Abstract
Long intervening noncoding RNAs (lincRNAs) are transcribed from thousands of loci in mammalian genomes and might play widespread roles in gene regulation and other cellular processes. This Review outlines the emerging understanding of lincRNAs in vertebrate animals, with emphases on how they are being identified and current conclusions and questions regarding their genomics, evolution and mechanisms of action.
Introduction
The conventional view of the mammalian genome was that ~20,000 protein-coding genes were dispersed within mostly repetitive and largely nontranscribed sequence. Over the past decade, this view has been challenged by increasingly thorough examinations of the RNA species in mammalian cells. These studies have revealed the fascinating complexity of the transcriptome, in which protein-coding genes produce many alternative products, and genomic regions previously thought to be transcriptionally silent give rise to a range of processed and regulated transcripts that do not appear to code for functional proteins. A few of these transcripts are precursors for small regulatory RNAs, such as microRNAs, but the vast majority have no recognizable purpose.
A sensible hypothesis is that most of the currently annotated long (typically >200 nt) noncoding RNAs are not functional, i.e., most impart no fitness advantage, however slight. Like all biochemical processes, the transcription machinery is not perfect and can produce spurious RNAs that have no purpose (Struhl, 2007). Due to the intrinsic properties of RNA, these transcripts would have a collapsed fold (Schultes et al., 2005). Because chromatin states vary across cell fates, cryptic promoters would be differentially accessible in different cellular contexts, and thus many spurious transcripts would also have tissue-specific expression. Because of the underlying transcriptional processes and chance occurrence of splice sites, many would also be capped, spliced, and polyadenylated. Thus, none of these features offer an informative indicator of function. Moreover, many of these spurious RNA species that confer no fitness advantage would also impose minimal fitness cost, in which case, simply tolerating them would be more feasible than evolving and maintaining more rigorous control mechanisms that could prevent their production. A second source of nonfunctional RNAs would be those generated during regulatory events in which the act of transcription matters, whereas the product of transcription does not. These would include RNAs generated during transcriptional interference, which involves transcription of noncoding loci that overlap regulatory regions and is known to regulate gene expression in both prokaryotes and eukaryotes (Shearwin et al., 2005). Against this backdrop of many nonfunctional transcripts, some long noncoding RNAs, including the Xist RNA, which is required for mammalian dosage compensation (Penny et al., 1996), clearly are functional, and the roster of biological processes in which long noncoding RNAs are reported to play key roles is rapidly growing and now includes cell-cycle regulation, apoptosis, and establishment of cell identity (reviewed in Ponting et al., 2009; Pauli et al., 2011; Rinn and Chang, 2012).
Despite general agreement that some long noncoding RNAs are functional and others are not, opinions vary widely as to the fraction that is functional (Kowalczyk et al., 2012). Because of their marginal sequence conservation and a sense that spurious transcripts would impose minimal fitness cost, we suspect that most are not functional. However, even a scenario in which only 10% are functional implies the existence of more than a thousand human loci generating noncoding transcripts with biological roles. These enigmatic RNAs will consume decades of effort for many labs undertaking molecular, mechanistic, and phenotypic analyses. And regardless of function, long noncoding RNAs might have diagnostic applications, with changes in their expression already associated with cancer and several neurological disorders (Prensner et al., 2011; Brunner et al., 2012; Ziats and Rennert, 2013).
To identify noncoding RNAs and their corresponding genes cleanly, and to simplify their analysis by avoiding the complications arising from overlap with other types of genes, recent focus has been on long intervening noncoding RNAs (lincRNAs, also called long “intergenic” noncoding RNAs even though the lincRNAs derive from genes and are thus genic), which do not overlap exons of either protein-coding or other non-lincRNA types of genes. Here, we also focus on this subgroup, as lincRNA gene expression patterns, sequence conservation and perturbation outcomes are easier to interpret than those of transcripts from loci overlapping other gene classes. We presume that the features of lincRNAs will also apply to many other long non-coding RNA transcripts that were excluded from lincRNA lists because of complicating (albeit, often functionally inconsequential) overlap with other annotations.
At the outset, we emphasize that lincRNA classification differs from that of other RNAs, in that lincRNAs are defined more by what they are not than by what they are. As is typical of stable RNA polymerase II products, lincRNAs are nearly always capped and polyadenylated, and are frequently spliced. But aside from this positive descriptor of being Pol II products, lincRNAs are defined using negative descriptors, i.e., not coding for proteins and not overlapping transcripts of certain other types of genes. Reliance on these negative descriptors risks grouping together a hodgepodge of transcripts with very diverse properties and mechanisms of action. In many ways the lincRNA field faces challenges similar to those faced by early biologists trying to categorize and contemplate the diverse array of life forms that were not plants and not animals. We suspect that there might be dozens of distinct functional noncoding RNA classes that have transcripts currently grouped into the catch-all class of lincRNAs. Until these classes are understood and differentiated, insights from the study of one lincRNA will be difficult to apply to others, and attempts to understand the general features of lincRNAs will at best reflect only the more populated classes. With these caveats in mind, we review the current understanding of vertebrate lincRNAs, focusing on their identification, genomics, evolution and mechanisms of action.
lincRNA Identification
lincRNAs and lincRNA candidates have been cataloged in human, mouse, zebrafish, frog, fly, nematode, Arabidopsis, maize, and Plasmodium (Table 1). Interrogation of lincRNA function or mechanisms depends on high-quality transcript models of lincRNA genes, including accurate genomic positions of the start site, splice sites, and polyadenylation site of each transcript. Useful collections of lincRNAs are those that capture full-length transcripts and avoid those encoding functional peptides. Methodological advances and increased throughput are continuously improving the ability to meet these goals and help explain the diversity of annotation criteria and cutoffs (Table 1), which in turn might be one of the reasons lincRNA lists from different studies do not have more overlap.
Table 1.
Large-Scale Efforts to Catalog lincRNA Loci and Transcripts
| Reference | Data for Transcript Reconstruction |
Genomic Features and Filters | Coding-Potential Filters | Number of lincRNAs |
|---|---|---|---|---|
| Mouse | ||||
| Ravasi et al., 2006 | cDNAs | Manual curation, ORF length, CRITICA |
13,502 transcripts | |
| Ponjavic et al., 2007 | cDNAs, CAGE | Manual curation, ORF length, BLAST, CRITICA |
3,122 transcripts | |
| Guttman et al., 2009 | Chromatin marks, tiling arrays |
Collection of approximate exonic regions, chromatin domain ≥5 kb |
CSF | 1,675 loci (1,250 conservatively defined) |
| Guttman et al., 2010 | RNA-seq | Multi-exon only | CSF | 1,140 lincRNA transcripts |
| Sigova et al., 2013 | RNA-seq, cDNAs, chromatin marks, |
Antisense overlap with mRNA introns allowed, ≥100 nt mature length |
CPC | 1,664 loci |
|
| ||||
| Human | ||||
| Khalil et al., 2009 | Chromatin marks, tiling arrays |
Collection of approximate exonic regions, chromatin domain ≥ 5 kb |
CSF | 3,289 loci |
| Jia et al., 2010 | cDNAs | Overlap with mRNAs allowed | 5,446 transcripts | |
| Ørom et al., 2010 | cDNAs | Restricted to loci >1 kb away from known protein-coding genes, ≥200 nt mature length |
Manual curation based on length, conservation and other characteristics of the ORFs |
3,019 transcripts from 2,286 loci |
| Cabili et al., 2011 | RNA-seq | Multi-exon only, ≥200 nt mature length |
PhyloCSF, Pfam | 8,195 transcripts (4,662 in the stringent set) |
| Derrien et al., 2012 | cDNAs | Overlap with mRNAs allowed (intergenic transcripts reported separately), ≥200 nt mature length |
Manual curation based on length, conservation and other characteristics of the ORFs |
14,880 transcripts from 9,277 loci, including 9,518 intergenic transcripts |
| Sigova et al., 2013 | RNA-seq, cDNAs, chromatin marks, |
Antisense overlap with mRNA introns allowed, ≥100 nt mature length |
CPC | 3,548 loci from embryonic stem cells, and 3,986 loci from endodermal cells |
|
| ||||
| Frog | ||||
| Tan et al., 2013 | RNA-Seq | >25 kb away from known protein- coding genes or on a different strand from the neighboring genes, ≥200 nt mature length |
ORF length, BLAST, Pfam | 6,686 transcripts from 3,859 loci |
|
| ||||
| Zebrafish | ||||
| Ulitsky et al., 2011 | RNA-seq, cDNAs, 3P-seq, chromatin marks |
Antisense overlap with mRNA introns allowed, ≥200 nt mature length |
CPC | 691 transcripts from 567 loci |
| Pauli et al., 2012 | RNA-seq | Stringent criteria for single exon, intron overlap with mRNA allowed, ≥160 nt mature length |
ORF length, PhyloCSF, BLAST, Pfam |
397 intergenic and 184 intronic overlapping transcripts |
|
| ||||
| Fly | ||||
| Tupy et al., 2005 | cDNA | Manual curation based on ORF length, conservation and other characteristics, Ka/Ks test, QRNA |
17 transcripts | |
| Young et al., 2012 | RNA-seq | ≥200 nt locus length | 1,119 trancripts | |
|
| ||||
| Nematode | ||||
| Nam and Bartel, 2012 | RNA-seq, 3P-seq | ≥100 nt mature length | CPC, RNAcode, ribosome profiling, polysome association |
262 lincRNA transcripts from 170 loci |
|
| ||||
|
Arabidopsis | ||||
| Liu et al., 2012a | cDNA, tiling arrays, RNA-seq |
In part a collection of approximate exonic regions, >500 bp away from protein-coding genes, no overlap with transposable elements allowed, ≥200 nt mature length |
ORF length | 6,480 transcription units from tiling arrays, 278 transcripts from RNA-seq |
|
| ||||
| Maize | ||||
| Boerner and McGinnis, 2012 | cDNA | Both sense overlap with introns and antisense overlap with mRNA or introns allowed, ≥200 nt mature length |
ORF length | 2,492 transcripts |
|
| ||||
|
Plasmodium falciparum
| ||||
| Broadbent et al., 2011 | Tiling arrays | Collection of approximate exonic regions, ≥200 nt mature length |
BLAST | 60 transcripts |
Transcripts overlapping protein-coding sequences on either strand were excluded unless noted otherwise. Coding-potential filters included: ORF length; similarity to known protein-coding regions (BLAST); substitution patterns in whole-genome alignments, quantified by CRITICA (Badger and Olsen, 1999), CSF (Lin et al., 2007), PhyloCSF (Lin et al., 2011), QRNA (Rivas and Eddy, 2001; Rivas et al., 2001), or RNAcode (Washietl et al., 2011), as indicated; the CPC algorithm, which evaluates ORF properties and similarity to known proteins (Kong et al., 2007); the HMMER algorithm, which tests for potential to encode a known protein domain (Pfam); ribosome profiling, and polyribosome association. Criteria used to define the lincRNA collection (and not those used only for characterization) are listed.
Because of their poly(A) tails and other mRNA-like features, lincRNAs are represented in typical cDNA cloning, tiling array, and RNA-seq data sets. The first large-scale catalog of putatively noncoding transcripts came from the FANTOM project (Okazaki et al., 2002; Carninci et al., 2005), which used cDNA cloning followed by Sanger sequencing and reported >34,000 long noncoding RNAs expressed in different mouse tissues, of which 3,652 had confident support (Ravasi et al., 2006). Subsequent studies refined EST- and cDNA-based lincRNA catalogs in mouse and human, which comprise the current RefSeq and Ensembl lincRNA annotations (Derrien et al., 2012; Pruitt et al., 2012). In parallel, tiling microarrays were used to detect transcribed regions (Bertone et al., 2004; Guttman et al., 2009; Khalil et al., 2009), which was potentially more sensitive than cloning but suffered from reduced dynamic range and difficulties in defining splice junctions and connecting transcribed regions into transcript models (Agarwal et al., 2010). More recently, high-throughput sequencing of millions of short RNA fragments (RNA-seq) is enabling transcript models to be reconstructed, either with the aid of a reference genome (Trapnell et al., 2010; Cabili et al., 2011) or without it (Grabherr et al., 2011). RNA-seq has yielded billions of strand-specific paired-end reads of ~100 nt each, and those can be sufficient for reconstruction of even very lowly expressed transcripts (Cabili et al., 2011; Pauli et al., 2012). Furthermore, even rarer transcripts can be specifically enriched using array-based capture methods prior to sequencing (Mercer et al., 2012).
Despite the advantages of RNA-seq in terms of sensitivity and accessibility, assembly of transcript models from short reads still has limitations, stemming primarily from the relatively small portion of the full transcript accounted for by each read and from sequence redundancies in the genome. It remains difficult to determine which exon combinations co-occur in long multiply spliced transcripts and to discriminate between independent lincRNAs and fragments of alternative mRNA isoforms or pseudogenes. Focusing only on spliced transcripts helps improve specificity (Cabili et al., 2011) but misses some bona fide single-exon lincRNAs, such as Malat1 and Neat1 (Hutchinson et al., 2007). Therefore, curated lincRNA databases (e.g., RefSeq and Ensembl) still rely primarily on cDNA sequences obtained using Sanger sequencing (Derrien et al., 2012), but we expect that this will change soon, as read lengths for high-throughput sequencing methods continue to improve and as multiple data sets are more effectively integrated to build models.
Additional data sets that can improve transcript models include chromatin maps and data from methods used to identify transcript start and polyadenylation sites (Figure 1A). Trimethylation of lysine 4 and lysine 36 in histone H3 (H3K4me3 and H3K36me3 marks), which characterize regions of Pol II transcription initiation and elongation, respectively, were used in conjunction with tiling arrays for building some lincRNA collections (Guttman et al., 2009; Khalil et al., 2009). These maps have limitations, however, as peaks of H3K4me3 can be broad and also occur at the first exon-intron junction (Bieberstein et al., 2012) (Figure 1A), and H3K36me3 enrichment is dependent on splicing and typically extends beyond the polyadenylation site (de Almeida et al., 2011) (Figure 1A). Other sources of supporting data have come from high-throughput sequencing experiments tailored to identify specific regions within RNA molecules. These include methods for high-resolution mapping of transcription start sites, e.g., using cap analysis of gene expression (CAGE) (Kodzius et al., 2006), and genome-wide annotation of polyadenylation sites, e.g., using 3P-seq (Jan et al., 2011; Ulitsky et al., 2012) (Figure 1A). A combination of independent evidence for transcription initiation, termination and exon-intron structure can enable confident identification of both multiple- and single-exon lincRNAs (Ulitsky et al., 2011).
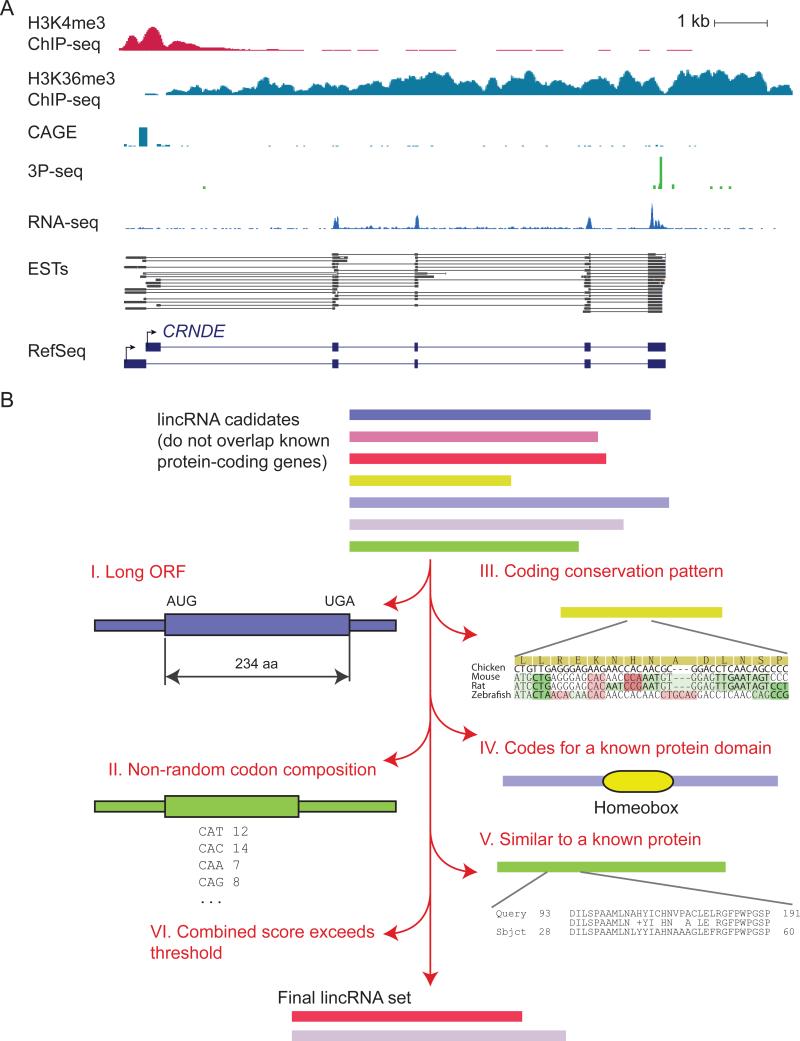
Figure 1. Assembling lincRNA Collections.
(A) Data sets useful for constructing lincRNA transcript models. Information from the indicated genome-wide data sets are plotted for the CRNDE lincRNA locus (chr16:54,950,197-54,963,922 in the human hg19 assembly). A subset of ESTs from GenBank and the corresponding RefSeq annotations are also shown. ChIP-seq and CAGE (ENCODE project, HeLaS3 cells), 3P-Seq (HeLa cells, C. Jan and D.P.B., unpublished data), RNA-seq (HeLa cells; Guo et al., 2010) were plotted using the UCSC genome browser.
(B) A generic lincRNA annotation pipeline, illustrating criteria used to filter potential mRNAs from the list of candidates.
Criteria for Distinguishing between Coding and Noncoding Transcripts
Perhaps the most challenging aspect of lincRNA discovery is that the concept of a noncoding RNA is loosely defined. Most long transcripts with known noncoding functions typically contain multiple potential open reading frames (ORFs). These ORFs might not be translated, might be translated inefficiently, or might be translated to produce a protein that has no functional consequences, e.g., because it is rapidly degraded. Due to their considerable lengths, many lincRNAs should by chance contain an ORF of at least 100 aa (Dinger et al., 2008). A clear binary separation between coding and noncoding transcripts is thus impossible, and the best that can be done is to use graded and imperfect criteria that preferentially identify transcripts that are unlikely to code for functional proteins.
Several features of bona fide protein-coding genes can be used as criteria to distinguish them from lincRNAs (Figure 1B,Table 1): (1) coding regions tend to be much longer than expected by chance (Dinger et al., 2008); (2) nucleotide frequencies of functional ORFs are dictated by nonrandom codon usage; (3) during evolution, selective pressures bias nucleotide substitutions in coding sequences (e.g., giving rise to a higher substitution rates in the silent positions of codons); (4) protein-coding genes typically contain known protein domains (e.g., present in the Pfam database); (5) coding regions are likely to bear sequence similarities to entries in protein databases. Different studies use different combinations of these five criteria in attempts to exclude protein-coding genes. The underlying assumption across these criteria is that short, recently evolved yet functional proteins are relatively rare. In support of this assumption, the current protein databases list very few functional peptides that originate from short ORFs—disregarding pseudogenes, Ensembl 68 lists only 11 human protein-coding genes that have a known function (described in Gene Ontology annotations) and an ORF < 50 aa, and none of these are shorter than 30 aa. (Note that most short peptides with known functions arise from longer ORFs because they are processed from longer precursors.)
Each of the criteria for predicting coding potential is of limited utility when used in isolation. For instance, presence of an ORF of at least 300 nt (100 aa) is commonly used for defining a transcript as coding. However, a transcript of 2 kb is expected to have an ORF of about 200 nt, and an ORF of 300 nt is only one standard deviation longer than expected (Dinger et al., 2008). Indeed, well characterized human lincRNAs, such as H19, Xist, Meg3, Hotair, and Kcnq1ot1 all have ORFs of at least 100 aa (Dinger et al., 2008). Even significant similarity to “known” protein-coding genes might be misleading, as protein databases contain large numbers of protein sequences predicted by translation of the longest ORF in sequenced cDNAs but without any further functional evidence. Using a combination of filters can address some of these problems (Badger and Olsen, 1999; Liu et al., 2006; Kong et al., 2007), though the scarcity of standards (in particular, long RNAs known to have exclusively noncoding functions) makes calibration of these difficult. An interim solution is to assemble two collections of transcript models, one with confidently predicted lincRNAs and another for which the evidence is less conclusive (referred to as transcripts of unknown coding potential or TUCPs) (Cabili et al., 2011).
Methods for focused experimental interrogation of the coding potential of a lincRNA include testing whether the transcript can yield peptides when translated in vitro (Lanz et al., 1999; Galindo et al., 2007), testing whether it associates with polysomes (Brockdorff et al., 1992), and checking if its ORFs can yield a protein when fused to a sequence coding for a peptide for which antibodies are available (Anguera et al., 2011). However, an ability to recruit the ribosome and be translated would not preclude a noncoding function. If the gene function can be assayed, the best approach is to introduce changes that perturb the ORF, such as those inducing frameshifts, and test for retention of the function (Hu et al., 2011; Ulitsky et al., 2011).
Global approaches can also show which transcripts are translated. Particularly useful is ribosome profiling, which utilizes high-throughput sequencing to map RNA regions associated with translating ribosomes (Ingolia et al., 2011). Analysis of ribosome profiling of mouse embryonic stem cells suggests that as many as half of the lincRNAs expressed in these cells are significantly associated with ribosomes (Ingolia et al., 2011). One interpretation of this observation is that the assumption of very few genes with short ORFs coding for functional peptides is wrong and that many of the currently annotated lincRNAs are in fact coding for short functional peptides. An example frequently cited in support of this interpretation is the Drosophila tarsal-less/polished rice transcript, which was originally thought to function as a long noncoding RNA but subsequently shown to code for very short functional peptides (Tupy et al., 2005; Kondo et al., 2010).
Although other examples of unrecognized functional peptides will undoubtedly be found, several lines of evidence suggest that this interpretation does not explain most of the ribosome association. First, as mentioned above, the algorithms used for generating lincRNA collections typically use sequence alignment to detect signatures of coding sequence conservation, and would detect at least those short coding regions that are highly conserved. Second, ribosomes are associated with some lincRNAs known to be enriched and function in the nucleus, such as Malat1 and Neat1, suggesting that those transcripts have some background engagement with ribosomes (presumably when they occasionally reach the cytoplasm) even though their known nuclear functions are noncoding. Third, a recent proteomics study that specifically focused on identifying short endogenous peptides detected peptides from only eight (0.4%) of the lincRNAs expressed in the human K562 cell line, and the extent to which even these peptides are functional is unknown (Slavoff et al., 2013). Fourth, and perhaps most important, is the concept of lincRNA upstream ORFs (uORFs; see below).
lincRNA uORFs
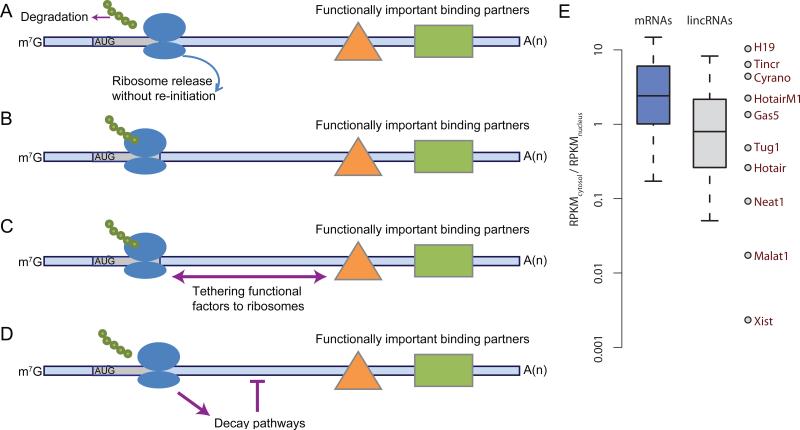
Engagement with the translating ribosome can serve purposes that have nothing to do with the translation product. Indeed, the ribosome profiling study that reported ribosome engagement in many lincRNAs reported similar engagement in annotated 5′UTRs of thousands of mRNAs, yet in contrast to translation in lincRNAs, translation of these short uORFs was not proposed to produce functional peptides (Ingolia et al., 2011). uORF translation typically plays regulatory roles, affecting translation of downstream ORFs or mRNA stability (Calvo et al., 2009; Wethmar et al., 2010). Consistent with the idea that the act of uORF translation, which can be the basis of the regulatory mechanism, is more important than the product of this translation, short peptides translated from uORFs are rarely conserved in sequence (Crowe et al., 2006), can be very unstable (Hackett et al., 1986) and are rarely detectable in mass-spectrometry-based proteomic data (Menschaert et al., 2013). We suggest that the same might be true for lincRNAs. The translated ORFs in lincRNAs might act as uORFs to prevent ribosome scanning or translation in downstream regions of the transcripts, thereby enabling the lincRNAs to perform noncoding functions in the cytoplasm without interference from the ribosome (Figures 2A and 2B). lincRNA uORFs might also tether factors to ribosomes (Figure 2C) or modulate the stability of the lincRNA by influencing RNA decay pathways, some of which depend on translation (Figure 2D).
Figure 2. Ribosomal Association and Subcellular Localization of lincRNAs.
(A) A potential role for a lincRNA uORF. Translation of a uORF into a peptide that is rapidly degraded would prevent ribosomal scanning of downstream regions, thereby protecting downstream binding factors from displacement by scanning ribosomes.
(B) Translating a nascent peptide sequence that induces ribosomal stalling would achieve an effect similar to that described in (A).
(C) The uORF can recruit a ribosome, which might be important for downstream lincRNA function.
(D) The translation of a uORF might influence the susceptibility of the lincRNA to different RNA decay pathways, such as nonsense-mediated decay (NMD).
(E) Relative subcellular localization of mRNAs and lincRNAs in MCF-7 cells. mRNA annotations were from Ensembl, and lincRNA annotations were from Ensembl, Refseq and (Cabili et al., 2011). RPKM (reads per kilobase per million mapped reads) values were computed with Cufflinks (Trapnell et al., 2010) using RNA-seq data for nuclear and cytoplasmic fractions of MCF-7 cells (Djebali et al., 2012). Ratios for selected lincRNAs are indicated.
At the molecular level, most lincRNAs appear indistinguishable from mRNAs, with 5′-m7GpppN cap structures, poly(A) tails, and exon-exon splice junctions, all of which stimulate mRNA translation (Shoemaker and Green, 2012). When considering these mRNA-like features, combined with the realization that most lincRNAs have a significant presence in the cytoplasm (see Subcellular Localization, below), the question is not: why are so many lincRNAs associated with ribosomes? The relevant question is: why are only half of the annotated lincRNAs associated with ribosomes? An important focus of future research will be determining how lincRNA export from the nucleus is regulated and how the cytoplasmic lincRNAs that do not depend on uORFs manage to avoid the translation machinery.
Bifunctional RNAs
The hypothesis that many lincRNAs have uORFs, which produce peptides, albeit nonfunctional ones, takes some liberties with the concept of noncoding RNA (although perhaps not as great as the liberties taken when speaking of uORFs falling in 5′UTRs, i.e., “untranslated regions”). Classification of noncoding transcripts is further complicated by the fact that some transcripts can have both coding and noncoding functions (Dinger et al., 2008). Xenopus and E. coli each provide an example in which the identical mature RNA embodies both coding and noncoding functions (Kloc et al., 2005; Wadler and Vanderpool, 2007). However, known examples of mRNAs moonlighting as long noncoding RNAs are still scarce, perhaps because of the challenges in identifying which mRNAs also have noncoding functions. When the coding and noncoding functions emerge at different times during evolution or when the noncoding function outlives the loss of ancestral coding potential of bifunctional mRNA, noncoding and coding transcripts with similar sequence might be found in different contemporary species, and the identification of such instances could potentially expedite the discovery of some bifunctional transcripts (Ulitsky et al., 2011; Marques et al., 2012).
lincRNA Genomics
As expected for a mixture of multiple classes of noncoding RNAs, lincRNAs lack defining sequence or structure characteristics. Nonetheless, several general features of lincRNAs in vertebrates are apparent in recent catalogs of human and zebrafish lincRNAs (Cabili et al., 2011; Ulitsky et al., 2011; Derrien et al., 2012; Pauli et al., 2012).
lincRNA genes are typically shorter than protein-coding genes (Ulitsky et al., 2011; Derrien et al., 2012; Pauli et al., 2012) and have fewer exons, typically only 2–3 (Cabili et al., 2011; Derrien et al., 2012; Pauli et al., 2012). Exons in lincRNA genes are on average slightly longer than exons in protein-coding genes (Ravasi et al., 2006; Derrien et al., 2012), presumably because the average estimate is skewed by typically longer first and last exons (Zhu et al., 2009). Transcriptional regulation, chromatinmodification patterns, and splicing signals of lincRNAs are similar to those of protein-coding genes (Ponjavic et al., 2007; Cabili et al., 2011; Derrien et al., 2012; Pauli et al., 2012), although lincRNA transcripts seem somewhat less efficiently spliced (Tilgner et al., 2012).
Most annotated lincRNAs are polyadenylated, although alternative 3′-end topologies are also occasionally observed. In humans, there are ~80 lincRNAs with circular isoforms—far fewer than the nearly 2,000 human mRNAs with circular isoforms identified in the same study (Memczak et al., 2013). A few other lincRNAs are stabilized by a triple-helical structure at their 3′ end (Brown et al., 2012; Wilusz et al., 2012) or by snoRNAs at both ends (Yin et al., 2012).
lincRNAs from human, mouse, and zebrafish are significantly more likely than mRNAs to overlap repetitive elements (Ulitsky et al., 2011; Kelley and Rinn, 2012), perhaps because lincRNA functions are more tolerant of retrotransposon insertions. Repetitive elements are also reported to play important mechanistic roles in lincRNAs, by facilitating base pairing with other RNAs containing repeats from the same family (Gong and Maquat, 2011) or through other, less understood mechanisms (Carrieri et al., 2012). Tandem repeats are also prevalent and occasionally functionally important in lincRNA genes: at least eight different tandem-repeat groups are found in Xist, seven in the first and functionally important exon (Nesterova et al., 2001; Elisaphenko et al., 2008; Zhao et al., 2008), and repetitive regions were also found within the functional domains of Miat (Tsuiji et al., 2011), DBE-T (Cabianca et al., 2012), CDR1as/ciRS-7 (Hansen et al., 2013; Memczak et al., 2013), and other lincRNAs.
lincRNA genes are preferentially found within 10 kb of proteincoding genes (Bertone et al., 2004; Ponjavic et al., 2009; Jia et al., 2010; van Bakel et al., 2010; Cabili et al., 2011; Sigova et al., 2013), which has led to the suggestion that many lincRNAs are byproducts of mRNA biogenesis (van Bakel et al., 2010). Countering this idea are analyses showing that (1) genomic colocalization persists in collections of lincRNAs supported by independent evidence for transcription initiation and termination, and (2) the distribution of distances between lincRNAs and their closest protein-coding genes resembles that of adjacent protein-coding genes (Ulitsky et al., 2011).
Studies in human, mouse, and zebrafish suggested that large gene deserts flanking transcription-factor (TF) genes, particularly those with roles in embryonic development, preferentially harbor lincRNAs (Mercer et al., 2008; Guttman et al., 2009; Ulitsky et al., 2011; Pauli et al., 2012; Wamstad et al., 2012). In vertebrates, developmental TF genes are preferentially surrounded by long intergenic regions (Ovcharenko et al., 2005), and these regions are enriched in regulatory elements, such as highly conserved noncoding elements (HCNEs), which frequently correspond to transcriptional enhancers (Ovcharenko et al., 2005). The extent to which lincRNAs found in gene deserts near developmental TFs are functional or fundamentally different from other lincRNAs is unclear. lincRNAs might preferentially fall in these regions because (1) these lincRNAs regulate gene expression in cis, as observed for HOTTIP (Wang et al., 2011) and Mistral (Bertani et al., 2011); (2) the colocalized lincRNA and TF genes might act in concert and thus benefit from coregulation, as observed for Six3 and Six3os (Rapicavoli et al. 2011); or (3) the multiplicity of enhancer elements around TFs might provide an accommodating environment for the emergence of new lincRNA genes. In offering the third possibility, we are not suggesting that a significant number of lincRNAs can be attributed to the transcription observed within many enhancer elements (De Santa et al., 2010; Kim et al., 2010); these enhancer transcripts are not typically polyadenylated, and lincRNA genes overlap enhancers no more frequently than do protein-coding genes (Cabili et al., 2011).
Secondary Structure
Secondary structure is important for most noncoding RNA classes, including some long noncoding RNA (Kino et al., 2010; Maenner et al., 2010; Novikova et al., 2012; Wilusz et al., 2012), but the prevalence of secondary structure-mediated roles in lincRNA biology remains unknown. Indeed, when the whole transcript is considered, lincRNAs are not predicted to be more structured than mRNAs. The fraction of paired nucleotides in the predicted optimal folds of the human and mouse lincRNA transcripts resembles that of mRNAs (Managadze et al., 2011). The amount of predicted secondary structure correlates positively with lincRNA expression levels, perhaps because more structured lincRNAs are more stable, or because both structure and expression correlate with G/C content (Kudla et al., 2006). In any case, no correlation is observed between the amount of predicted secondary structure and evolutionary conservation (Managadze et al., 2011).
If many lincRNAs contained short, highly structured regions critical for function, then these lincRNAs would have regions with evolutionary conserved secondary structures. Given alignable sequences, several computational tools (reviewed in Gorodkin et al., 2010) can detect such regions. Surprisingly, depending on the lincRNA set studied, such predicted structures are either depleted or only mildly enriched in lincRNA exons (Marques and Ponting, 2009; I.U. and D.P.B., unpublished data). As discussed below, it is unlikely that many additional conserved structures have been missed due to an inability to align their corresponding primary sequences. Conserved secondary structures thus seem to occupy only a small fraction of the vertebrate lincRNA transcriptome. Similar observations were made in C. elegans, where the overlap between a set of noncoding RNA candidates generated using predicted-structure-based criteria and a set of transcript models generated using RNA-seq data was even smaller than that expected by chance (Nam and Bartel, 2012).
These results should not be interpreted to indicate that lincRNAs are devoid of secondary structure. Even randomly generated RNA sequences have compact folds with secondary structure (Schultes et al., 2005), and there is no reason to suspect that lincRNAs would differ. Thus, the presence of a computationally predicted or an experimentally supported structured region in a lincRNA is not informative for judging whether the structure is functionally important. The emerging picture is that for most regions of most lincRNAs, the collapse characteristic of arbitrary RNA sequences is sufficient for lincRNA function, with specific, evolutionarily conserved structural elements occupying only a very small fraction of the lincRNA real estate. Known examples of such elements include the proposed PRC2-binding elements in Xist and the triple-helical elements that can impart lincRNA stability (Maenner et al., 2010; Brown et al., 2012; Wilusz et al., 2012). With additional study and improved tools, additional examples presumably will be found.
Expression Levels
Compared to mRNA expression, lincRNA expression is typically more variable between tissues (Cabili et al., 2011; Derrien et al., 2012; Pauli et al., 2012), with many lincRNAs preferentially expressed in brain and testis (Ravasi et al., 2006; Cabili et al., 2011; Derrien et al., 2012). Expression similarity between a lincRNA gene and its closest protein-coding neighbor is generally not greater than that between two adjacent protein-coding genes (Cabili et al., 2011; Ulitsky et al., 2011; Pauli et al., 2012).
The median lincRNA level is only about a tenth that of the median mRNA level (Ravasi et al., 2006; Guttman et al., 2009; Guttman et al., 2010; Cabili et al., 2011; Ulitsky et al., 2011; Derrien et al., 2012; Pauli et al., 2012; Sigova et al., 2013). The extent to which the lower level is caused by less efficient transcription or more efficient degradation of lincRNAs remains unknown. Two studies, one using a transcription inhibitor and the other using pulse-chase analysis, both concluded that mRNAs and long noncoding RNAs (including lincRNAs) have similar half-life distributions (Clark et al., 2012; Tani et al., 2012). Thus, at least the lincRNAs that accumulate to sufficient levels for quantification in such studies are not preferentially destabilized by pathways that degrade aberrant mRNA molecules. When comparing different lincRNAs, the characteristics associated with increased stability include those associated with increased mRNA stability, such as splicing, cytoplasmic localization and G/C-rich nucleotide composition (Clark et al., 2012).
Subcellular Localization
Perhaps the most common misperception of lincRNAs is that they are predominantly localized in the nucleus. Some of the best-studied lincRNAs, such as Xist, Malat1, Neat1, and Miat, are almost exclusively in the nucleus (Brown et al., 1992; Hutchinson et al., 2007; Sone et al., 2007) and even define specific nuclear domains (Hutchinson et al., 2007; Sone et al., 2007; Clemson et al., 2009). However, other studied lincRNAs are found mostly in the cytoplasm (Coccia et al., 1992; Kino et al., 2010; Yoon et al., 2012). When RNA is sequenced from nuclear and cytoplasmic fractions, lincRNAs have a ~2-fold enrichment in the nuclear fraction relative to mRNAs in five of the six human cell types examined (Derrien et al., 2012). In the remaining cell type, NHEK cells, the lincRNA distribution is no different than that of mRNAs. Similarly, we observe a 3-fold relative enrichment in the nucleus using data from MCF-7 cells (Figure 2E). However, because polyadenylated RNA species in the cell (dominated by cytoplasmic mRNAs) are not equally distributed between nucleus and cytoplasm, these relative enrichments do not accurately represent absolute enrichments. Therefore, although many lincRNAs are exclusively or predominantly nuclear (Figure 2E), the observed ~3-fold nuclear enrichments of lincRNAs relative to mRNAs refute the notion that as a group, currently annotated lincRNAs are predominantly localized in the nucleus. Consider, for example, cells in which the typical mRNA is six times more abundant in the cytoplasm than in the nucleus. With 3-fold relative nuclear enrichment, the typical lincRNA would still be two times more abundant in the cytoplasm than in the nucleus. Bearing in mind that some lincRNAs might act in the nucleus before making their way to the cytoplasm, the current picture is that most lincRNAs spend most of their time in the cytoplasm. The more specific localization of lincRNAs within either the cytoplasm or nucleus, as well as the factors and sequence elements that dictate this localization, remain largely unexplored.
lincRNA Evolution
Our understanding of other noncoding RNAs has been greatly advanced by studying conservation patterns within their genes and between the noncoding RNAs and their interaction partners (Woese et al., 1980; Michel and Westhof, 1990; Bartel, 2009). Likewise, analyzing the natural selection pressures acting on noncoding RNAs can identify elements and structures important for function. This analysis can also suggest which lincRNAs are functional, provide important clues to their modes of action and identify relevant model organisms for studying the biology of human lincRNAs.
Rapid Evolutionary Turnover of lincRNA Sequences
In stark contrast to mRNAs and many classes of noncoding RNAs, mammalian lincRNAs lack known orthologs in species outside of vertebrates. One possible exception is the Telomeric repeat-containing RNA (Terra), which is conserved between human and yeast but is a nonconventional lincRNA in that only a small fraction of its transcripts is polyadenylated (reviewed in Feuerhahn et al., 2010).
Compared to protein-coding sequences, most of which are highly conserved throughout vertebrates, lincRNA sequences evolve very rapidly. Less than 6% of zebrafish lincRNAs have detectable sequence conservation with human or mouse lincRNAs (Ulitsky et al., 2011), and only ~12% of human and mouse lincRNAs appear to be conserved in the other species (Church et al., 2009; Cabili et al., 2011). Within rodents, only ~60% of the lincRNAs (compared to >90% of mRNAs) expressed in Mus musculus liver have alignable counterparts expressed in the livers of Mus castaneus and rat (Kutter et al., 2012), which shared common ancestors with M. musculus only ~1 and ~15 million years ago, respectively. Interestingly, the presence of a lineage-specific lincRNA gene correlates with higher expression of adjacent protein-coding genes in that lineage (Kutter et al., 2012).
Despite their rapid evolution, lincRNA sequences display detectable, albeit weak, signatures of natural selection. Members of an initial lincRNA catalog in mouse (Okazaki et al., 2002) were poorly conserved when evaluated using mouse-rat and mouse-human genome alignments (Wang et al., 2004). More recently, improved identification and filtering of lincRNA candidates and improved methods for estimating conservation have led to evidence that lincRNA exons are more conserved than intergenic regions but significantly less than either coding or noncoding portions of mRNA exons (Ponjavic et al., 2007; Guttman et al., 2009; Khalil et al., 2009; Marques and Ponting, 2009; Ulitsky et al., 2011; Derrien et al., 2012). Interestingly, fly lincRNAs (which are much shorter than mammalian lincRNAs) appear better conserved at the sequence level, evolving faster than ORFs but slower than 3′UTRs and intergenic regions (Young et al., 2012) (I.U., unpublished data).
Is lincRNA Sequence Conservation Currently Overestimated or Underestimated?
Even the modest magnitude of the sequence conservation reported within lincRNA exons might be overestimated. Conservation scores and substitution rates used to evaluate lincRNA sequence conservation are derived from whole-genome alignments, which compare genome rather than lincRNA sequences. For example, the presence of a segment homologous to a human lincRNA exon in the chicken genome does not necessary imply that the homologous segment is part of a chicken lincRNA. In chicken, this segment might be transcribed as part of an mRNA or might not be transcribed at all. Indeed, when exons of human or mouse lincRNAs are traced to the zebrafish genome through whole-genome alignments, the corresponding regions rarely overlap zebrafish lincRNAs, and in about a third of the cases they overlap zebrafish mRNAs (Ulitsky et al., 2011). In another example, although both potentially functional regions in the human Hotair lincRNA appear to be conserved in the mouse genome (He et al., 2011) only the 3′ region appears to be part of the murine Hotair homolog (Schorderet and Duboule, 2011). Possible explanations for mapping to non-lincRNA annotations include annotation errors, interconversion between coding and noncoding transcripts during evolution (discussed below), or selective pressures on DNA elements, such as transcriptional enhancers, that overlap lincRNA genes. To the extent that any of these explanations are relevant, even the modest sequence conservation reported in lincRNA exons might overestimate the selective pressures acting to preserve lincRNA function. Obtaining more informative conservation estimates will require more comprehensive lincRNA catalogs in multiple vertebrate species so that lincRNAs can be compared to lincRNAs rather than to genomic alignments.
Why are lincRNA sequences so poorly conserved? Perhaps the fraction of lincRNAs that are nonfunctional is large, and thus changes in most lincRNA sequences exact no fitness cost. Alternatively, existing approaches for comparing genomic sequences, which rely heavily on stretches of high sequence conservation, might be poorly suited for detecting homology between lincRNAs. One idea is that lincRNAs might be under pressure to conserve structure but not sequence, and thus homologs would be missed with methods that focus on primary-sequence homology. However, pressures to conserve secondary structure also substantially slow down changes in the corresponding primary sequence, such that the evolutionary time needed to erase primary-sequence similarity within a conserved secondary structure is probably far too long to have occurred within the mammalian clade. Nonetheless, as illustrated below, detailed comparative analyses of specific lincRNAs supports the notion that lincRNA conservation has been systematically underestimated for other reasons.
Because finding optimal alignments between long sequences is time and resource consuming, the BLAST heuristic is typically used to identify sequence homologs or generate whole-genome alignments. BLAST accelerates search of similar sequences by identifying short regions of high sequence conservation and then refining the sequence alignments around these regions (Altschul et al., 1997). This approach is very powerful in many cases, and for the past 15 years BLAST has served as a major bioinformatics workhorse. However, BLAST as well as more sensitive tools often fail to identify sequence conservation in cases for which synteny and other genomic evidence strongly indicate that the corresponding lincRNAs are orthologous. Some improvements to BLAST designed to detect homology among RNA genes have been proposed (Bussotti et al., 2011), but more substantial increases in sensitivity await better understanding of the nature of selective pressures acting on lincRNA loci. Described below are case studies for six lincRNAs (Xist, Cyrano, Megamind, Miat, Malat1, and PAN), which illustrate the challenges of using existing methods for examining lincRNA evolution.
X-inactive specific transcript (Xist) is a master regulator of X chromosome inactivation in eutherian mammals (Brockdorff et al., 1992; Brown et al., 1992; Penny et al., 1996). Although poorly conserved throughout most of its sequence, Xist is conserved in its exon-intron structure, with a consensus of ten exons (Nesterova et al., 2001; Elisaphenko et al., 2008). Xist and at least three additional lincRNAs in the X-inactivation center descended from protein-coding genes still present in other amniotes (Duret et al., 2006). Although regions of sequence similarity are observed between at least four mammalian Xist exons and six chicken Lnx3 mRNA exons (Elisaphenko et al., 2008), none of these are evident in current whole-genome alignments. Xist sequences in contemporary species contain multiple ancient and conserved repeats alongside young and species-specific repeats originating from mobile elements, as the repetitive fraction of Xist increased from about 4.4% in the eutherian ancestor to as much as 12.4% in the human (Elisaphenko et al., 2008). Interestingly, the first exon of Xist, which contains most of the known functional repetitive elements (Beletskii et al., 2001; Wutz et al., 2002; Sarma et al., 2010), is characterized by low PhastCons scores, perhaps because some of these repeats contain short functional sequences interspersed among poorly conserved spacers (Wutz et al., 2002). In contrast, although the most obvious sequence conservation resides in exon 4, deleting this exon does not affect X inactivation (Caparros et al., 2002). Xist thus illustrates significant challenges for comparative analysis; due to its size and sequence divergence among mammals, and despite its functional importance, Xist appears quite poorly conserved when inspected through the lens of whole-genome alignments.
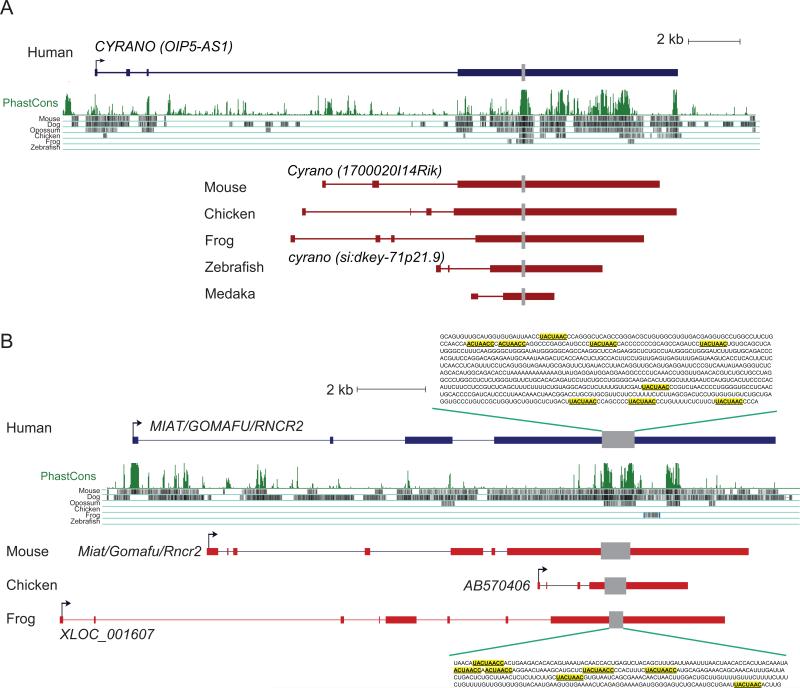
The Cyrano lincRNA is conserved throughout vertebrates (with the potential exception of lizards) and is required for proper morphogenesis and neurogenesis in zebrafish (Ulitsky et al., 2011). Within the most conserved region of Cyrano is a 26 nt site that pairs to the miR-7 miRNA and is perfectly conserved in at least 55 vertebrates from human to lamprey (Ulitsky et al., 2011). In addition to this conserved site, Cyrano orthologs share similar exon-intron architectures (Figure 3A) and multiple shorter (<10 nt) highly conserved sites (I.U. and D.P.B., unpublished data). Although the human ortholog can rescue the Cyrano knockdown in zebrafish, the human and fish genes do not align with each other in whole-genome alignments (Figure 3A). This alignment failure occurs because the signal for sequence similarity does not exceed detection thresholds when considered in the context of full-genome pairwise comparisons, even though BLASTN detects a conserved 67 nt segment when the human and zebrafish Cyrano genes are directly compared.
Figure 3. Evolution of Cyrano and Miat lincRNAs.
(A) Cyrano. Gene models from the indicated species are shown, together with the PhastCons track. The gray bar indicates a ~70 nt region of homology detected in a focused search, starting with the zebrafish ortholog.
(B) Miat. Gene models from the indicated species are shown, together with the PhastCons track. The gray box indicates a region in the last exon that contains multiple copies of the (U)ACUAAC(C) motif, as shown for human and frog.
Megamind is also conserved throughout vertebrates and required for proper brain development in zebrafish (Ulitsky et al., 2011). Unlike Cyrano, Megamind lacks stretches of consecutive highly conserved bases but instead contains 40 positions with at least 90% identity in over 50 vertebrates, which appear at phased positions within a 95 nt region. Even with the most permissive parameter settings, BLASTN fails to identify Megamind homologs in EST collections from some fish. These homologs are nonetheless identified with high statistical significance using a hidden Markov model trained using the Megamind conserved regions (Ulitsky et al., 2011). The reliance of BLAST on contiguous stretches of high conservation is thus a substantial limitation when comparing sequences in which highly conserved positions are intermingled with rapidly evolving ones.
Miat (also called Gomafu or Rncr2) was originally discovered as a lincRNA highly enriched in specific neurons in mouse retina (Blackshaw et al., 2004; Sone et al., 2007) and later found to be more widely expressed in the nervous system and cultured neurons, where it specifies cell identify (Sone et al., 2007; Rapicavoli et al., 2010). Miat sequence variants are also associated with increased risk of myocardial infarction (Ishii et al., 2006). Miat is retained in the nucleus in mammalian and avian cells, and defines a subnuclear domain that does not overlap with other nuclear bodies (Sone et al., 2007; Tsuiji et al., 2011). Although Miat appears to be restricted to mammals in whole-genome alignments based on the human and mouse genomes, orthologs are present in syntenic positions of chicken and frog (Figure 3B) (Rapicavoli et al., 2010; Tsuiji et al., 2011). These homologs all contain a relatively short region with multiple copies of the (U)ACUAAC(C) motif, which resembles the intron branch point and can bind to Splicing factor 1 (Sf1) (Rapicavoli et al., 2010; Tsuiji et al., 2011). This region maps to the last exon within Miat orthologs but is nested in rapidly evolving sequence, and apart from the motif repeats, sequence similarity within the region is sparse (Figure 3B). Indeed, BLASTN finds no significant similarity between human and frog Miat and only a short (<30 bp) region of similarity between human and chicken sequences.
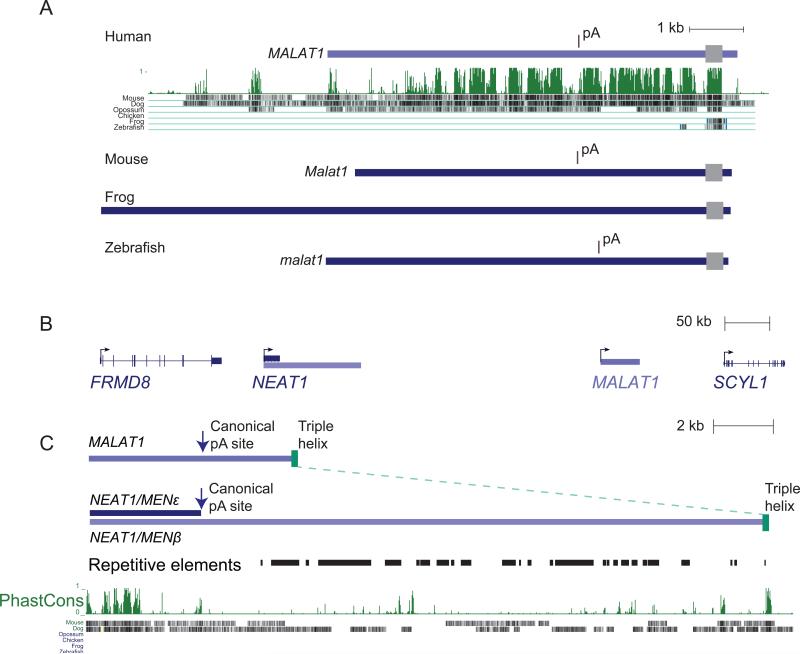
Malat1 is an exceptionally highly expressed, nuclear-retained, single-exon lincRNA that was originally identified in metastatic tumors (Ji et al., 2003). Although Malat1 helps organize nuclear speckle domains, which contain splicing factors (Tripathi et al., 2010), it is not essential for life and development in mouse (Eißmann et al., 2012; Nakagawa et al., 2012; Zhang et al., 2012). The most abundant Malat1 isoform is not polyadenylated, and its 3′ end instead forms a triple-helical RNA structure (Brown et al., 2012; Wilusz et al., 2012). This 3′ end is generated by RNase P cleavage of the nascent transcript, which releases the 61 nt Malat1-associated small cytoplasmic RNA (mascRNA). Malat1 was originally considered a mammalian-specific lincRNA (Hutchinson et al., 2007; Tripathi et al., 2010) and only more recently found in other vertebrates (Stadler, 2010; Ulitsky et al., 2011). Although the entire genomic region appears to have been lost in the avian clade, Malat1 orthologs appear in syntenic genomic positions near Scyl1 in mammals, frogs, and fish. The zebrafish malat1 shares striking features with the mammalian Malat1, including similar length of ~7 kb, very high expression levels, no apparent introns, a noncanonical 3′ end, and a canonical yet inefficient polyadenylation site ~4 kb after the transcription start site (Figure 4A). However, apart from its 3′ terminal region, which includes the mascRNA and another short (<70 bases) segment of homology (Figure 4A), the mammalian Malat1 gene has no recognizable sequence similarity with its fish counterpart.
Figure 4. Evolution of the Malat1 and Neat1 lincRNAs.
(A) Malat1 gene models from the indicated species are shown, together with the PhastCons track indicating homology to the human genome detected in the whole-genome alignments. The gray box corresponds to the region of sequence similarity at the 3′ end of Malat1.
(B) The human NEAT1/MALAT1 locus.
(C) Neat1 and its similarities with Malat1. The human gene models are shown, together with annotated repeats and the PhastCons track for Neat1.
Several features of the PAN lincRNA from Kaposi’s sarcoma-associated herpesvirus (KSHV) resemble those of Malat1 (Sun et al., 1996; Tycowski et al., 2012). Like Malat1, PAN is a long, unspliced, very abundantly expressed lincRNA that ends with triple-helical RNA element essential for its accumulation (Conrad et al., 2006; Mitton-Fry et al., 2010). A computational approach that relied on sequence and structure similarity identified homologous elements in six other viral genomes, including two additional gammaherpesviruses (Tycowski et al., 2012). Moreover, the elements in the other gammaherpesviruses occur at ends of lincRNAs that have similar lengths and syntenic positions with PAN but share little to no other detectable sequence similarity with PAN. These presumed homologs could be identified using a tailored bioinformatics approach but not a conventional sequence-homology search.
As the previous examples each illustrate, sequence-homology search tools often fail to detect known lincRNA orthologs. To the extent that orthologs are missed, metrics that depend on whole-genome alignments or other output from these tools will underestimate lincRNA conservation. Countering this underestimate are the false-positive orthologs arising from alignments to nonlincRNA sequences, described at the beginning of this section. Thus, the question as to whether lincRNA sequence conservation is currently overestimated or underestimated remains open, with the answer awaiting improved tools and more comprehensive lincRNA catalogs from more species.
lincRNA Synteny despite Undetectable Sequence Conservation
Some lincRNAs are at conserved genomic locations, with conserved exon-intron structures yet no detectable sequence conservation. For example, protein-coding genes adjacent to a lincRNA gene in zebrafish are more likely to have orthologs adjacent to lincRNA genes in human or mouse, even when all lincRNAs with sequence homology are excluded from the analysis (Ulitsky et al., 2011). Importantly, this enrichment remains significant after controlling for the fact that some genes (particularly those of developmental transcriptional regulators) tend to be far from other protein-coding genes and are therefore more likely to be adjacent to lincRNA genes. Perhaps these lincRNAs have conserved sequence-dependent functions, yet their sequences are too divergent to be detected with existing tools. The examples of conserved lincRNAs with limited sequence conservation listed above suggest that this scenario is relevant for at least some lincRNAs. Alternatively, the act of transcription rather than the identity of the transcribed RNA might be important, in which case, the inability to detect lincRNA sequence conservation would accurately reflect an absence of sequence-based posttranscriptional function.
Evolutionary Trajectories of lincRNA Genes
The low levels of sequence conservation observed in vertebrates point to either rapid sequence evolution or frequent gain and loss of lincRNA genes (Ulitsky et al., 2011). With respect to the gain of new genes, three evolutionary scenarios might be considered. New lincRNA genes might originate from either ancestral protein-coding genes; duplication and divergence of other lincRNA genes; or de novo, from intergenic DNA (Ponting et al., 2009). Although the origins of most mammalian lincRNAs are unknown, examples below illustrate the first two of these three evolutionary possibilities.
As mentioned previously, Xist evolved from a protein-coding gene Lnx3 that is still present in noneutherian vertebrates (Duret et al., 2006). Because pseudogenization is a rather common event, and many pseudogenes are transcribed (Pink et al., 2011; Pei et al., 2012), other lincRNAs might have similar origins. Because analyses of expression and conservation patterns of pseudogenes are complicated by their sequence-similar protein-coding relatives, pseudogenes are typically excluded from lincRNA collections. Nevertheless, the sequences of at least 68 human pseudogenes appear to be under selection in mammals (Khachane and Harrison, 2009), and an increasing number of pseudogenes are reported to have noncoding functions. Some contain inverted repeats or are transcribed in the antisense orientation, triggering RNAi-mediated repression of their protein-coding cousins in the oocyte (Tam et al., 2008; Watanabe et al., 2008). Others are proposed to influence mRNA regulation by binding and depleting trans-acting factors (reviewed in Pink et al., 2011), although this mechanism is often implausible when considering the unfavorable stoichiometry between the pseudogene transcripts and the factors (Ebert and Sharp, 2010). The emergence of new lincRNA genes from protein-coding genes might often occur through neofunctionalization of the pseudogene. In addition, the observation of transcripts possessing both coding and noncoding functions opens the alternative possibility for duplication and subfunctionalization of bifunctional ancestral genes.
New genes can also emerge from the opposite direction, with ancestral noncoding transcripts serving as raw material for the birth of novel protein-coding genes. Candidates for such an event include 24 predicted human protein-coding genes of at least 50 aa that in other primates have homologous genes that do not appear to code for sufficiently homologous proteins (Xie et al., 2012), with similar phenomena observed in other species (Cai et al., 2008; Carvunis et al., 2012). Although detecting most of the older protein-coding gene birthing events will be more difficult, examples might be detected if the coding transcript retained a noncoding function that constrained its sequence. Indeed, a zebrafish lincRNA gene conserved in teleosts and chondrichthyes appears to have acquired a functional protein-coding region in the tetrapod lineage (Ulitsky et al., 2011). The conserved noncoding region of these genes has a conserved predicted secondary structure (I.U. and D.P.B., unpublished data), which further supports the model of a conserved noncoding element residing within an ancient lincRNA that later evolved a short, functional protein-coding region to become a bifunctional mRNA.
Within a species, lincRNA sequences are rarely similar to each other (Ulitsky et al., 2011; Derrien et al., 2012), and with few exceptions (e.g., megamind; Ulitsky et al., 2011) most studied lincRNAs appear in single copies in vertebrate genomes. Thus, lincRNAs rarely originate from duplication of other lincRNAs, or their similarity becomes undetectable rapidly after duplication. Support for the latter explanation is found in one of the few clear examples of lincRNA duplication. In mammalian genomes, Neat1 appears immediately upstream of Malat1, in tandem orientation suggestive of an ancestral gene duplication (Figure 4B) (Stadler, 2010). Neat1 has two isoforms that resemble the two Malat1 isoforms (Figure 4C). These are the 3.7 kb Menε, which ends with a canonical polyadenylation site, and the 22.7 kb Menβ, which shares its 5′ end with Menε and the mechanism of its 3′-end formation and a triple-helical terminal structure with the longer Malat1 isoform (Brown et al., 2012; Wilusz et al., 2012). Malat1 and Neat1 lincRNAs each localize to specific nuclear domains, Malat1 to the nuclear speckles and Neat1 to the paraspeckles (Hutchinson et al., 2007). Despite these many lines of evidence for shared ancestry, comparison of the human Neat1 and Malat1 sequences reveals no homology beyond a short stretch at the very 3′ end, which includes the triple-helical element and downstream structure required for RNase P cleavage. Presumably other duplicated lincRNA genes also underwent similarly rapid divergence following their duplication, thereby obscuring their common origins.
Mechanisms of Action
Little is known about the biological roles of lincRNAs, and even less about how they carry out those roles, but several potential mechanisms for nuclear and cytoplasmic lincRNAs have been suggested based on the few relatively well-studied examples (Figure 5). lincRNAs might act through a broad array of mechanisms, which would be consistent with the wide variety of subcellular localizations, expression levels, and stabilities observed for lincRNAs in mammalian cells.
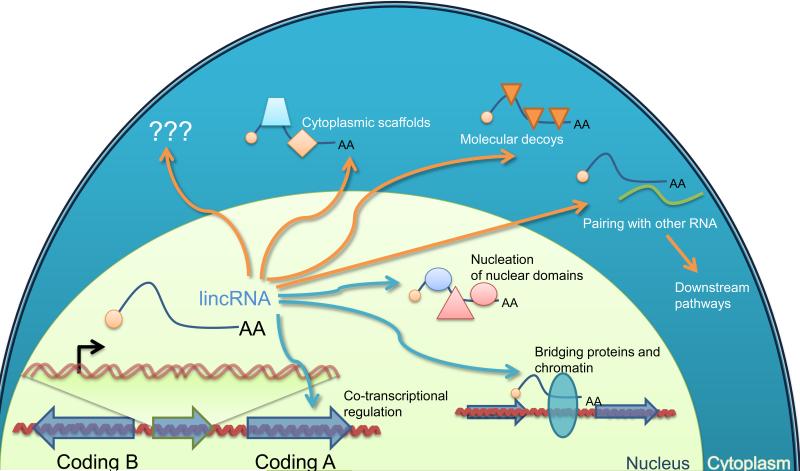
Figure 5. Diverse Mechanisms Proposed for lincRNA Function.
Modes of action include cotranscriptional regulation (e.g., through either the interaction of factors with the nascent lincRNA transcript or the act of transcribing through a regulatory region), regulation of gene expression in cis or in trans through recruitment of proteins or molecular complexes to specific loci, scaffolding of nuclear or cytoplasmic complexes, titration of RNA-binding factors, and pairing with other RNAs to trigger posttranscriptional regulation. The two latter mechanisms are illustrated in the cytoplasm (where they are more frequently reported) but could also occur in the nucleus. Additional mechanisms will presumably be proposed as additional functions of lincRNAs are discovered.
The potential mechanisms of lincRNA function can be divided into three groups: (1) those that rely solely on the act of transcription or on the nascent RNA; (2) those that require the processed RNA yet depend on the site of transcription; and (3) those that are independent of the site of transcription. A major difference between the first two groups and the last one is in whether the direct targets of the lincRNA activity are found only in proximity to the lincRNA gene (cis targets, groups 1 and 2), or anywhere in the cell (trans targets, group 3).
The well-studied examples of cis-acting chromatin-associated lincRNAs include some of the lincRNAs transcribed from and acting at the X-inactivation center (reviewed in Lee, 2009; Augui et al., 2011). Which features of these lincRNAs are unique to X-inactivation biology and which are relevant to other lincRNAs is unclear. Examples of other cis-regulatory lincRNAs include ncRNA-a1-7, Hottip, and Mistral, the perturbation of which leads to decreased expression of some nearby genes (Ørom et al., 2010; Bertani et al., 2011; Wang et al., 2011; Lai et al., 2013).
A single cis-acting molecule might be able to target a neighboring locus, which would explain the relatively low expression levels of many lincRNAs. A prevalence of cis-regulatory lincRNAs would also explain the significant synteny of lincRNA loci from distant vertebrates and their generally limited sequence conservation. A potential mechanism by which cis-acting lincRNAs might function without performing any sequence-specific activities would be for the nascent lincRNA transcripts to flag regions of open, transcriptionally competent chromatin through the recruitment of promiscuous RNA-binding proteins.
Despite known cis-acting examples and the above-mentioned arguments favoring the prevalence of cis-acting function, other observations challenge the notion that most lincRNAs act in cis-regulatory circuits. lincRNA knockdown in mouse embryonic stem cells rarely changes the expression of neighboring genes, with mRNA levels of one of the 20 closest neighbors of the lincRNA affected in <10% of the cases examined (Guttman et al., 2011). Moreover, only about 3% of the human lincRNAs have expression profiles strongly correlated with those of their neighbors (compared with 1.5% for mRNAs), and strong negative correlations are exceedingly rare (Derrien et al., 2012), arguing against widespread effects of lincRNA expression on neighboring regulatory programs. Further evidence favoring trans functions is the observation that most lincRNA are predominantly cytoplasmic (Figure 2E), which suggests that many might function in the cytosol and thus would not be cis-acting. More information on the relative prevalence of cis and trans mechanisms will come from genome-wide approaches to study lincRNA chromatin occupancy as well as focused studies of additional lincRNAs.
Interactions between lincRNAs and Other Cellular Factors
As expected, increasing evidence suggests that many lincRNAs function through specific interactions with other cellular factors, namely proteins, DNA, and other RNA molecules. Much effort is being devoted to finding these interacting partners as a strategy for gaining insight into molecular mechanism.
A popular view is that many lincRNAs regulate gene expression by directing chromatin-modification complexes to specific target regions (Rinn and Chang, 2012). This view is based on observations from some well-studied lincRNAs, such as Xist (Penny et al., 1996), Hotair (Tsai et al., 2010), Hottip (Wang et al., 2011), and Mistral (Bertani et al., 2011), and the mechanistic understanding of long RNAs that overlap the protein-coding regions of their targets (and hence are not classified as lincRNAs), such as Air (Sleutels et al., 2002), Kncq1ot1 (Pandey et al., 2008), and Anril (Yap et al., 2010). Accordingly, most studies of lincRNA-associated proteins have focused on chromatin factors. For example, lincRNAs are reported to associate with CTCF (Yao et al., 2010), YY1 (Jeon and Lee, 2011), Mediator (Lai et al., 2013), WDR5 (Wang et al., 2011; Gomez et al., 2013; Grote et al., 2013), LSD1 (Tsai et al., 2010), and the polycomb complexes PRC1 (Schoeftner et al., 2006) and PRC2 (Rinn et al., 2007; Zhao et al., 2008; Tsai et al., 2010; Grote et al., 2013; Klattenhoff et al., 2013), although the extent to which some of these interactions are direct and specific remains controversial (Brockdorff, 2013). Conversely, searches for transcripts associated with PRC2 detect significant fractions (~20% in human and ~10% in mouse) of annotated lincRNAs (Khalil et al., 2009; Zhao et al., 2010; Guttman et al., 2011). The functional outcomes of these binding events are unclear, as lincRNAs account for a relatively small fraction of the PRC2-RNA interactome, and lincRNAs reported to be associated with PRC2 in human and mouse have no overlap (Zhao et al., 2010). Another large-scale study found that as many as 30% of lincRNAs expressed in mouse embryonic stem cells are associated with at least one of 11 chromatin regulators (Guttman et al., 2011), although some of these interactions may be indirect and mediated by protein-protein interactions (Brockdorff, 2013). The nature of the lincRNA-protein recognition, and whether it relies primarily on RNA primary sequence or on structural features, remains largely unknown, as regions mediating lincRNA-protein interactions have been identified in only a few cases, and these regions are currently too large to suggest how binding specificity is achieved (Huarte et al., 2010; Murthy and Rangarajan, 2010).
Part of the appeal of lincRNAs acting to direct chromatin-modifying complexes to DNA is that it would help solve the mystery of how protein complexes without intrinsic sequence-specific DNA-binding ability, such as the polycomb complex, find their DNA targets. However, this model pushes to the fore the questions of how these proteins recognize RNA, how the low abundance of most lincRNAs can be reconciled with roles in recruiting protein complexes to hundreds or thousands of genomic loci, and how lincRNAs might recognize DNA targets.
lincRNAs might recognize specific regions in genome through direct interactions with the DNA. One way to do this would be to act as a nascent transcript, while still tethered to the DNA by the RNA polymerase, as occurs for transcripts targeted by the endogenous small interfering RNAs (siRNAs) that direct chromatin silencing in fission yeast (Moazed, 2009). In theory, lincRNAs might also directly recognize DNA by other mechanisms, either through triplex interactions with the Hoogstein face of purine stacks within the DNA duplex (Frank-Kamenetskii and Mirkin, 1995) or through base-pairing interactions with single strands within an unwound region of the DNA. Such interactions might be facilitated by proteins that could either help stabilize the base triples or help melt the DNA to enable RNA pairing. Alternatively, lincRNAs might recognize specific genomic regions through indirect interactions, either base pairing with nascent transcripts or interacting with DNA-binding proteins or complexes. Identification of principles that guide lincRNAs to specific chromatin regions will benefit from methods for high-throughput identification of target regions akin to the recent genome-wide isolation and sequencing of DNA associated with an RNA of interest (Chu et al., 2011; Simon et al., 2011).
Many lincRNAs presumably have functions unrelated to chromatin modification. An appealing way for these lincRNAs to form interactions is through base pairing with other RNA molecules, as this is the way that members of other classes of noncoding RNAs (e.g., tRNAs, snRNAs, snoRNAs, and microRNAs) interact with their targets and partners. For example, antisense Uchl1 regulates Uchl1 translation by pairing to a segment of its 5′UTR (transcribed from an overlapping genomic region) (Carrieri et al., 2012), and the TINCR lincRNA is reported to pair with and stabilize mRNAs containing a 25 nt motif (Kretz et al., 2013). Formation of double-stranded RNA by a lincRNA and its target might also activate downstream pathways. For example, a group of Alu repeat-containing RNAs are reported to repress targets with sequence-similar complementary Alu elements in their 3′UTRs via the Staufen 1 (STAU1)-mediated mRNA decay pathway (Gong and Maquat, 2011). Another proposed function of mammalian lincRNAs is to pair to microRNAs and titrate them away from their mRNA targets, as can be done using artificial “sponge” RNAs (Ebert et al., 2007) and as observed for select plant and viral RNAs (Franco-Zorrilla et al., 2007; Cazalla et al., 2010). In mammals, however, nearly all of the proposed “competing endogenous RNAs” fail to reach levels sufficiently high to achieve consequential miRNA titration. The most notable exception is CDR1as/ciRS-7, a highly expressed circular RNA with more than 70 conserved miR-7 target sites (Hansen et al., 2013; Memczak et al., 2013). The paucity of other highly expressed noncoding RNAs with many target sites argues against the widespread function of lincRNAs as microRNA sponges. Nonetheless, Cyrano illustrates that lincRNA function can require microRNA pairing, presumably for purposes other than titration (Ulitsky et al., 2011).
A compelling idea is that many lincRNAs might make use of interactions with protein, DNA, and other RNAs to act as scaffolds to bring together different proteins or bridging protein complexes and specific chromatin regions (Guttman and Rinn, 2012). For example, Neat1/Menβ and Malat1 bind multiple proteins localizing to the paraspeckles and nuclear speckles, respectively, and Menβ is essential for paraspeckle formation (Clemson et al., 2009; Sunwoo et al., 2009; Murthy and Rangarajan, 2010; Souquere et al., 2010; Tripathi et al., 2010). With the recognition that most lincRNAs are mostly cytoplasmic, we suggest that this scaffolding mechanism might also play important roles in the cytosol. The binding of a lincRNA to a protein might also regulate the protein activity. For example, lincRNA binding was shown to affect the action of some transcription regulators, including Tsl (Wang et al., 2008) and Nfat (Willingham et al., 2005). One possible mechanism is for the lincRNA to act as a decoy that titrates the protein away from its potential targets, as has been reported for lincRNA Gas5 and glucocorticoid receptor (Kino et al., 2010), PANDA and NF-Y (Hung et al., 2011), sno-lncRNAs and Fox2 (Yin et al., 2012), and Gadd7 and TDP-43 (Liu et al., 2012b). However, when considering that most proteins accumulate to many more molecules per cell than do their corresponding mRNAs and that the typical mRNA is still expressed at higher levels than the typical lincRNA, the titration mechanism seems possible for only a small subset of lincRNAs.
Concluding Remarks
lincRNA research is at a very interesting juncture—thousands of lincRNA genes have been identified, and the diverse functional and mechanistic underpinnings of a few well-studied examples suggest that many of these (hundreds, if not more) might participate in important and diverse aspects of biology. Recent observations regarding lincRNA genomics and evolution, such as their frequently cytoplasmic accumulation or their frequently syntenic loci despite undetectable sequence conservation, only add to the mysteries of lincRNA function and mechanism. With all this intrigue, biologists with diverse interests and backgrounds are exploring how lincRNAs might participate in the biological processes that they study. To do so, some are also expanding the experimental toolbox for interrogating lincRNA function and mechanism by developing improved tools for comparative genomics and for high-throughput identification of binding partners. The insights on the horizon will help separate this rag-tag set of transcripts into coherent, well-defined subclasses, thereby enabling the information gained from the study of one lincRNA to be more reliably leveraged for the understanding of many others, and ultimately providing a firm grasp on how many of the thousands of lincRNA genes found in the cell are functional.
ACKNOWLEDGMENTS
We thank A. Shkumatava, M. Cabili, B. Kleaveland, L. Boyer, and other colleagues for stimulating discussions and for helpful comments on this manuscript. Our work on lincRNAs is supported by NIH grant GM067031. D.B. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- Agarwal A, Koppstein D, Rozowsky J, Sboner A, Habegger L, Hillier LW, Sasidharan R, Reinke V, Waterston RH, Gerstein M. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics. 2010;11:383. doi: 10.1186/1471-2164-11-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ, 3rd, Lee JT. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011;7:e1002248. doi: 10.1371/journal.pgen.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- Badger JH, Olsen GJ. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beletskii A, Hong YK, Pehrson J, Egholm M, Strauss WM. PNA interference mapping demonstrates functional domains in the noncoding RNA Xist. Proc. Natl. Acad. Sci. USA. 2001;98:9215–9220. doi: 10.1073/pnas.161173098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol. Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. First exon length controls active chromatin signatures and transcription. Cell Rep. 2012;2:62–68. doi: 10.1016/j.celrep.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner S, McGinnis KM. Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS ONE. 2012;7:e43047. doi: 10.1371/journal.pone.0043047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent KM, Park D, Wolf AR, Van Tyne D, Sims JS, Ribacke U, Volkman S, Duraisingh M, Wirth D, Sabeti PC, Rinn JL. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol. 2011;12:R56. doi: 10.1186/gb-2011-12-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. USA. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussotti G, Raineri E, Erb I, Zytnicki M, Wilm A, Beaudoing E, Bucher P, Notredame C. BlastR—fast and accurate database searches for non-coding RNAs. Nucleic Acids Res. 2011;39:6886–6895. doi: 10.1093/nar/gkr335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhao R, Jiang H, Wang W. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics. 2008;179:487–496. doi: 10.1534/genetics.107.084491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparros ML, Alexiou M, Webster Z, Brockdorff N. Functional analysis of the highly conserved exon IV of XIST RNA. Cytogenet. Genome Res. 2002;99:99–105. doi: 10.1159/000071580. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. Proto-genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA. Down-regulation of a host micro-RNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, et al. Mouse Genome Sequencing Consortium Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia EM, Cicala C, Charlesworth A, Ciccarelli C, Rossi GB, Philipson L, Sorrentino V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol. Cell. Biol. 1992;12:3514–3521. doi: 10.1128/mcb.12.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol. Cell. 2006;24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Crowe ML, Wang XQ, Rothnagel JA. Evidence for conservation and selection of upstream open reading frames suggests probable encoding of bioactive peptides. BMC Genomics. 2006;7:16. doi: 10.1186/1471-2164-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr. Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eißmann M, Gutschner T, Hämmerle M, Günther S, Caudron-Herger M, Groß M, Schirmacher P, Rippe K, Braun T, Zörnig M, Diederichs S. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisaphenko EA, Kolesnikov NN, Shevchenko AI, Rogozin IB, Nester-ova TB, Brockdorff N, Zakian SM. A dual origin of the Xist gene from a protein-coding gene and a set of transposable elements. PLoS ONE. 2008;3:e2521. doi: 10.1371/journal.pone.0002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010;584:3812–3818. doi: 10.1016/j.febslet.2010.07.032. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu. Rev. Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the inter-feron-g locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodkin J, Hofacker IL, Torarinsson E, Yao Z, Havgaard JH, Ruzzo WL. De novo prediction of structured RNAs from genomic sequences. Trends Biotechnol. 2010;28:9–19. doi: 10.1016/j.tibtech.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett PB, Petersen RB, Hensel CH, Albericio F, Gunderson SI, Palmenberg AC, Barany G. Synthesis in vitro of a seven amino acid peptide encoded in the leader RNA of Rous sarcoma virus. J. Mol. Biol. 1986;190:45–57. doi: 10.1016/0022-2836(86)90074-4. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol. Biol. 2011;11:102. doi: 10.1186/1471-2148-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large inter-genic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Rinn JL. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachane AN, Harrison PM. Assessing the genomic evidence for conserved transcribed pseudogenes under selection. BMC Genomics. 2009;10:435. doi: 10.1186/1471-2164-10-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, Sasaki D, Imamura K, Kai C, Harbers M, et al. CAGE: cap analysis of gene expression. Nat. Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–9. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Higgs DR, Gingeras TR. Molecular biology: RNA discrimination. Nature. 2012;482:310–311. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4:e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, Odom DT, Marques AC. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O’Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, Wan KH, Schroeder AJ, Gramates LS, St Pierre SE, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17:1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gough J, Rost B. Distinguishing protein-coding from non-coding RNAs through support vector machines. PLoS Genet. 2006;2:e29. doi: 10.1371/journal.pgen.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012a;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012b;31:4415–4427. doi: 10.1038/emboj.2012.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianférani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Managadze D, Rogozin IB, Chernikova D, Shabalina SA, Koonin EV. Negative correlation between expression level and evolutionary rate of long intergenic noncoding RNAs. Genome Biol. Evol. 2011;3:1390–1404. doi: 10.1093/gbe/evr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Tan J, Lee S, Kong L, Heger A, Ponting CP. Evidence for conserved post-transcriptional roles of unitary pseudogenes and for frequent bifunctionality of mRNAs. Genome Biol. 2012;13:R102. doi: 10.1186/gb-2012-13-11-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Menschaert G, Van Criekinge W, Notelaers T, Koch A, Crappe J, Gevaert K, Van Damme P. Deep proteome coverage based on ribosome profiling aids MS-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics. 2013 doi: 10.1074/mcp.M113.027540. Published online February 21, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy UM, Rangarajan PN. Identification of protein interaction regions of VINC/NEAT1/Men ε RNA. FEBS Lett. 2010;584:1531–1535. doi: 10.1016/j.febslet.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JW, Bartel DP. Long noncoding RNAs in C. elegans. Genome Res. 2012;22:2529–2540. doi: 10.1101/gr.140475.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, Zakian SM. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001;11:833–849. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group Phase I & II Team Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 anti-sense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, Schier AF. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu XJ, Harte R, Balasubramanian S, Tanzer A, Diekhans M, et al. The GENCODE pseudogene resource. Genome Biol. 2012;13:R51. doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17:792–798. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40(Database issue):D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc. Natl. Acad. Sci. USA. 2010;107:22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes EA, Spasic A, Mohanty U, Bartel DP. Compact and ordered collapse of randomly generated RNA sequences. Nat. Struct. Mol. Biol. 2005;12:1130–1136. doi: 10.1038/nsmb1014. [DOI] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference—a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell. 2010;21:4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler PF. Evolution of the Long Non-coding RNAs MALAT1 and MEN. In: Ferreira CE, Miyano S, Stadler PF, editors. Advances in Bioinformatics and Computational Biology. Springer; Rio de Janeiro, Brazil: 2010. [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013;23:201–216. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigó R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells. 2011;16:479–490. doi: 10.1111/j.1365-2443.2011.01502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE, Rubin GM. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2005;102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012;2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, Bartel DP. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong GK. Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431 1 p following 757; discussion following 757. [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Findeiss S, Müller SA, Kalkhof S, von Bergen M, Hofacker IL, Stadler PF, Goldman N. RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA. 2011;17:578–594. doi: 10.1261/rna.2536111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wethmar K, Smink JJ, Leutz A. Upstream open reading frames: molecular switches in (patho)physiology. Bioessays. 2010;32:885–893. doi: 10.1002/bies.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, Noller HF. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, Zhang M, Zhang R, Wei L, Li CY. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 2012;8:e1002942. doi: 10.1371/journal.pgen.1002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. Mol. Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL, Ponting CP. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol. Evol. 2012;4:427–442. doi: 10.1093/gbe/evs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Zhang W, Yang S, Chen JQ, Tian D. Patterns of exon-intron architecture variation of genes in eukaryotic genomes. BMC Genomics. 2009;10:47. doi: 10.1186/1471-2164-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. J. Mol. Neurosci. 2013;49:589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]