (Figures 3A and 4A) (Motola et al., 2006). These assignments were confirmed by comparison of spectroscopic data and GC/MS retention times with those of synthetic samples of Δ1,7- and Δ7-DA (Figures 3B, 3C, and S2D–S2I). To determine the relative configuration of the chiral centers at position 25 in the sidechains of Δ7-DA and Δ1,7-DA, we compared the NMR spectra of synthetic samples of (25S)- and (25R)-diastereomers with those of the natural samples. These analyses established the configuration of natural Δ7-DA and Δ1,7-DA as (25S) (Figure 4). Δ1-unsaturated steroids are rare in nature, and we are aware of only one other example in animals (Wang et al., 2009a).
Figure 3. Detection and identification of endogenous DAF-12-ligand candidates via 2D NMR-based comparative metabolomics and SIM-GC/MS.
(A) dqfCOSY spectrum of inactive daf-9;daf-12 metabolome fraction corresponding to active region I (Figures 2C–2E). Enlarged section (upper left) compares daf-9;daf-12 with the corresponding section of the daf-22 spectrum, showing one of the differential crosspeaks (red) that led to identification of Δ1,7-DA, next to non-differential signals representing a metabolite present in both daf-9;daf-12 and daf-22, an epidioxy sterol (blue). Enlarged section (lower right) shows example crosspeaks from the comparison of the spectra of strm-1 (vide infra) and daf-9;daf-12 metabolomes, showing signals (green) characteristic for Δ7-DA in the strm-1 spectrum but not the daf-9;daf-12 spectrum.
(B) SIM-GC/MS of active daf-22 metabolome fraction indicating the presence of Δ1,7-DA. The additional peak at ~29.4 min in the ion trace m/z = 426 belongs to an unrelated compound.
(C) SIM-GC/MS of synthetic Δ1,7-DA confirm retention times and fragmentation patterns.
(D) Major EI-MS fragments of Δ1,7-DA used in Figures 3B and 3C.
See also Figure S2
Figure 4. Structures and 1H-NMR-based assignment of absolute configuration of DAF-12 ligands.
(A) Structures of, Δ1,7-DA (dafa#1), Δ7-DA (dafa#2), 3α-OH-Δ7-DA (dafa#3), Δ4-DA (dafa#4), Δ0-DA (dafa#5), and 3β-OH-Δ7-DA (dafa#6). See Experimental Procedures for compound nomenclature.
(B) NMR-spectroscopic determination of the relative configuration at C-25 in natural Δ7-DA. C-18 singlet region of 1H NMR spectra (600 MHz, pyridine-d5) of natural Δ7-DA and mixtures with synthetic (25R)-Δ7-DA and (25S)-Δ7-DA.
(C) NMR-spectroscopic determination of the relative configuration at C-25 in natural Δ1,7-DA. C-18 singlet region of 1H NMR spectra (600 MHz, pyridine-d5) of natural Δ1,7-DA and mixtures with synthetic (25R)-Δ1,7-DA and synthetic (25S)-Δ1,7-DA.
Comparative analysis of active region II led to the identification of another daf-9-dependent cholestenoic acid derivative, featuring unusual 3α-hydroxylation: (25S)-3α-hydroxy cholest-7-enoic acid (“3α-OH-Δ7-DA”) (Figures 4A and S2J–S2O). The GC/MS fragmentation pattern of 3α-OH-Δ7-DA suggests that it is identical to the heretofore unidentified isomer of (25S)-cholest-3β-enoic acid previously reported as a DAF-12-ligand by Held et al. (Figure S2K) (Held et al., 2006). As all 3-hydroxylated steroids previously described from C. elegans feature 3β-configuration, the identification of 3α-OH-Δ7-DA was surprising. We then inspected both active and inactive C. elegans metabolome fractions for the presence of the corresponding 3β-stereoisomer, 3β-OH-Δ7-DA (Figure 4A); however, 3β-OH-Δ7-DA could not be detected in either WT or daf-22 metabolomes, indicating that production of 3α-OH-Δ7-DA is highly selective. Active region III, representing a series of metabolome fractions with weak daf-9(dh6) dauer rescue activity, revealed trace quantities of glucosides of DAs that were not characterized further.
Δ1,7-DA and Δ7-DA are potent DAF-12-ligands
Next we investigated the biological properties of synthetic samples of the identified daf-9-dependent steroids (Figures 5A–5D). Synthetic Δ1,7-DA activated DAF-12 in mammalian cells (EC50 = 146 nM) and its potency in the daf-9(dh6) dauer rescue assay was similar (EC50 = 2 nM) or slightly higher than that of Δ7-DA (Figure 5B and Table S3). Similarly, 3α-OH-Δ7-DA was active in both the in vivo and in vitro assays, whereas its 3β-stereoisomer did not activate DAF-12 in mammalian cells at any tested concentration and rescued the daf-9(dh6) dauer phenotype only at very high concentrations (Figure 5B). We also assayed the two most abundant daf-9-dependent compounds for their effect on lifespan in germline-deficient glp-1 mutant worms. Germline-deficient glp-1 mutant worms live up to 60% longer than WT, and this lifespan extension has been shown to depend on functional DAF-12 and DAF-12-ligand biosynthetic enzymes (Gerisch et al., 2001; Hsin and Kenyon, 1999; Yamawaki et al., 2010). Correspondingly, ligand-deficient glp-1;daf-36 double mutant worms lack the glp-1 lifespan phenotype (Gerisch et al., 2007; Rottiers et al., 2006). We found that both Δ7-DA and Δ1,7-DA fully restore glp-1-dependent lifespan extension to glp-1;daf-36 worms (Figures 5C and S3A).
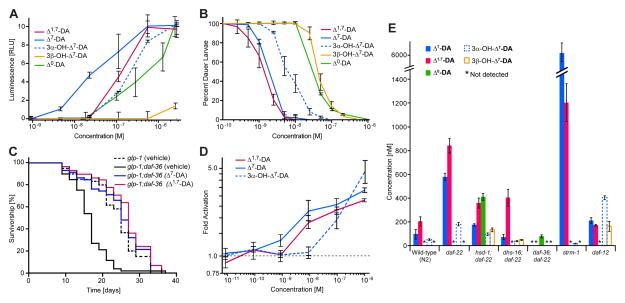
Figure 5. Biological activities of DAF-12 ligands and quantification of DAF-12 ligands in C. elegans WT, steroid metabolism mutant, and daf-12(0) mutant worms.
(A) DAF-12 transcriptional activation in HEK-293T cells by the identified endogenous DAF-12-ligand candidates. Luciferase assays were measured in triplicates (error bars, ±SD).
(B) daf-9(dh6) dauer rescue with the identified endogenous DAF-12-ligand candidates at 27 °C. For each data point there were two replicates with 100 animals per replicate (error bars, ±SD).
(C) Both Δ7-DA (100 nM) and Δ1,7-DA (100 nM) restore lifespan extension to glp-1 animals in daf-36 null mutant background.
(D) Alphascreen assay for ligand-dependent recruitment of SRC1-4 peptide by DAF-12, showing fold activation of DAF-12 with different ligand candidates over ethanol control (error bars, SEM).
(E) In vivo concentrations of the identified endogenous dafachronic acids in WT (N2), daf-22, daf-12, and steroid metabolism mutants (error bars, ±SD) grown in mixed stage liquid cultures, as determined from SIM-GC/MS analysis of metabolome fractions (for distribution of life stages at the time of harvesting see Figure S3R). Detection limits for SIM-GC/MS-based quantification of dafachronic acids in these experiments were around 2.5 nM.
See also Figure S3.
For quantification of the identified daf-9-dependent steroids, we employed SIM-GC/MS detection of characteristic MS fragments of volatile methylated or silylated derivatives (Supplemental Experimental Procedures). SIM-GC/MS showed that Δ1,7-DA is slightly more abundant than Δ7-DA in daf-22 worms, whereas in WT animals Δ1,7-DA is more than twice as abundant as Δ7-DA, with concentrations (averaged over the worm bodies) of 93 nM and 197 nM for Δ7-DA and Δ1,7-DA, respectively (Figure 5E). 3α-OH-Δ7-DA occurs at about 3–5-fold lower concentrations than Δ7-DA in both WT and daf-22 mutants. Based on the specific activities determined for synthetic samples of Δ7-DA, Δ1,7-DA, and 3α-OH-Δ7-DA it appears that these three compounds account for all of the activity in regions I and II in both the WT and daf-22 metabolomes. Using SIM-GC/MS, we also checked the daf-22 and WT metabolomes for the presence of the previously reported Δ4-DA. We were unable to detect Δ4-DA in any of our C. elegans metabolome fractions, whereas fractions spiked with trace quantities of synthetic Δ4-DA confirmed the sensitivity of our detection methods (Figure S2P). We then considered that our growth conditions may have affected production of the putative precursor of Δ4-DA, 4-cholesten-3-one (Motola et al., 2006; Patel et al., 2008). However, analysis of WT metabolome samples by GC-MS and NMR spectroscopy revealed that 4-cholesten-3-one is as abundant as lathosterone (Figures S2Q-S2V), a putative precursor of Δ7-DA (Motola et al., 2006; Wollam et al., 2012), suggesting that absence of Δ4-DA is not the result of a lack of suitable precursors. Therefore, it appears that Δ4-DA may not play a significant role as a DAF-12 ligand, although its transient or very low-level production cannot be excluded.
To test whether the identified daf-9-dependent compounds constitute bona-fide ligands of DAF-12, we measured ligand-dependent binding of DAF-12 with the SRC1-4 peptide containing the nuclear receptor box (“NR box”) motif of mammalian coactivator SRC-1 (Heery et al., 1997). We found that Δ7-DA, Δ1,7-DA and 3α-OH-Δ7-DA effect concentration-dependent recruitment of SRC1-4, with EC50 values of 8 nM and 15 nM for Δ7-DA and Δ1,7-DA, respectively, whereas affinity of 3α-OH-Δ7-DA was lower (EC50 = 200 nM, Figure 5D and Table S3). These relative potencies of Δ7-DA, Δ1,7-DA and 3α-OH-Δ7-DA are similar to relative activities observed in the luciferase assay (Figure 5A).
Taken together, these results indicate that Δ1,7-DA and Δ7-DA are high-affinity ligands of DAF-12 that promote reproductive development and adult longevity, whereas 3α-OH-Δ7-DA is of lower potency in promoting these phenotypes and Δ4-DA may not be present at physiologically relevant concentrations. Therefore, previous hypotheses about DAF-12-ligand structures and their biosynthetic pathways must be revised.
Comparative metabolomics suggests tissue-specific ligand biosynthesis
Using our comparative metabolomics strategy, we reinvestigated the roles of four additional enzymes that have been proposed to participate in DAF-12-ligand biosynthesis. In a recent model (Figure 1B), DAF-12-ligand biosynthesis begins with oxidation of cholesterol by the Rieske-like oxygenase, DAF-36, yielding 7-dehydrocholesterol (Wollam et al., 2011; Yoshiyama-Yanagawa et al., 2011). 7-dehydrocholesterol is converted by an unknown enzyme to lathosterol, which is oxidized to the corresponding ketone, lathosterone, by the short-chain dehydrogenase, DHS-16 (Wollam et al., 2012). Lathosterone is then converted to Δ7-DA by DAF-9 (Gerisch et al., 2007; Motola et al., 2006). In addition, a parallel pathway for the biosynthesis of Δ4-DA involving the putative hydroxysteroid dehydrogenase, HSD-1, has been proposed (Dumas et al., 2010; Patel et al., 2008), although recent evidence suggests that HSD-1 has no role in the production of the putative precursor of Δ4-DA, 4-cholesten-3-one (Wollam et al., 2012), leaving its role in DAF-12 ligand biosynthesis undetermined. Therefore, we began our biosynthetic analysis with profiling hsd-1 mutants and hsd-1;daf-22 double mutants. Whereas hsd-1 fractions contained only very small amounts of dauer rescuing activity, the activity profile of hsd-1;daf-22 fractions was similar to that of daf-22 fractions, consistent with the hypothesis that dauer pheromone biosynthesis via DAF-22 downregulates DAF-12-ligand production (Figure S3D). However, comparative 2D NMR analysis of hsd-1;daf-22 region I revealed production of large quantities of an additional steroid in hsd-1;daf-22 worms that is not produced in either daf-22 or daf-9;daf-12 mutants. Comparison with a synthetic sample led to the identification of this hsd-1-specific steroid as (25S)-3-keto-cholestanoic acid (“Δ0-DA”, Figures 4A and S3G-S3K). This fully saturated DA derivative is active in both the DAF-12 luciferase assay and the daf-9(dh6) dauer rescue assay, although its activity is lower than that of Δ1,7-and Δ7-DA (Figures 5A and 5B). Δ0-DA was absent in WT and daf-22 metabolomes, as determined by NMR and SIM-GC/MS. Analysis of hsd-1;daf-22 activity region II revealed a second hsd-1-specific steroid, 3β-OH-Δ7-DA (Figure S3L). In hsd-1;daf-22 mutants, 3β-OH-Δ7-DA is as abundant as the 3α-isomer, whereas 3β-OH-Δ7-DA is absent in both WT and daf-22 mutants. GC/MS analysis of hsd-1;daf-22 activity region I further revealed that amounts of both Δ1,7- and Δ7-DA are slightly reduced compared to daf-22 worms, whereby production of Δ7-DA may be more strongly affected than that of Δ1,7-DA (Figure 5E).
The identification of 3β-OH-Δ7-DA and Δ0-DA in hsd-1;daf-22 mutant worms suggests that HSD-1 may directly or indirectly participate in the biosynthesis of Δ7-DA and possibly Δ1,7-DA. HSD-1 has homology to mammalian 3β-hydroxysteroid dehydrogenases (Patel et al., 2008), which suggested its participation in introducing the 3-keto functionality in Δ4-DA. The absence of Δ4-DA and our identification of Δ0-DA as a shunt metabolite in hsd-1;daf-22 mutants may indicate that the hsd-1 pathway includes introduction of the 7,8-double bond in a saturated precursor, for example cholestanol, 3β-OH-Δ0-DA, or Δ0-DA, by an unknown enzyme, or that HSD-1 itself has 7-dehydrogenase activity. Notably, HSD-1 is expressed mainly in the neuron-like XXX cells, which lack expression of the oxygenase, DAF-36, required for biosynthesis of most 7,8-unsaturated steroids in C. elegans (Wollam et al., 2011; Yoshiyama-Yanagawa et al., 2011). Taken together, our results suggest that HSD-1 contributes to Δ7-DA biosynthesis in the XXX cells, but that Δ7-DA and Δ1,7-DA are produced via a separate pathway in other tissues.
Next, we investigated the DAF-12-ligand profile of mutants of the Rieske oxygenase, DAF-36 (Figure 1B) (Wollam et al., 2011; Yoshiyama-Yanagawa et al., 2011). Mutants of daf-36, which is expressed primarily in the worm intestine, exhibit phenotypes consistent with strongly reduced DAF-12-ligand biosynthesis, such as a higher tendency to enter dauer (Rottiers et al., 2006). As in the case of hsd-1 mutants, daf-36 mutant metabolome fractions were largely inactive in the dauer rescue and luciferase assays, whereas daf-36;daf-22 double mutants produced significant dauer rescue activity in region I (Figure S3E). GC/MS analysis revealed the presence of saturated Δ0-DA in this region (Figures S3M-S3O), which we had previously found only in hsd-1;daf-22 mutants. In contrast to hsd-1;daf-22 mutants, Δ0-DA appears to be responsible for all activity observed in daf-36;daf-22 region I, and neither Δ7- nor Δ1,7-DA were detectable in this mutant. Total DAF-12-ligand amounts were much lower in the daf-36;daf-22 metabolome than in hsd-1;daf-22 (Figure 5E). These results indicate that the intestinally expressed 7,8-dehydrogenase, DAF-36, is required for Δ7- and Δ1,7-DA biosynthesis, whereas HSD-1 contributes to additional production of Δ7-unsaturated DAs in the XXX cells. Thus it appears that biosynthesis of Δ7- and Δ1,7-DA relies on partially redundant and tissue-specific pathways.
Biosynthesis of different ligands is differentially regulated
Whereas the hsd-1 mutation did not significantly affect the relative abundance of Δ1,7- and Δ7-DA, mutations of two other genes involved in DAF-12-ligand biosynthesis and regulation, the short-chain dehydrogenase, dhs-16 (Wollam et al., 2012), and the methyltransferase, strm-1 (Hannich et al., 2009), showed strongly altered ligand ratios. Similar to what we observed for daf-36 and hsd-1 single mutants, bioassays of metabolome fractions from dhs-16 single mutants showed very low activity, whereas dhs-16;daf-22 double mutant fractions showed levels of activity similar to those of daf-22 fractions (Figure S3F). Chemical analysis of the dhs-16;daf-22 fractions revealed that production of Δ7-DA was greatly reduced compared to daf-22 single mutants, whereas Δ1,7-DA levels were only slightly reduced (Figures 1B and 5E). These results indicate that Δ1,7- and Δ7-DA may be derived from partially divergent biosynthetic pathways or that deletion of dhs-16 indirectly affects regulation of the Δ1,7-DA to Δ7-DA ratio. Only trace quantities of 3α-OH-Δ7-DA were detected in dhs-16;daf-22 worms, suggesting that production of both 3α-OH-Δ7-DA and Δ7-DA are DHS-16-dependent (Figure 5E). Next, we characterized the DAF-12-ligand profile of strm-1 mutants (Figure S3F), which had previously been shown to produce elevated levels of DAF-12-ligands (Hannich et al., 2009). The methyltransferase, STRM-1, regulates DAF-12-ligand levels by converting cholesterol-derived intermediates of ligand biosynthesis into 4-methylated steroids, thereby rendering them unsuitable as ligand precursors (Hannich et al., 2009). NMR spectroscopy and SIM-GC/MS analyses revealed more than 100-fold increased levels of Δ7-DA in strm-1 metabolomes, whereas Δ1,7-DA levels increased only about 7-fold compared to WT (Figure 5E).
We also investigated the DAF-12 ligand profile of daf-12 null mutants, which previously had been shown to produce increased dauer rescuing activity (Held et al., 2006). We found that daf-12 mutants produced roughly six-fold higher amounts of 3α-OH-Δ7-DA than WT, whereas amounts of Δ1,7- and Δ7-DA were only slightly increased (Figure 5E). In addition, daf-12 mutants produce large quantities of 3β-OH-Δ7-DA, whereas this compound could not be detected in WT. These observations indicate that DAF-12 ligand biosynthesis is dysregulated in daf-12 null mutants, possibly as a result of changes in the expression levels of additional ligand biosynthetic (or catabolic) enzymes.
Given that DAF-12 ligands regulate both larval development and germline-dependent adult longevity, it seemed possible that different life stages may produce different ligand profiles. SIM-GC/MS analysis of metabolite extract from synchronized larvae at the L2/L3 stages showed a Δ7- to Δ1,7-DA ratio of about 1:1, whereas in worms at the L4/young adult stages the ratio is shifted toward Δ1,7-DA (Figures S3P and S3Q). More detailed analysis of the ligand profiles of different life stages and under different environmental conditions will have to await development of more sensitive detection techniques, especially for the chemically somewhat unstable Δ1,7-DA. Taken together, our results indicate that the biosynthesis of different DAF-12 ligands is differentially regulated, in part via feedback through DAF-12.
DISCUSSION
Identification of the endogenous ligands of NHRs is central to understanding their role as transcriptional regulators in metazoans. Screening of synthetic ligands of the vitamin D receptor and other mammalian NHRs has demonstrated that even small changes in ligand structures can strongly affect gene transcription (Brown and Slatopolsky, 2008; Singarapu et al., 2011; Wollam and Antebi, 2011). Yet there are few approaches to comprehensively identify the endogenous NHR ligands from complex animal metabolomes, whose chemical annotations remain largely incomplete. In this study, we demonstrate the use of comparative metabolomics to identify the endogenous ligands of DAF-12, a central regulator of development and adult lifespan in C. elegans and an important model for ligand-regulated NHRs in higher animals. In contrast to earlier work that, primarily based on screening candidate structures, proposed Δ4-DA and Δ7-DA as endogenous DAF-12 ligands, our approach revealed Δ1,7-DA as the most abundant ligand in WT worms, in addition to smaller amounts of Δ7-DA and 3α-OH-Δ7-DA.
Steroids featuring a Δ1-double bond have been described from very few natural sources (Wang et al., 2009a), although it is well known that introduction of Δ1-unsaturation in natural 3-keto sterols, e.g. testosterone, can have pronounced effects on their activities (Counsell and Klimstra, 1962). Analysis of X-ray structures of the DAF-12-ligand binding domain complexed with DAs have demonstrated that small structural changes in the A and B rings of the bound steroid have significant effects on the ligand’s affinity to DAF-12 (Wang et al., 2009b; Zhi et al., 2012), suggesting that specific modifications in the steroid A-ring may serve to fine-tune DAF-12 transcriptional regulation. Identification of the enzyme(s) introducing the Δ1-double bond will play an important role in elucidating functional differences between Δ1,7- and Δ7-DA and may motivate re-analysis of mammalian metabolomes for the presence of endogenous Δ1-steroids. The additional identification of the 3α-OH-Δ7-DA creates a striking parallel to mammalian bile acid metabolism, which produces predominantly sterols with 3α configuration (Figure 6A) (Russell, 2003). Given its stereoselective biosynthesis by an as-yet unknown enzyme, it is likely that 3α-OH-Δ7-DA serves specific functions in DAF-12 signaling. Notably, 3β-OH-Δ7-DA, which was absent in WT worms but accumulated in hsd-1; daf-22 and daf-12 mutant worms, is much less active in both the in vivo and in vitro assays. However, it should be noted that both the transcriptional activation assay in mammalian cell-culture (Figure 5B) and the Alphascreen assay (Figure 5D) have limited cogency for judging the relative potency of different DAF-12 ligands in vivo, as both assays depend on recruitment of mammalian coactivators such as SRC-1, whereas in-vivo function of DAF-12 is thought to involve ligand-dependent dissociation of the endogenous C. elegans corepressor DIN-1 followed by binding of yet unidentified co-activators (Ludewig et al., 2004). The identification of multiple endogenous small molecule regulators of DAF-12 in this study will accelerate the pursuit of yet elusive DAF-12 interactors and other components of DAF-12-dependent dauer and lifespan regulation.
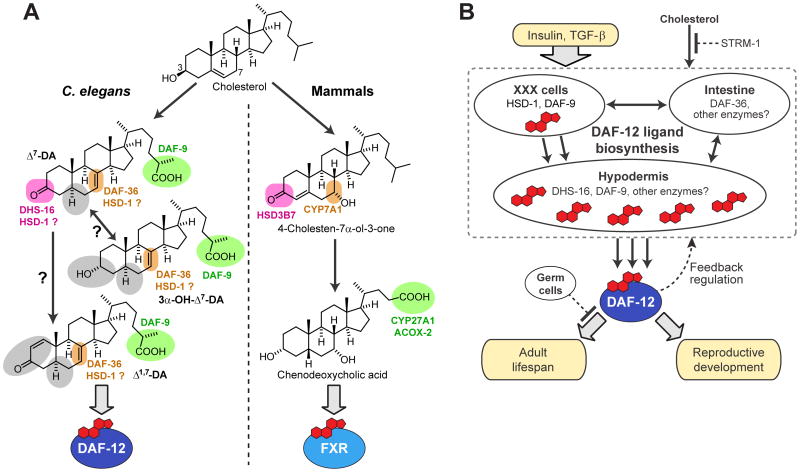
Figure 6. Comparison of NHR signaling in nematodes and mammals and a model for DAF-12 signaling in C. elegans.
(A) Comparison of NHR signaling in nematodes and mammals. In nematodes, oxidation/epimerization in position 3, oxidation in position 7, and side chain oxidation produces ligands of DAF-12, whereas similar modification of the steroid skeleton in mammals produces bile acids that serve as ligands of farnesoid X receptor (FXR) (Russell, 2003) and possibly LXR (Theofilopoulos et al., 2013). Colors are used to highlight the functions of known enzymes in this pathway, whereas enzymes introducing the structural features highlighted in gray have not been described.
(B) Biosynthesis model for DAF-12-ligands regulating development and lifespan. Enzymes in the XXX cells downstream of insulin and TGF-β signaling produce small quantities of DAF-12 ligands, which trigger additional biosynthesis of DAF-12 ligands via upregulation of DAF-9 expression in the hypodermis, dependent on intestinally expressed DAF-36 and possibly other tissues. Although DAF-9 has been assumed to act directly upstream of DAF-12, our results suggest that DAF-9 may act on a variety of different substrates, including both 3-keto and 3-hydroxy sterols. Ligand biosynthesis is additionally regulated by STRM-1 and feedback through DAF-12. The lifespan-increasing effects of DAF-12 ligands depend on germ cell removal.
Whereas it is well established that DAF-12-ligands are ultimately derived from dietary cholesterol, identification of Δ1,7- and 3α-OH-Δ7-DA and the absence of Δ4-DA necessitates revision of DAF-12-ligand biosynthesis models (Figures 1B and 6A). Our results indicate that DAF-36 as well as HSD-1 participate in the biosynthesis of Δ7-DA and possibly Δ1,7-DA, whereas previously, HSD-1 had been assumed to function in Δ4-DA biosynthesis. The finding that mutation of dhs-16 affects Δ7-DA much more strongly than Δ1,7-DA production suggests that introduction of the 3-keto moiety in Δ1,7-DA may involve a different enzyme. Therefore, it appears that different DAF-12-ligands are produced via partially separate biosynthetic pathways, which is also supported by the finding that in strm-1 mutants Δ7-DA production is increased to a much greater extent than that of Δ1,7-DA. Taken together, our identification of Δ1,7-DA, 3α-OH-Δ7-DA, and Δ0-DA indicates that several DAF-12-ligand biosynthetic enzymes remain to be identified.
C. elegans offers a unique opportunity to study the role of tissue-specific NHR ligand biosynthesis for endocrine signaling in a simple model system. Expression of DAF-12-ligand biosynthetic enzymes is thought to be controlled by insulin/IGF and TGF-β signaling, and thus ligand biosynthesis connects DAF-12 transcriptional regulation with these two highly conserved pathways (Figure 1A) (Wollam et al., 2012). The biosynthesis of multiple DAF-12-ligands via partially separate pathways suggests that different ligands may serve different functions (Arda et al., 2010). Notably, a recent study showed that DAF-12-ligands or ligand derivatives produced in the neuroendocrine XXX cells function as signaling molecules that trigger abundant additional ligand biosynthesis in the hypodermis, locking in organism-wide commitment to reproductive development (Figure 6B) (Gerisch and Antebi, 2004; Schaedel et al., 2012). Since the XXX cells are the primary sites of HSD-1 expression (Dumas et al., 2010; Patel et al., 2008), our finding that HSD-1 may contribute to Δ7-DA biosynthesis supports a model in which XXX-produced Δ7-DA (or derived 3α-OH-Δ7-DA) triggers biosynthesis of large quantities of additional Δ7-DA and Δ1,7-DA via daf-36, dhs-16, and hypodermal daf-9 (Figure 6B). Ultimately, the elucidation of DAF-12-ligand biosynthetic pathways will require combining comparative metabolomics with tissue-specific manipulation of candidate genes. This will entail consideration of life-stage specific aspects of ligand functions and biosynthesis, especially with regard to the intriguing role of DAF-12-ligands for adult longevity, which depends on additional signaling from the germline (Figure 6B) (Yamawaki et al., 2010).
DAF-12 ligands regulate adult longevity by activating microRNA targets that ultimately increase conserved DAF-16/FOXO transcriptional activity, thereby extending lifespan (Shen et al., 2012). Although the enzymes in DA biosynthesis are not strict orthologs of functionally corresponding enzymes in mammalian bile-acid biosynthesis, the striking similarities of steroidal NHR-ligand biosynthetic pathways in nematodes and humans demonstrate the utility of C. elegans as a model organism for endocrine signaling (Figure 6A). Further functional characterization of the identified DAF-12-ligands will advance understanding of the roles of ligand-dependent NHRs in organism-wide coordination of metazoan development and aging. In addition, our study shows that NMR-based comparative metabolomics can provide detailed insight into metazoan small molecule signaling pathways, and that this approach can reveal signaling molecules and biosynthetic functions not suspected based on classical genetics and biochemical approaches. Finally, the DAF-12 ligands discovered here will yield important insights to combat parasitic infections, as parasitic nematodes use the DAF-12/DA signaling mechanism to regulate emergence from the infective stage (Ogawa et al., 2009; Wang et al., 2009b).
EXPERIMENTAL PROCEDURES
C. elegans Strains and Maintenance
Nematode stocks were maintained on Nematode Growth Medium (NGM) plates made with Bacto agar (BD Biosciences) and seeded with bacteria (E. coli OP50) at 20 °C (http://www.wormbook.org/). C. elegans strains: WT (N2, Bristol), daf-22(m130), daf-22(ok693), daf-9(dh6), daf-9(dh6);daf-12(rh411rh61), hsd-1(mg345), hsd-1(mg433), hsd-1(mg433);daf-22(ok693), daf-36(k114), daf-36(k114);daf-22(ok693), dhs-16(tm1890), dhs-16(tm1890);daf-22(m130), strm-1(tm1781), glp-1(e2141), glp-1(e2141);daf-36(k114), daf-12(rh411rh61). Compound mutants were constructed using standard techniques. Worms were grown at 20 °C for at least two generations under replete conditions prior to liquid cultures.
Liquid Cultures
Worms from four 10 cm NGM agar plates were washed using M9-medium into a 100 mL S-complete medium pre-culture where they were grown for 4 days at 22 °C on a rotary shaker. Concentrated bacteria from 1 L of E. coli OP50 culture was added as food at days 1 and 3. Subsequently, the pre-culture was divided equally into sixteen 500 mL Erlenmeyer flasks each containing 100 mL of S-complete medium on day 4, which were grown for another5 days at 22 °C on a rotary shaker and continuously fed with concentrated bacteria, avoiding depletion of bacterial food at all times. The cultures were harvested on day 5 and centrifuged to separate supernatant media and worm pellets. At harvest, liquid cultures contained approximately 50–70% L1–L3 worms. In liquid cultures grown for ~5 days at 22 °C (at time of harvest), we observed 15–20% dauers for N2, 2–5% for daf-22 and none for daf-9;daf-12 and daf-12 mutants. daf-36, dhs-16, and hsd-1 mutant worm cultures show 70–95% dauers, whereas we observed less than 10% dauers in daf-36;daf-22, dhs-16;daf-22, and hsd-1;daf-22 double mutant cultures. The worm pellets were stored at −20 °C until needed. Each 100 mL liquid culture yielded 2–3 mL of worm pellet all of which was used for one replicate of metabolite extraction and analysis. At least three independent sets of cultures for daf-22 and N2 and two for each of the steroid metabolism mutants were used.
For DA analysis of synchronized cultures, gravid adults from 540 10 cm plates were washed with M9 buffer and treated with alkaline hypochlorite solution to isolate eggs. Eggs were washed twice with M9 buffer and hatched in fresh M9 for 24 h. The synchronized L1 larvae were divided into two sets and transferred to S-complete medium containing OP50. One set was allowed to grow for 25 h at 20 °C and 220 rpm, at which point the cultures were predominantly a 1:1 mixture of L2 and L3 larvae. A second set was grown for 48 h and harvested when the culture contained a 9:1 mixture of L4 and young adults (YA). At the end of the specified growth period, the cultures were centrifuged at 4 °C and the worm pellets stored at −20 °C until processing for further analyses.
Preparation of Metabolome Extracts
The frozen worm pellets were added to pre-cooled (−78 °C) methanol (200 mL for each liquid culture and 20 mL for each staged culture) in a Waring blender and blended until no chunks remained. Methanol was evaporated in vacuo at 0–20 °C and the residue resuspended in water (300 mL for liquid cultures and 25 mL for staged cultures). The resulting suspension was then lyophilized and the residue crushed to a fine powder using a mortar and pestle over 8 g granular sodium chloride. The powder was then extracted twice with a 9:1 ethyl acetate:ethanol mixture (250 mL for samples obtained from liquid cultures and 25 mL for samples from staged liquid cultures) over 12 h. The resulting suspension was filtered and the filtrate evaporated in vacuo at room temperature to produce the worm pellet metabolome extract used for chromatographic separations and analysis. (For details please see Supplemental Experimental Procedures).
NMR Spectroscopic Instrumentation and Analysis
NMR spectra were recorded on a Varian 900 MHz NMR spectrometer equipped with a 5 mm 1H (13C/15N) cryogenic probe, a Varian INOVA 600 MHz NMR spectrometer equipped with an HCN indirect detection probe, and a Varian INOVA 500 MHz NMR spectrometer equipped with an DBG broadband probe. Each spectrum was manually phased, baseline corrected and calibrated to solvent peaks (CHCl3 singlet at 7.26 ppm; CHD2OD pseudoquintet at 3.31 ppm). Non-gradient phase-cycled dqfCOSY spectra were acquired using the following parameters: 0.8 s acquisition time, 500–900 complex increments, 16–64 scans per increment. dqfCOSY spectra were zero-filled to 8k–16k × 4k and a cosine bell-shaped window function was applied in both dimensions before Fourier transformation. NMR spectra were processed using Varian VNMR, MestreLabs’ MestReC, and MNova software packages. Dynamic range of the resulting spectra ranged from 300:1 to 500:1. For example, coupling constants could be determined for characteristic steroidal crosspeaks from dqfCOSY spectra containing as little as 5 μg Δ1,7-DA in a 2.5 mg metabolome fraction.
daf-9(dh6) Dauer Rescue Assay
Plate based assay: Metabolome fractions were resuspended in ethanol, mixed with 40 μL of 5 x concentrated OP50 bacteria (from an overnight culture in LB media) and plated on 3 cm plates containing 3 mL NGM agar without added cholesterol. For rescue, ~100 eggs from a 4–8 hour egg lay were transferred onto the bacterial lawn, and scored for dauer arrest at 27 °C after 60 h. For rescue experiments with synthetic steroids (0.1 nM – 500 nM tested), 10 μL compounds in ethanol (or ethanol alone) were mixed with 40 μL 5X concentrated OP50 bacteria and plated. Final concentrations include the total volume of agar (3 mL). 100 nM Δ7-DA was used as positive control. Additional rescue assays in liquid cultures were also carried out (For details please refer to Supplemental Experimental Procedures). Data obtained from both plate based and liquid culture assays are comparable. Figures in this paper are based on results from the plate based assay.
Luciferase Assay for DAF-12 Transcriptional Activation
Luciferase assays to determine transcriptional activation of DAF-12 were performed as described earlier (Bethke et al., 2009). Briefly, HEK-293T cells were seeded and transfected in 96-well plates with (per well) 30 ng transcription factor vector, 30 ng of gfp expression vector, 30 ng of luciferase reporter, and 5 ng β-galactosidase expression vector using the calcium phosphate precipitate method. Ethanol or ethanol solutions of ligands (synthetic DAs, 1 nM – 3125 nM tested and metabolome fractions) were added 8h after transfection and the luciferase and β-galactosidase activities were measured by a Synergy 2 Biotek LC Luminometer, 16 h after compound addition. 100 nM Δ7-DA was used as positive control. Data was processed using GEN5 software. Individual fractions were dried in vacuo and resuspended in 500 μL EtOH. 1 μL per 100 μL of media solution was added to each well.
Alphascreen Assay for Direct Binding of DAF-12 Ligand Candidates
Direct binding of ligand candidates to DAF-12 was assessed using the Alphascreen assay kit (Perkin Elmer) as described previously (Motola et al., 2006). Also see Supplemental Experimental Procedures. Supplemental
Statistical Analyses
All data except for Figure 5D presented as mean ±SD. Figure 5D presented as mean ±SEM.
Nomenclature of DAF-12 Ligands
Both the IUPAC names for dafachronic acids as well as semi-rational constructs such as “3α-hydroxy-Δ7-dafachronic acid” are cumbersome, unlikely to be used consistently, and thus pose problems for text search algorithms and database curation. Therefore, we have obtained, for each of the identified dafachronic acid derivatives, unique small molecule identifiers (SMIDs), from the C. elegans Small Molecule Identifier Database (SMID DB, http://smid-db.org/), an affiliate of Wormbase (http://wormbase.org). SMIDs can be obtained for all newly identified C. elegans metabolites from SMID-DB.org upon request. Thus, Δ1,7-DA, Δ7-DA, 3α-OH-Δ7-DA, Δ4-DA, Δ0-DA, and 3β-OH-Δ7-DA have been named dafa#1, dafa#2, dafa#3, dafa#4, dafa#5, and dafa#6, respectively, as shown in Figure 4. To ensure effective curation, SMID-DB recommends that the dafachronic acids be referred to by their respective SMIDs in subsequent publications.
Supplementary Material
HIGHLIGHTS.
Comparative metabolomics provides a blueprint for identification of NHR-ligands
Most abundant endogenous DAF-12-ligand has a structural moiety uncommon in animals
Ligand biosynthesis involves several different tissue-specific pathways
Acknowledgments
We thank the Caenorhabditis Genetics Center (P40 OD010440) and Shohei Mitani (Tokyo Women’s Medical University) for nematode strains. We thank D. N. Drechsel (MPI-CBG) for the purification of DAF-12. We thank A. S. Edison, J. Srinivasan, C. Coburn, and P. Sternberg for valuable suggestions. This work was supported in part by the National Institutes of Health (GM088290 and GM085285 to FCS, AG027498 to AA, and T32GM008500 to JCJ), Cornell/Rockefeller/Sloan-Kettering Tri-Institutional Training Program in Chemical Biology (to NB), and the DFG (to AB). AA was additionally supported by the Ellison Medical Foundation, the MPG, and CECAD. KJD received support from the Cellular and Molecular Biology Program at the UM Medical School and the UM Training Grant in the Biology of Aging, and PH from the American Cancer Society (10-132-01). This study made use of NMRFAM, which is supported by NIH grants P41RR02301 (BRTP/NCRR) and P41GM66326 (NIGMS).
Footnotes
Supplemental Information includes four figures, three tables, and Supplemental Experimental Procedures and can be found with this article online at http://
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antebi A. Nuclear hormone receptors in C. elegans. WormBook; 2006. pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Taubert S, MacNeil LT, Conine CC, Tsuda B, Van Gilst M, Sequerra R, Doucette-Stamm L, Yamamoto KR, Walhout AJ. Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol Syst Biol. 2010;6:367. doi: 10.1038/msb.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Slatopolsky E. Vitamin D analogs: therapeutic applications and mechanisms for selectivity. Mol Aspects Med. 2008;29:433–452. doi: 10.1016/j.mam.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci U S A. 2009;106:1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counsell RE, Klimstra PD. Anabolic agents: derivatives of 2-Halo 5alpha-Androst-1-Ene. J Med Pharm Chem. 1962;91:477–483. doi: 10.1021/jm01238a007. [DOI] [PubMed] [Google Scholar]
- Dumas KJ, Guo C, Wang X, Burkhart KB, Adams EJ, Alam H, Hu PJ. Functional divergence of dafachronic acid pathways in the control of C. elegans development and lifespan. Dev Biol. 2010;340:605–612. doi: 10.1016/j.ydbio.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forseth RR, Schroeder FC. NMR-spectroscopic analysis of mixtures: from structure to function. Curr Opin Chem Biol. 2011;15:38–47. doi: 10.1016/j.cbpa.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Antebi A. Hormonal signals produced by DAF-9/cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development. 2004;131:1765–1776. doi: 10.1242/dev.01068. [DOI] [PubMed] [Google Scholar]
- Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, Mangelsdorf DJ, Antebi A. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci U S A. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18668–18673. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich JT, Entchev EV, Mende F, Boytchev H, Martin R, Zagoriy V, Theumer G, Riezman I, Riezman H, Knolker HJ, et al. Methylation of the sterol nucleus by STRM-1 regulates dauer larva formation in Caenorhabditis elegans. Dev Cell. 2009;16:833–843. doi: 10.1016/j.devcel.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Held JM, White MP, Fisher AL, Gibson BW, Lithgow GJ, Gill MS. DAF-12-dependent rescue of dauer formation in Caenorhabditis elegans by (25S)-cholestenoic acid. Aging Cell. 2006;5:283–291. doi: 10.1111/j.1474-9726.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hu PJ. Dauer. WormBook; 2007. pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Albert PS, Riddle DL. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development. 2002;129:221–231. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci. 2010;1204:156–162. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig AH, Kober-Eisermann C, Weitzel C, Bethke A, Neubert K, Gerisch B, Hutter H, Antebi A. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 2004;18:2120–2133. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Ogawa A, Streit A, Antebi A, Sommer RJ. A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr Biol. 2009;19:67–71. doi: 10.1016/j.cub.2008.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and β-oxidation. Cell Metabolism. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DS, Fang LL, Svy DK, Ruvkun G, Li W. Genetic identification of HSD-1, a conserved steroidogenic enzyme that directs larval development in Caenorhabditis elegans. Development. 2008;135:2239–2249. doi: 10.1242/dev.016972. [DOI] [PubMed] [Google Scholar]
- Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, Schroeder FC. A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:7708–7713. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D, Swanson M, Albert P. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Albert PS. Genetic and environmental regulation of dauer larva development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C elegans II. Cold Spring Harbor (NY): 1997. [PubMed] [Google Scholar]
- Rottiers V, Motola DL, Gerisch B, Cummins CL, Nishiwaki K, Mangelsdorf DJ, Antebi A. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev Cell. 2006;10:473–482. doi: 10.1016/j.devcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Schaedel ON, Gerisch B, Antebi A, Sternberg PW. Hormonal signal amplification mediates environmental conditions during development and controls an irreversible commitment to adulthood. PLoS Biol. 2012;10:e1001306. doi: 10.1371/journal.pbio.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp M, Lazar MA. Endogenous ligands for nuclear receptors: digging deeper. J Biol Chem. 2010;285:40409–40415. doi: 10.1074/jbc.R110.182451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Wollam J, Magner D, Karalay O, Antebi A. A steroid receptor-microRNA switch regulates life span in response to signals from the gonad. Science. 2012;338:1472–1476. doi: 10.1126/science.1228967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singarapu KK, Zhu J, Tonelli M, Rao H, Assadi-Porter FM, Westler WM, DeLuca HF, Markley JL. Ligand-specific structural changes in the vitamin D receptor in solution. Biochemistry. 2011;50:11025–11033. doi: 10.1021/bi201637p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Ward JD, Yamamoto KR. Nuclear hormone receptors in nematodes: evolution and function. Mol Cell Endocrinol. 2010;334:49–55. doi: 10.1016/j.mce.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilopoulos S, Wang Y, Kitambi SS, Sacchetti P, Sousa KM, Bodin K, Kirk J, Saltó C, Gustafsson M, Toledo EM, et al. Brain endogenous liver X receptor ligands selectively promote midbrain neurogenesis. Nat Chem Biol. 2013;9:126–133. doi: 10.1038/nchembio.1156. [DOI] [PubMed] [Google Scholar]
- von Reuss SH, Bose N, Srinivasan J, Yim JJ, Judkins JC, Sternberg PW, Schroeder FC. Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small molecule signals in C. elegans. J Am Chem Soc. 2012;134:1817–1824. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lee JS, Nakazawa T, Ukai K, Mangindaan RE, Wewengkang DS, Rotinsulu H, Kobayashi H, Tsukamoto S, Namikoshi M. (25S)-cholesten-26-oic acid derivatives from an Indonesian soft coral Minabea sp. Steroids. 2009a;74:758–760. doi: 10.1016/j.steroids.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhou XE, Motola DL, Gao X, Suino-Powell K, Conneely A, Ogata C, Sharma KK, Auchus RJ, Lok JB, et al. Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes. Proc Natl Acad Sci U S A. 2009b;106:9138–9143. doi: 10.1073/pnas.0904064106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TW, Dumas KJ, Hu PJ. EAK proteins: novel conserved regulators of C. elegans lifespan. Aging (Albany NY) 2010;2:742–747. doi: 10.18632/aging.100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollam J, Antebi A. Sterol regulation of metabolism, homeostasis, and development. Annu Rev Biochem. 2011;80:885–916. doi: 10.1146/annurev-biochem-081308-165917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollam J, Magner DB, Magomedova L, Rass E, Shen Y, Rottiers V, Habermann B, Cummins CL, Antebi A. A novel 3-hydroxysteroid dehydrogenase that regulates reproductive development and longevity. PLoS Biol. 2012;10:e1001305. doi: 10.1371/journal.pbio.1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollam J, Magomedova L, Magner DB, Shen Y, Rottiers V, Motola DL, Mangelsdorf DJ, Cummins CL, Antebi A. The Rieske oxygenase DAF-36 functions as a cholesterol 7-desaturase in steroidogenic pathways governing longevity. Aging Cell. 2011;10:879–884. doi: 10.1111/j.1474-9726.2011.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki TM, Berman JR, Suchanek-Kavipurapu M, McCormick M, Gaglia MM, Lee SJ, Kenyon C. The somatic reproductive tissues of C. elegans promote longevity through steroid hormone signaling. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama-Yanagawa T, Enya S, Shimada-Niwa Y, Yaguchi S, Haramoto Y, Matsuya T, Shiomi K, Sasakura Y, Takahashi S, Asashima M, et al. The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J Biol Chem. 2011;286:25756–25762. doi: 10.1074/jbc.M111.244384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi X, Zhou XE, Melcher K, Motola DL, Gelmedin V, Hawdon J, Kliewer SA, Mangelsdorf DJ, Xu HE. Structural conservation of ligand binding reveals a bile acid-like signaling pathway in nematodes. J Biol Chem. 2012;287:4894–4903. doi: 10.1074/jbc.M111.315242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.