Abstract
Purpose of review
Lipase maturation factor 1 (LMF1) is a membrane-bound protein located in the endoplasmic reticulum (ER). It is essential to the folding and assembly (i.e., maturation) of a select group of lipases that include lipoprotein lipase (LPL), hepatic lipase (HL) and endothelial lipase (EL). The purpose of this review is to examine recent studies that have begun to elucidate the structure and function of LMF1, and to place it in the context of lipase folding and assembly.
Recent findings
Recent studies identified mutations in LMF1 that cause combined lipase deficiency and hypertriglyceridemia in humans. These mutations result in the truncation of a large, evolutionarily conserved domain called DUF1222, which is essential for interaction with lipases and their attainment of enzymatic activity. The structural complexity of LMF1 has been further characterized by solving its topology in the ER membrane. Recent studies indicate that in addition to LPL and HL, the maturation of EL is also dependent on LMF1. Based on its apparent specificity for dimeric lipases, LMF1 is proposed to play an essential role in the assembly and/or stabilization of head-to-tail lipase homodimers.
Summary
LMF1 functions in the maturation of a select group of secreted lipases that assemble into homodimers in the ER. These dimeric lipases include LPL, HL and EL, all of which contribute significantly to plasma triglyceride and HDL cholesterol levels in human populations. Future studies involving genetically engineered mouse models will be required to fully elucidate the role of LMF1 in normal physiology and disease.
Keywords: Protein folding, endoplasmic reticulum, lipase maturation factor 1, lipoprotein lipase, hepatic lipase, endothelial lipase
Introduction
Lipoprotein lipase (LPL), hepatic lipase (HL) and endothelial lipase (EL) are lipolytic enzymes involved in plasma lipid metabolism. They are part of a lipase gene superfamily [1], and have emerged as important genetic determinants of plasma triglyceride (TG) and high density lipoprotein (HDL)- cholesterol levels in humans [2]. Given the critical role of lipases in lipid homeostasis, it is of interest to identify mechanisms and factors that influence their expression and activity. While transcriptional control is certainly an important component of lipase regulation, it is becoming increasingly clear that post-translational factors play an essential role as well [3]. In this review, we will discuss recent studies on Lipase maturation factor 1 (LMF11), a newly discovered protein involved in the post-translational maturation of secreted homodimeric lipases (LPL, HL and EL). Moreover, we will place LMF1 in the context of the folding and assembly of this select group of lipase proteins in the ER.
LMF1 mutations and disease
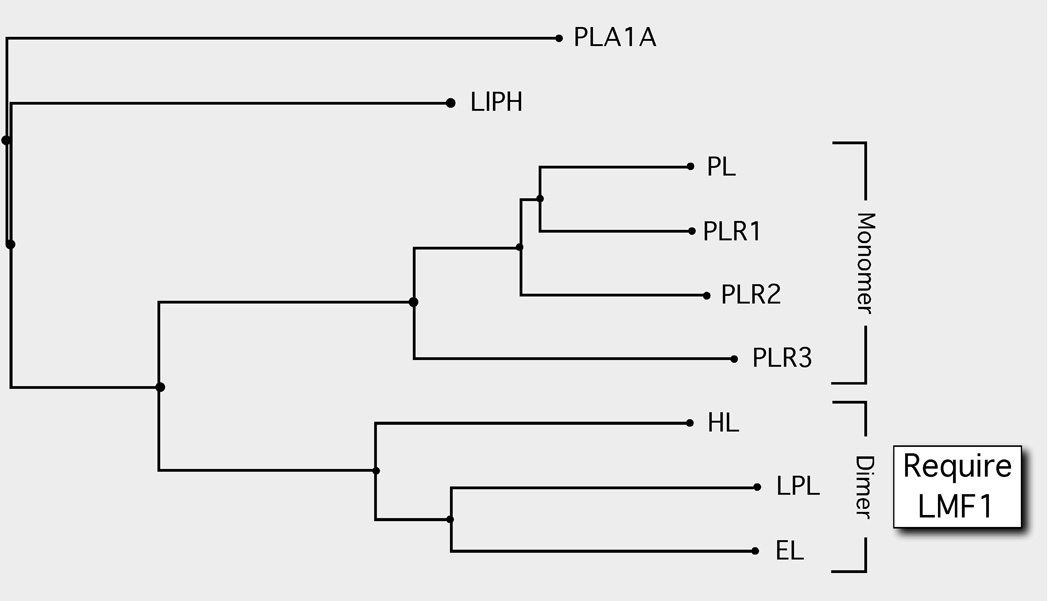
In 1983, a mouse mutation called combined lipase deficiency (cld) was described [4]. Whereas heterozygous mice appeared normal, homozygous neonates exhibited massive hypertriglyceridemia and died several days after birth due to the virtual absence of enzyme activity for both LPL and HL. Interestingly, lipase mRNA and protein expression was normal in cld tissues indicating that the mutation affected a gene critically required for the post-translational attainment of enzymatic activity. Some 24 years after its initial description, the cld mutation was found to affect a previously uncharacterized gene, which was named Lipase maturation factor 1 (Lmf1) [5]. Recently, we have found that the cld mutation also disrupts maturation of a third member of the lipase protein family, EL (Peterfy M, unpublished data). Thus, all known homodimeric lipases (LPL, HL, EL), but not a monomeric lipase, pancreatic lipase (PL) [6], require Lmf1 to mature into active, secreted enzymes (Fig. 1).
Figure 1. The LMF1-dependent dimeric lipases are a select group within the larger lipase gene family.
Phylogenetic tree of the lipase gene family. Members of the family are secreted enzymes, and thus all mature within the ER. The members include: PLA1A, Phospholipase A1 member A (Q53H76); LIPH, Lipase member H (Q8WWY8); PL, Pancreatic triacylglycerol lipase (P16233) and the three PL-related lipases PLR1 (P54315), PLR2 (P54317) and PLR3 (Q17RR3); HL, Hepatic triacylglycerol lipase (P11150); LPL, Lipoprotein lipase (P06858); and EL, Endothelial lipase (Q9Y5X9). The monomeric state of the PL-related lipases 1, 2 and 3 are inferred based on the parental PL enzyme. The homodimeric state of HL, LPL and EL has been experimentally determined. The subunit structure and requirement for LMF1 is not known for either PLA1A or LIPH. Protein sequence alignments and phylogenetic tree construction was conducted using COBALT [23].
Subsequent to the cloning of Lmf1, a rare nonsense mutation (Y439X) was also documented in the orthologous human LMF1 gene [5]. Consistent with the mouse phenotype, a patient homozygous for the Y439X mutation exhibited combined lipase deficiency associated with severe hypertriglyceridemia. In addition, the patient suffered from repeated episodes of pancreatitis, developed tuberous xanthomas and acquired partial lipodystrophy in conjunction with type 2 diabetes. More recently, a second homozygous nonsense mutation (W464X) has also been described in an individual with hypertriglyceridemia, pancreatitis and combined lipase deficiency [7]. In addition to LPL and HL, post-heparin plasma EL activity was also severely diminished in this patient (Peterfy M, unpublished data). Interestingly, lipase activities appear to be somewhat less affected compared to the Y439X patient, while tuberous xanthomas and lipodystrophy were not evident. Although both mutant transcripts may be subject to nonsense-mediated RNA decay, it is intriguing to speculate that the apparently stronger phenotype of Y439X is due to the larger C-terminal truncation of the protein (Fig. 2). Indeed, it has been demonstrated that Y439X completely abolishes LMF1 activity, whereas W464X is a hypomorphic mutant with ~40% of wild-type activity retained [7].
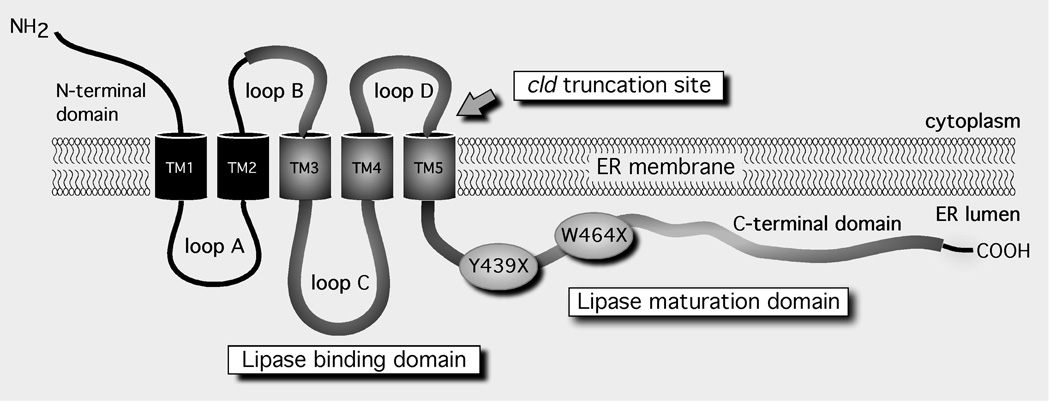
Figure 2. The membrane topology of LMF1.
Five alpha-helical transmembrane (TM) segments divide the protein into six separate domains. The approximate size of each domain in mouse Lmf1 is 49 aa, N-terminal domain; 56 aa, loops A, B; 71 aa, loop C; 46 aa, loop D; and 188 aa, C-terminal domain (aa = amino acids). The evolutionarily conserved domain of unknown function (DUF1222) is depicted as a thick line. The domains involved in lipase binding and maturation are indicated. Y439X and W464X are nonsense mutations that cause combined lipase deficiency in human subjects.
The structure and function of LMF1
LMF1 is a polytopic membrane-spanning protein of the ER membrane (Fig. 2). It has five transmembrane segments, which separate the protein into six domains, half of which face either the ER lumen or cytoplasm [8]. Nearly 70% of the LMF1 sequence is comprised of an evolutionarily conserved Domain of Unknown Function (referred to as DUF1222 in the Pfam database). Importantly, all known LMF1 mutations associated with combined lipase deficiency in mouse and humans truncate the large C-terminal domain of DUF1222 (Fig. 2), which indicates an essential role of this region in lipase maturation. Notably, while cld and Y439X cause a complete loss of function [5], the downstream W464X mutation is less severe, resulting in the loss of ~60% function [7]; thus, the proximal region of the C-terminal domain upstream of W464X may be particularly important in lipase maturation. Indeed, this region exhibits a remarkable degree of amino acid conservation across different species (Doolittle M, unpublished data). In addition to decreasing LMF1 specific activity, these mutations also diminish LMF1 protein expression levels [5, 7, 9]. This is a potential consequence of improper membrane integration and non-native topologies resulting in rapid degradation of mutant LMF1 proteins.
Besides the large C-terminal domain, another DUF1222 domain facing the ER lumen is loop C (Fig. 2), which also appears to play a central role in lipase folding and assembly. Notably, its ER lumenal localization places it in proximity to newly synthesized lipase polypeptides. Using a series of truncated mouse Lmf1 constructs, loop C was identified as a lipase-binding site [8]. Among the lipase family members examined in this study, LPL and HL, but not pancreatic lipase (PL), were found to bind to loop C. Recently, EL has also been shown to interact with this domain (Peterfy M, unpublished data). Importantly, whereas LPL, HL and EL are homodimers [10–12], PL is a monomer and does not require LMF1 for its maturation [6]. Thus, loop C binding appears to be specific for dimeric lipases, consistent with the specificity of the lipase maturation function of LMF1 (Fig. 1). Loop C also represents the most highly conserved domain within LMF1, suggesting that its lipase-binding function tolerates very little sequence divergence. Moreover, the involvement of loop C and the C-terminal domain in the related functions of lipase binding and maturation suggests that these two adjacent regions may physically interact during the course of LMF1 function.
The role of LMF1 in lipase folding and assembly
Maturation of LPL, HL and EL requires the lipase polypeptides to first attain a proper tertiary fold, followed by their assembly into homodimers arranged in a head-to-tail orientation [10–12]. In vitro refolding experiments have indicated that properly folded LPL monomers do not exist free of the homodimer [13], although monomer subunits can readily exchange between homodimers [14]. When LPL dimers disassemble, which occurs readily because they represent a high free energy state, the monomers misfold and lose their ability to re-assemble. Thus, it is likely that a fully folded monomer is thermodynamically unstable in solution, perhaps due to the presence of exposed hydrophobic regions that in the dimer form contact points holding the subunits in place. Indeed, the major LPL intermediate form that is detected during refolding in vitro is a monomer exhibiting a partially folded N-terminal domain in a “molten globule” state with a fully folded C-terminal domain [13].
LPL refolding to a fully assembled state takes hours in vitro [13], but only minutes in vivo with a relatively high efficiency [15]. Thus, it is clear that lipase maturation in vivo proceeds with the aid of chaperones, increasing not only the kinetics of folding and assembly, but protecting nascent chains from inappropriate intra- and inter-chain interactions; this latter function increases productive folding while discouraging protein aggregation. Indeed, chaperones and associated folding factors generally assist most nascent polypeptide chains as soon as they emerge from the Sec61 translocon into the ER lumen. They provide surveillance of maturation, a process called ER quality control [16], ensuring that all nascent chains remain in the ER until their folding is complete. Moreover, they direct terminally misfolded proteins to pathways of ER-associated degradation (ERAD), and provide safeguards against massive protein misfolding by regulating the ER stress response [17]. As many chaperones assist a wide array of proteins, they are considered “general” maturation factors. However, some nascent proteins engage with specialized chaperones when folding and/or assembly requires the attainment of unique structural characteristics; these are often called “client-specific” factors. Indeed, lipase maturation requires both: the ER chaperone calnexin (CNX) is a general factor that is required for efficient lipase maturation [15, 18, 19], as is the client-specific factor LMF1 (Fig. 3).
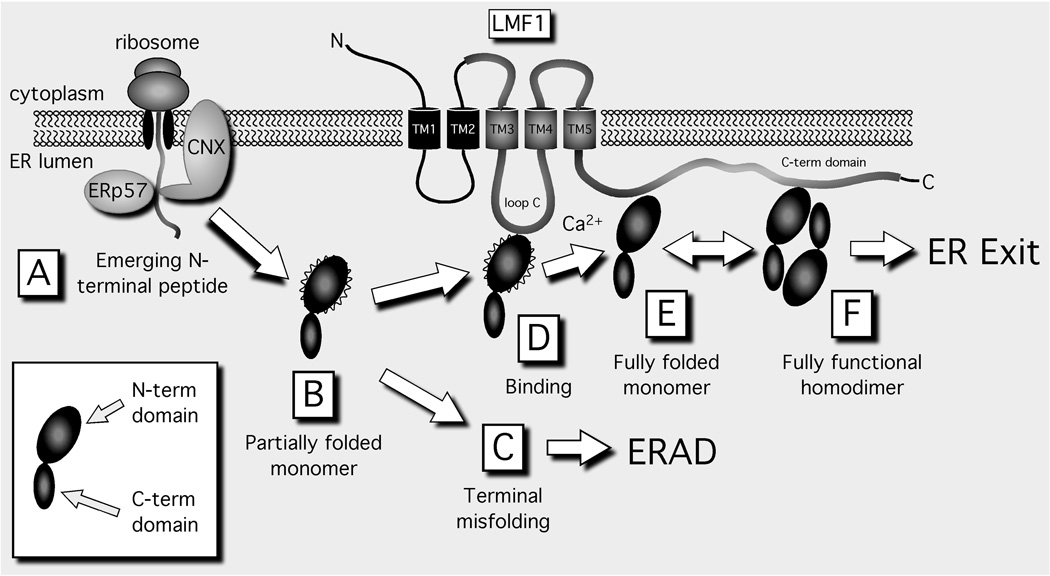
Figure 3. The proposed role of LMF1 in lipase folding and assembly.
The inset illustrates the N- and C-folding domains comprising the lipase monomer; wavy lines indicate an N-terminal domain that is partially folded. Only the homodimer exhibits enzyme activity and exits the ER; all other lipase forms are inactive and are retained in the ER. Terminally misfolded forms are destined for ER-associated degradation (ERAD). CNX, calnexin; ERp57, 57 kDa endoplasmic reticulum protein; LMF1, Lipase maturation factor 1.
While LMF1 likely promotes later stages of lipase folding and assembly (Fig. 3, D–F), CNX plays an equally important role early on, stabilizing the emerging lipase N-terminal portion by delaying premature folding until translation is complete (Fig. 3A). Association with CNX occurs through a glycan chain that is attached to a conserved asparagine site within the lipase N-terminal domain; the attached glycan chain acts as a ligand that binds CNX through its lectin domain [16]. After CNX release, the lipase polypeptide begins to fold, possibly aided by the BiP chaperone complex that includes a peptidyl-prolyl cis-trans isomerase (PPIase); both BiP and PPIase were recently shown to associate with HL during its maturation in the ER [20]. The PPIase converts cis peptidyl-prolyl bonds into the favored trans configuration that has been shown to increase the kinetics of LPL refolding in vitro [13]. While the lipase C-terminal domain likely folds efficiently and quickly, the N-terminal domain remains in a molten-globule state, with a native-like secondary structure but a less-ordered tertiary structure [13]. We consider such an intermediate as a “partially folded” monomer (Fig. 3, B and D) with the potential to form either a fully folded monomer (Fig. 3E) or to undergo misfolding (Fig. 3C). Since fully folded lipase monomers are likely thermodynamically unstable in solution, LMF1 may bind and stabilizes this form (Fig. 3E); in contrast, lipase misfolding would prevent LMF1 association and result in ERAD (Fig. 3C). In fact, in combined lipase deficiency, LPL homodimers decline severely while LPL misfolded aggregates accumulate [21]. Thus, without functional LMF1, maturation steps D–F (Fig. 3) appear to occur very inefficiently. In contrast, early lipase maturation steps occurring through CNX seem to be independent of LMF1. For example, LPL in cld cells is properly glycosylated and processed [21], indicating that these early maturation steps remain unaffected. Thus, LMF1 most likely functions in later stages of lipase maturation (Fig. 3, D–F), when lipase monomers and homodimers may need to be shielded from an ER environment conducive to their misfolding. LMF1 may also stabilize the homodimer from undergoing disassembly in the ER prior to its secretion (Fig. 3F).
Open questions and future perspectives
The elucidation of LMF1 function remains a challenging undertaking, particularly in defining its role as a maturation factor and as a candidate in lipase regulation. As a maturation factor, a number of questions remain unanswered. What is the precise role of LMF1 in the mechanism of lipase folding; does it stabilize the partially folded monomer and homodimer as proposed in Fig. 3? What are the functions of the various LMF1 domains in this process? The polytopic nature of LMF1 suggests a complexity of functions involving associations with both cytoplasmic and ER lumenal proteins; what is the identity of these binding partners and how do they assist in lipase maturation? Is LMF1 also involved in the effective exit of homodimers from the ER, possibly by associating with the cytosolic coat protein II vesicles budding from the ER?
The essential role of LMF1 in the maturation of all three dimeric lipases suggests that LMF1 deficiency may have complex and unique consequences on plasma and tissue lipid metabolism. An interesting observation in this regard is the diminished adiposity in the Y439X patient [5]. Is lipodystrophy a consequence of combined lipase deficiency and, if so, what is the underlying etiology? Unfortunately, the cld mouse model has limited utility to answer these questions, because of the lethal nature of LPL deficiency occurring shortly after birth. Clearly, inducible and tissue-specific models of LMF1 deficiency are needed to overcome the lethality and investigate the role of individual tissues.
The involvement of LMF1 in the post-translational activation of lipases raises the possibility of a regulatory role in lipase activity and lipoprotein metabolism. Is LMF1 rate-limiting in the expression of active lipases? Is LMF1 expression and/or activity regulated by physiological states affecting lipase activities (e.g., feeding and fasting)? In this regard, a recent study found that metformin, an activator of the 5’-adenosine monophosphate-activated kinase, caused a substantial increase in LMF1 mRNA levels in heart tissue from Zucker rats, although its impact on lipase activity levels was not explored [22].
Conclusion
The functional loss of LMF1 results in combined lipase deficiency with a profound impact on plasma lipid metabolism and related pathophysiological phenotypes, such as hypertriglyceridemia. LMF1 is required for the post-translational maturation of LPL, HL and EL, a select group of lipases that are known to form homodimers. Lipase maturation occurs in the ER and involves the folding and assembly of newly synthesized (nascent) lipase polypeptides into fully functional enzymes. The structural similarities of LPL, HL and EL, particularly in their requirement for a homodimer configuration, suggest that LMF1 plays a role in the assembly of partially folded monomers and/or stabilization of the lipase homodimers. While questions abound concerning LMF1 function, expression and regulation, it is abundantly clear that it plays a pivotal role in lipase maturation.
Acknowledgements
The authors acknowledge funding from NIH grant HL-24841 and the Cedars-Sinai Medical Center.
Footnotes
By convention, LMF1 and Lmf1 refers to the human and mouse protein, respectively. However, in this review, LMF1 is also used as a general term without regard to species; Lmf1 will only be used when referring to the mouse protein.
Contributor Information
Mark H. Doolittle, Department of Medicine, David Geffen School of Medicine, University of California at Los Angeles, and VA Greater Los Angeles Healthcare System, 11301 Wilshire Blvd., Bldg. 113, Rm. 312, Los Angeles, CA 90073, USA, Tel.: 661-433-6349, Fax: 310-268-4981, markdool@ucla.edu
Nicole Ehrhardt, Medical Genetics Institute, Cedars-Sinai Medical Center, 8700 Beverly Blvd., Los Angeles, CA 90048, USA, Tel.: 310-423-3862, Fax: 310-423-0299, Nicole.Ehrhardt@cshs.org.
Miklós Péterfy, Medical Genetics Institute, Cedars-Sinai Medical Center, and Department of Medicine, David Geffen School of Medicine, University of California at Los Angeles, 8700 Beverly Blvd., Los Angeles, CA 90048, USA, Tel.: 310-478-3711 x42153, Fax: 310-268-4981, mpeterfy@ucla.edu.
References and recommended reading
- 1.Wong H, Schotz MC. The lipase gene family. J Lipid Res. 2002;43:993–999. doi: 10.1194/jlr.r200007-jlr200. [DOI] [PubMed] [Google Scholar]
- 2.Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Eckel RH. Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab. 2009;297:E271–E288. doi: 10.1152/ajpendo.90920.2008. [DOI] [PubMed] [Google Scholar]
- 4.Paterniti JR, Jr, Brown WV, Ginsberg HN, Artzt K. Combined lipase deficiency (cld): a lethal mutation on chromosome 17 of the mouse. Science. 1983;221:167–169. doi: 10.1126/science.6857276. [DOI] [PubMed] [Google Scholar]
- 5.Peterfy M, Ben-Zeev O, Mao HZ, et al. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat Genet. 2007;39:1483–1487. doi: 10.1038/ng.2007.24. [DOI] [PubMed] [Google Scholar]
- 6.Scow RO, Schultz CJ, Park JW, Blanchette-Mackie EJ. Combined lipase deficiency (cld/cld) in mice affects differently post-translational processing of lipoprotein lipase, hepatic lipase and pancreatic lipase. Chem Phys Lipids. 1998;93:149–155. doi: 10.1016/s0009-3084(98)00039-5. [DOI] [PubMed] [Google Scholar]
- 7. Cefalu AB, Noto D, Arpi ML, et al. Novel LMF1 nonsense mutation in a patient with severe hypertriglyceridemia. J Clin Endocrinol Metab. 2009;94:4584–4590. doi: 10.1210/jc.2009-0594. This is the first report of a hypomorphic LMF1 mutation (W464X). The residual activity of W464X, when compared to the complete loss of function caused by Y439X, suggests the presence of important structural determinants of lipase folding in the 439–464 region of LMF1.
- 8. Doolittle MH, Neher SB, Ben-Zeev O, et al. Lipase maturation factor LMF1, membrane topology and interaction with lipase proteins in the endoplasmic reticulum. J Biol Chem. 2009;284:33623–33633. doi: 10.1074/jbc.M109.049395. This study solves the complex membrane topology of LMF1, and identifies a lipase interaction site within DUF1222 (loop C).
- 9.Yin F, Doolittle MH, Peterfy M. A quantitative assay measuring the function of lipase maturation factor 1. J Lipid Res. 2009;50:2265–2269. doi: 10.1194/jlr.M900196-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong H, Yang D, Hill JS, et al. A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc Natl Acad Sci U S A. 1997;94:5594–5598. doi: 10.1073/pnas.94.11.5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill JS, Davis RC, Yang D, et al. Hepatic lipase: high-level expression and subunit structure determination. Methods Enzymol. 1997;284:232–246. doi: 10.1016/s0076-6879(97)84015-3. [DOI] [PubMed] [Google Scholar]
- 12. Griffon N, Jin W, Petty TJ, et al. Identification of the active form of endothelial lipase, a homodimer in a head-to-tail conformation. J Biol Chem. 2009;284:23322–23330. doi: 10.1074/jbc.M109.037002. This study demonstrates that similar to LPL and HL, the active conformation of EL is a head-to-tail homodimer, which has important implications for the molecular function of LMF1.
- 13.Zhang L, Lookene A, Wu G, Olivecrona G. Calcium triggers folding of lipoprotein lipase into active dimers. J Biol Chem. 2005;280:42580–42591. doi: 10.1074/jbc.M507252200. [DOI] [PubMed] [Google Scholar]
- 14.Lookene A, Zhang L, Hultin M, Olivecrona G. Rapid subunit exchange in dimeric lipoprotein lipase and properties of the inactive monomer. J Biol Chem. 2004;279:49964–49972. doi: 10.1074/jbc.M407419200. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Zeev O, Mao HZ, Doolittle MH. Maturation of lipoprotein lipase in the endoplasmic reticulum. Concurrent formation of functional dimers and inactive aggregates. J Biol Chem. 2002;277:10727–10738. doi: 10.1074/jbc.M108128200. [DOI] [PubMed] [Google Scholar]
- 16.Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19:515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 18.Briquet-Laugier V, Ben-Zeev O, Doolittle MH. Determining lipoprotein lipase and hepatic lipase activity using radiolabeled substrates. Methods Mol Biol. 1999;109:81–94. doi: 10.1385/1-59259-581-2:81. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Zeev O, Doolittle MH. Maturation of hepatic lipase. Formation of functional enzyme in the endoplasmic reticulum is the rate-limiting step in its secretion. J Biol Chem. 2004;279:6171–6181. doi: 10.1074/jbc.M310051200. [DOI] [PubMed] [Google Scholar]
- 20.Doolittle MH, Ben-Zeev O, Bassilian S, et al. Hepatic lipase maturation: a partial proteome of interacting factors. J Lipid Res. 2009;50:1173–1184. doi: 10.1194/jlr.M800603-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briquet-Laugier V, Ben-Zeev O, White A, Doolittle MH. cld and lec23 are disparate mutations that affect maturation of lipoprotein lipase in the endoplasmic reticulum. J Lipid Res. 1999;40:2044–2058. [PubMed] [Google Scholar]
- 22. Forcheron F, Basset A, Del Carmine P, Beylot M. Lipase maturation factor 1: its expression in Zucker diabetic rats, and effects of metformin and fenofibrate. Diabetes Metab. 2009;35:452–457. doi: 10.1016/j.diabet.2009.05.004. This is the first demonstration that LMF1 mRNA expression is regulated in a physiological setting.
- 23.Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]