Abstract
Significance: Three signaling systems, chemical, electrical, and mechanical, ubiquitously contribute to cellular activities. There is limited information on the mechanical signaling system because of a lack of tools to measure stress in specific proteins. Although significant advances in methodologies such as atomic force microscopy and laser tweezers have achieved great success in single molecules and measuring the mean properties of cells and tissues, they cannot deal with specific proteins in live cells. Recent Advances: To remedy the situation, we developed a family of genetically encoded optical force sensors to measure the stress in structural proteins in living cells. The sensors can be incorporated into specific proteins and are not harmful in transgenic animals. The chimeric proteins distribute and function as their wild-type counterparts, and local stress can be read out from changes in Förster resonance energy transfer (FRET). Critical Issues: Our original sensor used two mutant green fluorescence proteins linked by an alpha helix that served as a linking spring. Ever since, we have improved the probe design in a number of ways. For example, we replaced the helical linker with more common elastic protein domains to better match the compliance of the wild-type hosts. We greatly improved sensitivity by using the angular dependence of FRET rather than the distance dependence as the transduction mechanism, because that has nearly 100% efficiency at rest and nearly zero when stretched. Future Directions: These probes enable researchers to investigate the roles of mechanical force in cellular activities at the level of single molecules, cells, tissues, and whole animals. Antioxid. Redox Signal. 20, 986–999.
Introduction
Three to four billion years ago when life first began as single cells, they were exposed to environments containing a vast variety of physical and chemical challenges. Chemical factors included ions, pH, water, oxygen, photons, and electrons (16, 57, 94), while the physical modulators included hydrostatic pressure (104), fluid shear stress (18), gravity (34), friction (36), and so on. Cells evolved many sophisticated mechanisms for dealing with the chemical factors, and they also succeeded with the physical challenges. However, we have only limited information of how they did that, as probes have not been available to extract the force information from proteins in living cells. The mechanisms that generate, transmit, and sense forces are intimately involved in the entire spectrum of biology, and these are integrated into traditional biochemistry. There are a lot of data on the macroscopic mechanics of cells and tissues, and there are single-molecule data on the effects of physical forces on gene expression, protein folding, membrane potential, enzymatic activity, cell morphology, motility, bone and muscle homeostasis, embryonic development, and stem cell replication and differentiation. However, we have not been able to examine which proteins are involved and to what extent each contributes to the observed responses. Knockout and mutant effects on interesting proteins only say that the target gene product influences the reaction under study, but not how it does so. Mechanobiology is at the core of changes in cell shape and motility, and there are probably no diseases that do not involve changes in cell shape. Thus, we need to understand cell mechanics to understand pathology.
Advances in mechanobiology can be attributed to new technologies such as microfabricated substrates with variable compliance (28, 82), atomic force microscopy (AFM) (49), laser tweezers (45), and magnetic tweezers (96). However, these tools cannot explore stresses in vivo. To ask questions about the generation and transmission of forces in living cells, we developed genetically encoded fluorescence force sensors (29, 63, 86). These sensors can be integrated into specific intra- or extracellular structural proteins and can report mechanical forces in the host protein in real time in living cells. The probes do not interfere with normal cell physiology and since the mechanical signals are derived from fluorescence signals, they are noninvasive. We have introduced these sensors into the genome of cell lines and animals to create stable cell lines and transgenic animals, reliable data showing that the probes are not toxic. In this review, after a brief overview of the role of mechanical forces in biology and current available techniques, we will discuss more specifically the fluorescence-based force sensors, addressing the basic principles, different versions, limitations, and possible future improvements. We will also discuss potential applications and their impact on biomedicine.
Mechanical Force in Physiology and Pathology
All cells generate forces and are mechanically sensitive. These properties span prokaryotes, fungi, higher plants, insects, and mammals, and the list is not all inclusive. All biological systems utilize three sources of free energy: chemical potential, electrical potential, and mechanical potential and life involves the flux between these sources of energy. Mechanical potential influences differentiation, mitosis, meiosis, motility, apoptosis, and homeostasis. Tissue and organ physiology involve the integration of these activities. Inappropriate forces, too much or too little, lead to pathological conditions in all organs, clearly including bone, lung, heart, and skeletal muscle.
Mechanical force in physiology
Mechanical forces affect all biological systems, including angiogenesis, bone growth, lung and heart physiology, differentiation, embryogenesis, and organogenesis (23, 102). During embryogenesis and organogenesis, the mechanical stress generated by individual cells and their neighbors modulates gene regulation and other biochemical signals affecting differentiation and tissue morphogenesis (58). To give an extreme example, the polygonal scales covering the face and jaws of crocodilians are adjoining and nonoverlapping, but these polygons are irregular and their spatial distribution seems largely random. The organ is generated by the physical cracking of highly keratinized skin, and this is enhanced by forces from the fast growth of the skeleton in the head. This process is primarily physical, rather than genetically controlled. In this case, mechanical forces play a dominant role in organogenesis (67). In lower organisms such as Drosophila, sea urchins, and horseshoe crabs, mechanical forces also play significant roles from the earliest stages of embryo development. The penetration of sperm to eggs is obviously driven by forces. The physical penetration activates a calcium influx affecting downstream pathways for cell division (58, 85). The application of mechanical stress or osmotic pressure can trigger similar activities in Drosophila embryos and sea urchin eggs (38, 39). The mechanical signal is usually the primary factor that determines cell linage (62).
Muscle stem cells differentiate optimally on substrates that imitate the stiffness of muscle tissue (24). Matrices with different elasticity influence human mesenchymal stem cells to assume neuronal, muscle, or bone lineages (25). Fluid shear stress has a dominant effect during early cardiogenesis and angiogenesis (40, 77). In the developing zebra fish heart, high-shear flow is present, and occluding this flow results in abnormal heart chamber development and errors in valve formation (40).
In adults, differentiated and specialized organs utilize mechanical loading for organ and tissue homeostasis. A common example is the muscle and bone, where patients with extended bed rest, or astronauts at zero gravity, suffer from atrophy. Physical inactivity leads to mechanical unloading and that slows protein synthesis and increases muscle protein degradation (83). In one pathway, mechanical stress increases bone growth by inhibiting the production of sclerostin (55, 79, 99). Physically active individuals have lower serum sclerostin, minimizing inhibition of bone formation. Statistical data confirm that higher levels of bone formation occur in athletes performing high-impact exercise relative to nonimpact athletes (69). Similarly, the continuously changing mechanical stress and strain from cyclic contraction and expansion of the lungs influences lung physiology from development to maturation (101). Cells can convert biochemical signals into mechanical signals, a process that is most distinctive in the muscle. The reverse, signal transduction can convert forces to biochemical signals (notably in the ear) using tools such as mechanosensitive ion channels (60, 80), cytoskeletal remodeling, and force-modulated enzymes (46, 87). Mechanical and biochemical systems collectively orchestrate cell physiology.
Mechanical force in pathology
All cells are mechanosensitive, so inappropriate mechanical inputs and disturbed distribution of stress contribute to pathology. Jaalouk and Lammerding published a review summarizing the role of mechanical forces in cancer and the pathology of skeletal and cardiac muscle that results from muscular dystrophy or cardiomyopathies (44). Cardiomyocytes respond directly to mechanical stress through several mechanosensors such as mechanosensitive channels, integrins, and G-protein-coupled receptors. Multiple downstream signal pathways are triggered by these mechanosensors and activate the expression of hypertrophic genes, resulting in an increase in myocyte size, eliciting hypertrophy (5, 15). Mutations in structural proteins can lead to hypertension, aortic stenosis, and myocardial infarction that cause abnormal stress loading in the heart and which results in arrhythmias–mechanoelectric coupling (47). Force loading exerts similar influences on skeletal muscle and bone. Mutations in dystrophin disrupt the force transmission between the cytoskeleton and the extracellular matrix (ECM) of muscle, leading to excessive membrane stress that activates stress-sensitive ion channels (92) and muscle degeneration, a pathological condition named Duchenne muscular dystrophy (35, 51).
Abnormal mechanotransduction is also involved in the response of vascular endothelium to shear stress, resulting in attenuated dilation of arteries and progressive muscle loss (56). Stress gradients and elevated shear stress are the leading cause of atherosclerosis (37). Regions of arteries that branch and bifurcate or change curvature are exposed to low stress and oscillatory shear stress, and these regions are more likely to develop lesions (13).
Another major pathological condition involving mechanics is carcinoma. Cancer cells respond to mechanical stresses during tumor inception, transformation, proliferation, and metastasis (42, 93). Metastasis is the major cause of cancer death, accounting for 90% of fatalities. Mechanical stress regulation occurs at each step of cancer metastasis, including local invasion, intravasation, circulation, arrest and extravasation, proliferation, and colonization (52, 76, 100). Although not completely understood, there is a close correlation between physical inputs and metastasis. For example, using a microfluidic optical cell stretcher, Guck et al. studied the deformability of nonmalignant and malignant human breast cancer cells and showed that higher deformability is related to increased metastatic efficiency (30). AFM indentation revealed similar mechanical properties in immortalized ovarian surface epithelial cells, noninvasive ovarian cancer HEY cells, and invasive HEY A8 cell lines, again showing a correlation of metastatic potential and deformability (31, 103). Mechanical cues in microenvironments also contribute to cancer progression and metastasis (107). The ECM is a major component of the microenvironment, and it is responsible for its mechanical remodeling and cancer malignancy (52, 98). Breast tumorigenesis is characterized with collagen cross-linking and ECM stiffening. Experimentally induced collagen cross-linking stiffened the ECM and enhanced the invasion of transformed epithelial cells (53).
Techniques Used for Mechanical Force Measurements in Biology
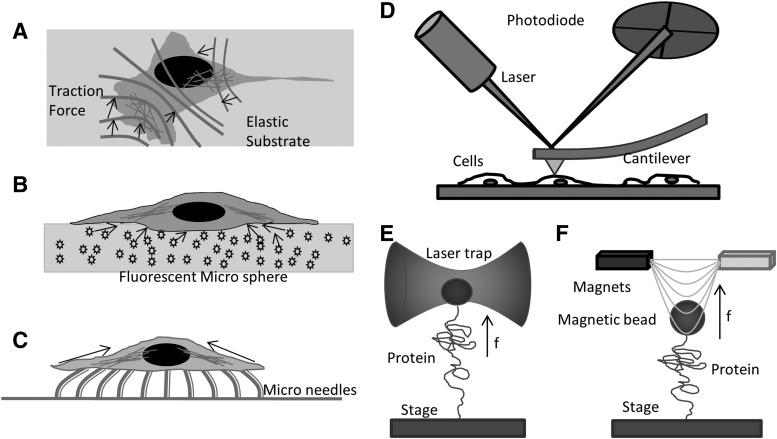
Five traditional techniques are used to study cell mechanics: elastic substrates (70, 84), microfabricated surfaces, bendable fibers (28, 82), AFM (49), laser tweezers (45), and magnetic tweezers (Fig. 1). Cells growing on soft elastic substrates, typically thin silicone rubber, reveal the net mechanical forces exerted by cells on the substrate. The first elastic substrate introduced by Harris in the early 1980s used a thin sheet of cross-linked silicone rubber (33). The viscous liquid, polydimethylsiloxane, was cross-linked at the surface by exposing it to heat. The traction forces exerted by cells are visible through the distortion and wrinkling of the substrate (Fig. 1A). By tuning their compliance, the substrates can be made suitable for force detection in the range of nanonewtons (nN) and micronewtons (μN) (10). Polyacrylamide gel provides another material to make soft substrates. A series of soft substrates with a wide range of flexibility can be created by varying the concentration of bis-acrylamide while maintaining a constant concentration of acrylamide (75). To more precisely track and measure the traction forces, the elastic substrates were further modified by microfabrication. Coated with an ECM protein and embedded with fluorescence microspheres, the rubber substrate enables quantitative measures of traction forces (Fig. 1B) (70). The surface can be micropatterned by standard lithographic techniques. The regularity of such a pattern simplifies analysis and increases versatility (Fig. 1B) (84). Combining micropatterning and fluorescence imaging of focal adhesions in live cells expressing green fluorescence protein (GFP) tagged vinculin, the elastic substrate technique shows that we can measure traction forces at a single focal adhesion site (4). Chen's lab has made further modifications by microfabricating closely spaced arrays of elastomeric vertical microfibers as cells' substrate (Fig. 1C) (95, 105). The microfibers have different heights and geometry yielding different rigidities. The tips can be functionalized with ECM proteins through contact printing.
FIG. 1.
Technologies for cellular force measurements and mechanobiology. (A) On an elastic substrate, migrating cells exert forces that wrinkle the substrate or change the location of embedded fiducial beads; (B) elastic substrate with fluorescence beads embedded measures cell-generated extracellular stress; (C) microfabricated posts/needles measure cell generated stress by bending the posts; (D) atomic force microscopy measures the cortical stiffness and topology; (E) laser trap measures the unfolding force of an isolated protein; and (F) magnetic tweezers pulling on an isolated protein.
The atomic force microscope is another major technique that is used for in vitro and in vivo mechanobiology studies, although the term “atomic” is a misnomer in biological applications. AFM applies forces through micron-sized cantilevers of known stiffness. By measuring the deflecting of the cantilever and the distance moved, one can derive the force/distance relationship of the sample or in raster mode, it can create topographic maps of cell height and stiffness (Fig. 1D) (32, 59). It can be used for single-molecule mechanical stress measurements with a resolution of a few pN. The AFM is widely applied to whole cell biomechanics and creates a mean measurement of the mechanical properties of small cortical domains (6, 7, 90). The cantilever tips can be functionalized with antibodies for recognition of specific proteins. However, AFM applications are limited to in vitro single-molecule measurements or by applying forces to the extracellular surface of cells over areas of about 100 nm2. Similarly, laser tweezers (2) and magnetic tweezers (89, 91) can apply calibrated forces to micron-sized beads that are bound to surface protein molecules (Fig. 1E, F). Laser tweezers/traps are created by focusing a laser to a diffraction-limited spot with a high numerical aperture (NA) microscope objective. The force exerted on a particle in a laser trap depends on the steep energy gradient at focus, the laser power, and the polarizability of the bead. Within a range of ∼150 nm, the force is proportional to the displacement of the particle from the equilibrium position. Laser tweezers have been used to study particles ranging from 20 nm to several micrometers and even whole cells (72) and lipid vesicles (14). Mechanical properties of isolated proteins can be studied with the laser trap when the proteins are linked to polystyrene or silica microspheres and the substrate (8). Magnetic tweezers consist of a pair of magnets that are placed above a magnetic bead (Fig. 1F). Magnetic tweezers are similar to laser traps and can exert 1–200 pN forces typically utilizing particles ranging in size from 0.5 to 5 μm. Magnetic tweezers have been employed for in vitro biomechanical studies pulling on single molecules of DNA and proteins (11, 43). Coated with antibodies or other ligands, the technique can be applied to many experiments, including membrane proteins of live cells (48).
The main drawback of these techniques is that the measurements are confined to purified proteins or extracellular domains of cells where the beads or cantilevers can be attached. The elastomeric substrate only measures the mean traction force at a cellular or subcellular scale. To examine the intracellular stresses in specific proteins in a noninvasive manner in intact cells, we need a biological gauge that can be integrated into individual proteins in the cytoskeleton, ECM, and the nuclear matrix. Then, mechanical stresses can be measured at molecular, sub-cellular, cellular, tissue and the whole animal level. We and other research groups have combined fluorescence energy transfer (FRET) and the elastic spring properties of protein domains to create genetically encoded force sensors for live cell imaging of mechanical forces.
Principles of FRET-Based Force Sensors
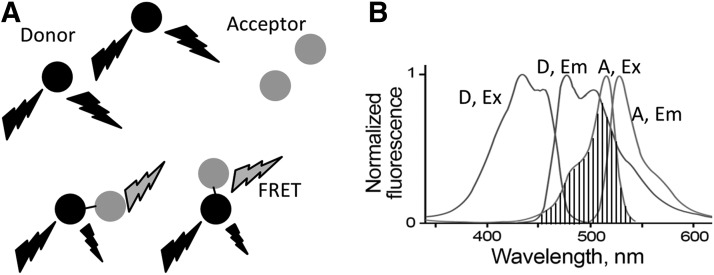
Förster resonance energy transfer (FRET) refers to the dipole–dipole interactions of the electromagnetic fields of two fluorescent dyes or commonly two GFP variants with overlapping excitation and emission wavelengths. FRET was introduced by Förster in 1948 (26, 27) using two fluorescent dyes, one as the energy donor the other one as the acceptor. There are many fluorescence dyes and fluorescence proteins (FPs) that are suitable for FRET pairs. The key factors for an optimal FRET pair are the spectral overlap between donor emission and acceptor excitation with the donor emission well separated from acceptor emission (Fig. 2). This separation of the emission spectrum reduces bleed-through corrections. Most FRET pairs need to be placed closer than 100Å to have nonradioactive interactions, and it has proved useful in measuring distances of 10–100Å. These distances are comparable to the dimensions of most biological molecules, making FRET a widely used technique for biological applications. To give a few examples, Redoxfluor uses FRET to detect the redox state in yeast and Chinese hamster ovary cells. A redox sensor created with a FRET pair senses the redox state through reduction of its internal disulfide bond, resulting in a conformational change in the sensor and corresponding changes in FRET (106). Another good example is Cameleon, an FRET-based calcium sensor that consists of an FRET pair attached to calmodulin. Binding of Ca2+ makes calmodulin wrap around the M13 domain, increasing FRET efficiency (68). There are numerous FRET biosensors for enzymes such as GTPases (50), caspases (54), and protein kinases (19). These sensors rely on the capability of FRET to monitor the distance (R) variations between the donor and acceptor, arising from a protein conformational change, linker cleavage, or protein–protein interactions. As shown in Equation 1, the efficiency of energy transfer E depends on the distance R separating the donor and acceptor, and R0 is called the characteristic distance of a particular FRET pair. At R0, there is 50% transfer efficiency and that is the location of steepest distance dependence.
 |
FIG. 2.
The principles of FRET. (A) FRET donor and acceptor, when free in solution and far apart yield no FRET. When brought closer than 10 nm, FRET occurs and is visible with donor excitation and donor emission. (B) Spectra of donor and acceptor of a typical FRET pair. The shadowed area shows the spectral overlap between donor emission and acceptor excitation. D, donor; A, acceptor; Ex, excitation; Em, emission; FRET, Förster resonance energy transfer.
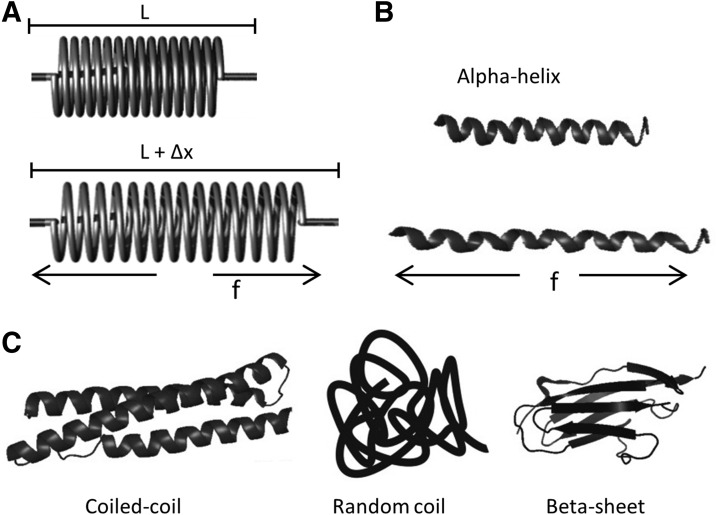
Mechanical force leads to deformation of protein structures, and many protein domains, alpha-helix, beta sheet, and random-coiled protein structures are biological analogs to mechanical springs and possess similar elastic properties (Fig. 3). At physiological forces, a biological spring can deform and extend in length. Based on Hooke's law, force F=−kx, where x is the displacement and k is the spring constant. FRET is well equipped to measure the length extension (10–100Å) of the protein domains. Thus, if we connect a GFP FRET pair with a biological spring, we can create a genetically encoded stress-sensitive cassette and this cassette can be integrated into the middle of structural proteins. The interpretation of the stress data is simplest when the protein is linear, so that tension is the relevant force. Tension loading will extend the spring and increase of distance between the donor and acceptor producing a decrease of FRET. Figure 4 A shows the first such cassette we created, and we named it “stretch sensitive FRET” or stFRET (66). We used a stable alpha-helix (11) as the spring linking a well-defined FRET pair of mutant GFPs called Cerulean (76) and Venus (71). At rest, stFRET has a robust energy transfer, with strong emission at 527 nm and a quenched donor emission at 475 nm (Fig. 4B). The alpha helix mimics well a mechanical spring. Force loading at the ends of the cassette stretches the helix, increasing the distance and reducing FRET. Since the GFP beta barrels are very stiff (withstanding approximately 100 pN without unfolding), forces less than 50 pN will only elongate the alpha helix and not affect the fluorescence of the GFPs. In the next section, we will discuss individual FRET force sensors and their in vitro force calibration, proper controls for stress-free conformations, and the applications to live cells biomechanics.
FIG. 3.
Biological analogs of mechanical spring. (A) Mechanical spring with and without force (f) loading where L is the resting length and Δx is the displacement with stress; (B) biological analog of the metal spring, alpha-helix with and without force loading; and (C) other protein domains that may be treated as springs.
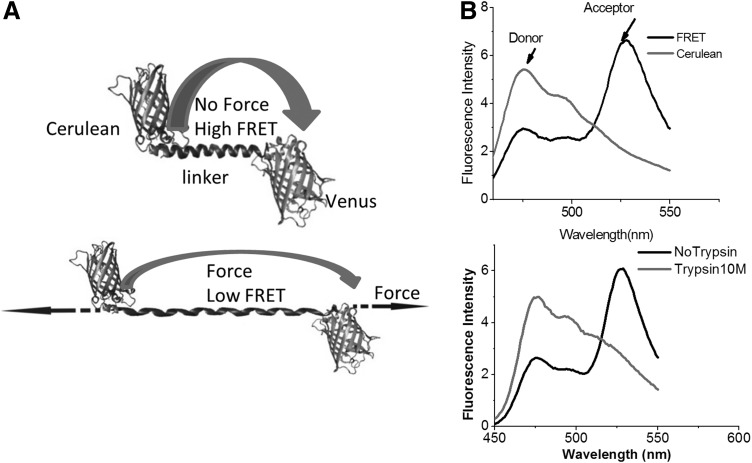
FIG. 4.
Stretch-sensitive FRET sensor. (A) stFRET force sensor (upper panel). Force loading stretches the alpha-helix linker (lower panel); (B) Upper panel, spectra of donor cerulean and stFRET with 433 nm donor excitation. stFRET shows FRET peak at 527 nm, while cerulean shows 475 nm emission; Lower panel, trypsin cuts the linker in stFRET and eliminates FRET. Trypsin 10 μM, 10 min trypsin digestion. stFRET, stretch sensitive FRET.
Fluorescence-Based Force Sensors
Cy3/Cy5 force sensor
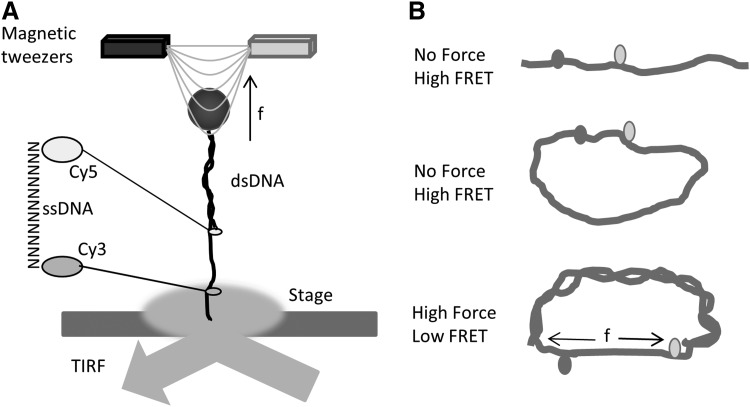
FRET was first used by Shroff et al. to assess the force loading in single-stranded DNA (86). They developed the nN sensors consisting of a single-stranded DNA oligomer, with cy5 and cy3 fluorescent dyes covalently attached as FRET donor and acceptor (Fig. 5A). DNA acted as the spring. The optical readout of the FRET efficiency constitutes a direct measurement of the mechanical force loading on the sensor. Applications of such sensors include measurements of various material properties and stresses in nanoscale machines, including programmable DNA assembly, polymer meshes, and DNA tiles (61, 73, 74). Before applying the sensors to biological machines and nanoscale device studies, they used magnetic tweezers and single molecule fluorescence microscopy to characterize the force sensitivity and dynamic range of the sensors. To create a force sensor that is responsive to the displacements of the linker between the Cy3/Cy5, there are two premises. One, the linker should be compliant at the force scale of interest, typically less than 100 pN in biology. Second, the spacer resting length should be near R0, where the strain sensitivity is maximum. R0 ∼6 nm for Cy3/Cy5. The authors designed the sensors with a 15-base ssDNA linker. Single-stranded DNA has a persistence length of 1–4 nm, while double-stranded DNA (dsDNA) is much stiffer with a persistence length of 50 nm (88). At 6 nm, dsDNA behaves similar to a rigid rod and there is negligible distance change over the applied force. Even though 15 bp ssDNA has a contour length of 9.5 nm, at zero force, the end-to-end distance would be less than 9.5 nm because of folding (worm-like chain effects). The force and FRET efficiency (E) correlations of sensors need to be characterized at the single-molecule level before applying absolute values to other force measurements.
FIG. 5.
Cy3/Cy5 single strand DNA force sensor. (A) Diagram of FRET force sensor with a DNA linker and magnetic force application. The force sensor was composed of Cy3/Cy5 dyes that were linked with single-stranded DNA of variable lengths. One end was attached to the substrate; (B) FRET force sensor calibration using DNA springs.
Shroff et al. immobilized the sensor on the surface of a flow cell with biotin-streptavidin and excited the dyes using internal reflection fluorescence microscopy (Fig. 5A). They first determined the FRET efficiency at zero force and then applied 0–20 pN force ramp to the sensor using magnetic tweezers. Sensors with spacers varying from 10 bp, 15 bp to 20 bp were tested for their sensitivity to the force ramp. The longer ssDNA spacer gave the lowest FRET as expected. The data showed a force resolution of all three sensors at ∼5 pN. They are able to distinguish between high (15–20 pN) and low (0–5 pN) forces. The first application of the sensor was to determine the force in a 37-nucleotide DNA loop (Fig. 5B). A single-stranded DNA (ssDNA) loop is constructed with Cy3/Cy5 integrated 10 bp apart, and complementary ssDNA is added to the system to form dsDNA. Theoretical predictions of a bendable rod predict a 25 pN force at the ends. However, the experimentally measured force was more similar to 6 pN, much less than the prediction probably due to the development of kinks, underlining the necessity of confirming theoretical simulation data with experimental results. The applications of such sensors are limited to in vitro systems but established the foundation of using FRET to detect pN forces. We have adapted the DNA stretching method for the in vitro calibration of our FPs-based force sensors.
Stretch-sensitive FRET
We created the first genetically encoded forces sensor stFRET for live cell force imaging (Fig. 4) (65, 66). We used GFP variants Cerulean (78) and Venus (71) as the donor and acceptor, respectively, and a stable alpha-helix (12) as the linker. Usually, a single alpha-helix structure is not stable in solution, but after searching the database, we found a stable alpha-helix ∼5 nm long at rest, the characteristic distance (R0) of Cerulean and Venus. Cyan fluorescence protein and yellow fluorescence protein have been optimized for brightness and resistance to bleaching. The strain sensitivity, the derivative of E with regard to the distance, is maximal at R=R0. When inserted into host proteins, the force loading in the host t creates strain in the sensor and elongates the linker, reducing FRET. After transfecting cells with the chimera genes, we were able to derive the force loading in the host protein from the optical readout of FRET from the images. However, before conducting the in vivo work, we conducted extensive in vitro characterizations.
To characterize stFRET itself, we inserted the stFRET gene into a prokaryotic vector and expressed the protein in bacteria. We measured the spectra of purified protein in a spectrometer and measured the FRET efficiency in the fluorescence microscope. At rest, stFRET had robust FRET (Fig. 4B). Excited by 433 nm, the sensor showed increased acceptor (Venus) emission at 527 nm and quenched donor (Cerulean) emission at 475 nm. Cleavage of the linker by trypsin or Proteinase K eliminated the 527 nm peak and the 475 nm emission peak recovered (Fig. 4B). Similar results were observed on the microscope with stFRET in solution. FRET efficiency is affected not only by the distance between dipoles, but also by the relative orientation. We estimated the absolute geometry of the sensor by varying the length of the linker by adding or subtracting single residues. Deleting one amino acid from the helix makes the structure rotate 100° with little change in distance. Deleting a full turn of the helix reduces the distance and rotates the structure by 360°, producing no change in angle. We created six mutants with different orientations and linker length and measured the FRET efficiency. Some nonlinear equations and six independent measures were more than sufficient to resolve the three unknown angles that determine the orientation factor κ2 as well as the global configuration of the protein.
All FRET force sensor measurements are based on a key assumption: The fluorophore spectra are unaffected by force loading. GFPs can withstand approximately 100 pN forces without unfolding (20). Physiological forces are typically in the range of 0–20 pN; therefore, the fluorophores will remain intact during measurements in living cells, and FRET changes can faithfully represent the distance changes caused by the stretching the linker (these probes are not useful for compression due to the possibility for buckling the host). Using steered molecular dynamic simulation, Saeger et al. suggested that 20–70 pN forces led to substantial structural changes at the N terminal of EGFP, reducing its brightness (81). We inserted Cerulean/Venus, the FRET pair of our sensors, into the middle of spectrin and examined the fluorescence changes. There were no detectable fluorescence changes with forces experienced by spectrin in living cells (data not published). GFPs have a very flexible N terminal that can change structure at a few pN, but Cerulean and Venus, two much more stable GFP variants, are not sensitive to forces at that level. However, other factors than mechanical forces, environmental factors, may also influence GFP fluorescence.
We tested the linker integrity under the conditions of urea denaturing, heating, and cleavage with proteinase K and trypsin. Urea of approximately 8 M melts the alpha-helix, increasing the end–end spacing in the sensor and significantly reducing FRET efficiency. Remarkably, the fluorescence spectra of both GFPs were unaffected. These data confirm the robust energy transfer of the sensor at the ground state. A second test elevated the temperature to 80°C. Both the donor and acceptor emission declined slightly as expected from the increases in thermal fluctuations, but there was no significant change in the energy transfer efficiency. This suggests that the alpha-helix linker did not melt. Trypsin and proteinase K cleaved the linker, producing a drop in the FRET efficiency within 20 s. Again, the donor and acceptor spectra were surprisingly unchanged after 30 min digestion with either enzyme. These experiments suggest use of the probes as detectors of proteinases in living cells and whether stress in host proteins can expose cryptic proteolytic sites.
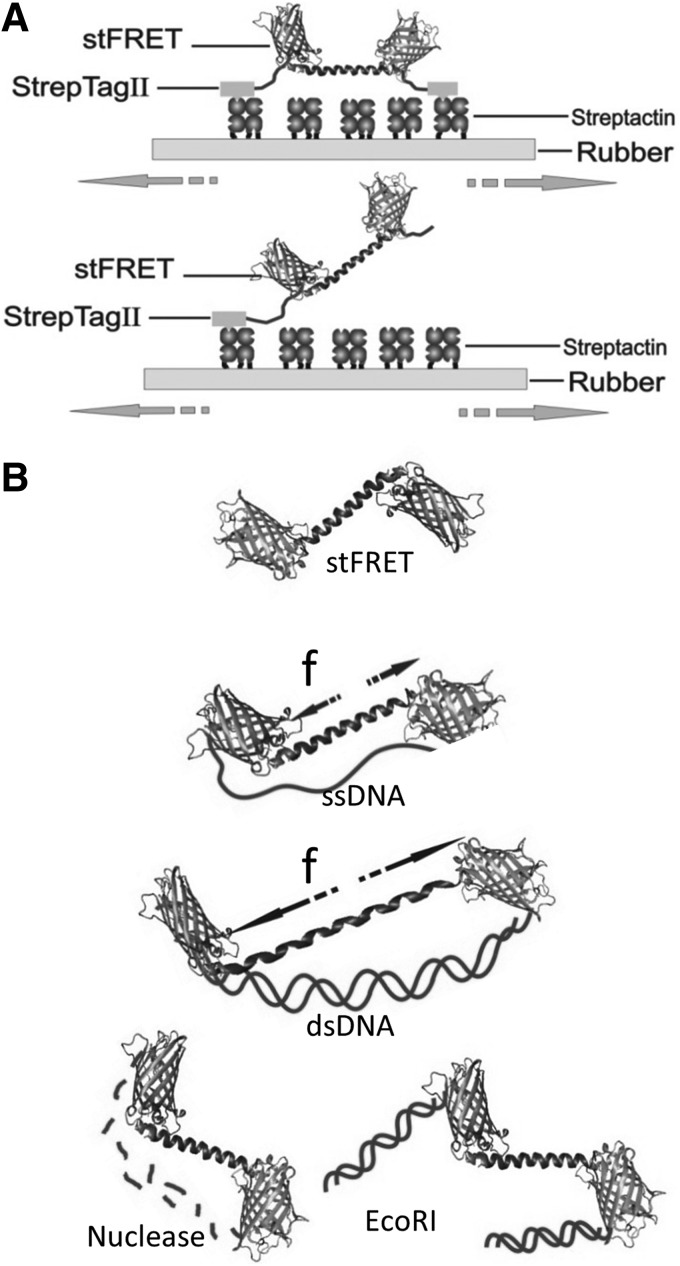
We performed in vitro stretching of the sensors to display its strain sensitivity (Fig. 6A). We bonded the ends of derivatized stFRET to a silicone rubber sheet using StreptagII-Streptactin™ and stretched the sheet biaxially on the fluorescence microscope. When the C and N termini of stFRET are attached to the rubber, the probe will be stretched with the sheet. We observed a reversible 11% FRET decrease with stretch and no change when only one end was attached to the rubber. Nonspecific binding of double-tagged stFRET to an untreated silicone surface produced no significant change in FRET. Thus, stFRET actually monitors strain as expected from the design of the sensor.
FIG. 6.
stFRET in vitro calibration. (A) Rubber sheet stretching of stFRET to verify strain-dependence of the linker. The rubber sheet was coated with streptactin, and stFRET was flanked with StrepTagII. stFRET flanked by one StrepTagII as a zero force control; (B) DNA springs stretching stFRET.
Another method we developed to test the stress sensitivity was to covalently attach a 60mer DNA oligonucleotide to the two naturally occurring free cysteines in the donor and acceptor (65) (Fig. 6B). The cysteines at position 48 of the donor and acceptor are in reduced form, providing anchor sites for the DNA that had chemically modified N and C termini. Given the 1–4 nm persistence length of ssDNA (88), the ssDNA did not apply much force to the sensor but we did observe some drop in efficiency, hinting that the entropic spring forces from the floppy ssDNA are not insignificant. However, adding the complementary DNA that anneals to the ssDNA formed dsDNA with a persistence length of 50 nm and pushes the ends apart. Such transition corresponds to a 5–7 pN force increase (97), leading to further FRET reduction. Forces from the DNA could be released either by cleavage with nuclease or by cutting the dsDNA in the middle with EcoRI. The ground state (zero force) FRET is fully restored after cutting the DNA. These in vitro characterizations demonstrated the force sensitivity of stFRET. We then verified its use in cells.
stFRET expresses well in 3T3, HEK-293, bovine aortic endothelial cells (BAEC) cells, and Caenorhabditis elegans (65). We first integrated the sensor gene near the middle of host proteins genes, then introduced these chimeric genes into cells. stFRET data are only easily interpretable when they are in series with linear fibers, but not in globular proteins such as beta-actin and tubulin. A possible drawback to chimeric proteins is the potential interference to function and distribution of the host protein. We have utilized a few strategies to test for interference.
Positioning the insertion site near the center of the host was best; second, avoiding known functional domains with the optimum sites being linker domains; third, testing several insertion sites. Host proteins tagged with a single GFP are widely accepted standards for inert marking of proteins, although generalization is not reliable and functional perturbations should be checked. We compared the cell expression pattern of chimeric proteins to single terminal GFP-tagged versions and in general, but not always, they proved inert to altering structure. A force-free control construct is required to distinguish force-induced FRET changes from chemically induced changes (such as Ca2+ levels or pH) in live cells. A host protein tagged by stFRET on the C or N terminal is not expected to be subjected to tension, because the dangling end frees stFRET from force-induced deformation. These negative controls suggested that FRET change in the centrally tagged host would arise from stress/strain but not other chemical signals.
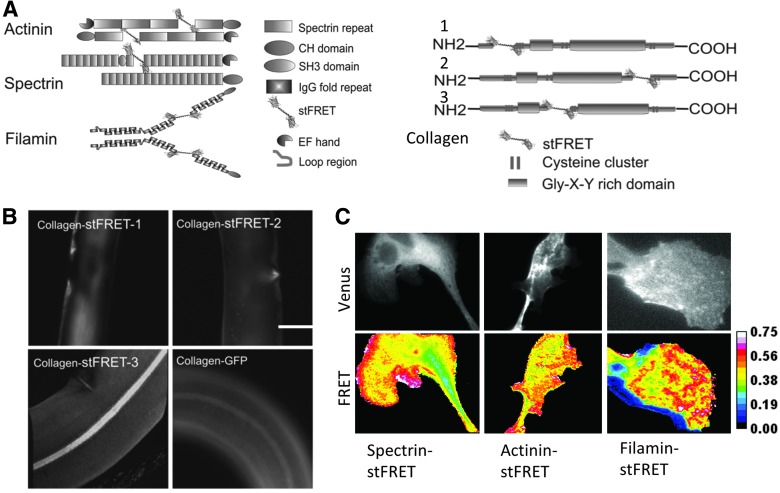
We have inserted stFRET into a variety of host proteins, including nonerythroid spectrin, alpha-actinin, filamin, and collagen (Fig. 7A). The spectrin, actinin, and filamin constructs showed cellular distributions similar to those observed from a host protein tagged at the C terminal by GFP. For collagen19 constructs in worms, we picked three different positions to insert the sensor and created three transgenic lines. Only one location created worms with the proper protein assembly and the typical striated pattern in the cuticle (Fig. 7B) (65, 66). All worm lines reproduced and behaved normally, showing that the chimeric constructs posed no toxicity to cells or whole animals. When we stretched the worms with micromanipulators, the labeled collagen showed acute reversible changes in FRET associated with tension and relaxation. stFRET integrated in actinin, filamin, and spectrin revealed resting tension (prestress) in these hosts (Fig. 7C). We examined migrating 3T3 and BAEC cells that have characteristic leading and lagging edges. stFRET actinin and filamin-expressing cells revealed that during migration, the leading edge showed higher stress in both proteins at the leading edge and lower stress at the lagging edge. stFRET provides a powerful tool for real-time biomechanical studies in living cells. Sensors similar to stFRET will open new gateways to the study of many biological processes.
FIG. 7.
Force measurements in live cells and animals. (A) Diagram of actinin-stFRET, filamin-stFRET, spectrin-stFRET, and collagen-stFRET; (B) collagen-stFRET and collagen-GFP distribution pattern in C. elegans cuticles. Only construct 3 (as shown in A) was indistinguishable from Collagen-GFP; (C) 3T3 Cells expressing actinin, filamin, and spectrin stFRET constructs. The images of Venus display the probe distribution, the FRET ratio 16 color map shows constitutive stress in these proteins and uneven force distribution in subcellular domains, and higher FRET ratio reflects higher constitutional stress. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Spectrin stFRET
The linker in stFRET is stiffer than many of the cytoskeletal proteins, and we wondered whether the change in compliance of the host might introduce some changes in function, so we introduced a more natural linker. When the linker is compliant to the host protein, there is a better impedance match and more efficient energy transfer. As a start we substituted the alpha-helix with a spectrin repeat and named the new construct spectrin-stFRET (sstFRET) (Fig. 8A) (63). Spectrin repeat domains are composed of three-folded alpha-helices and are conserved in spectrin superfamily proteins, including alpha-actinin, dystrophin, utrophin, and kalirin (22). The most common actin cross-linker proteins in cytoskeleton use spectrin repeats.
FIG. 8.
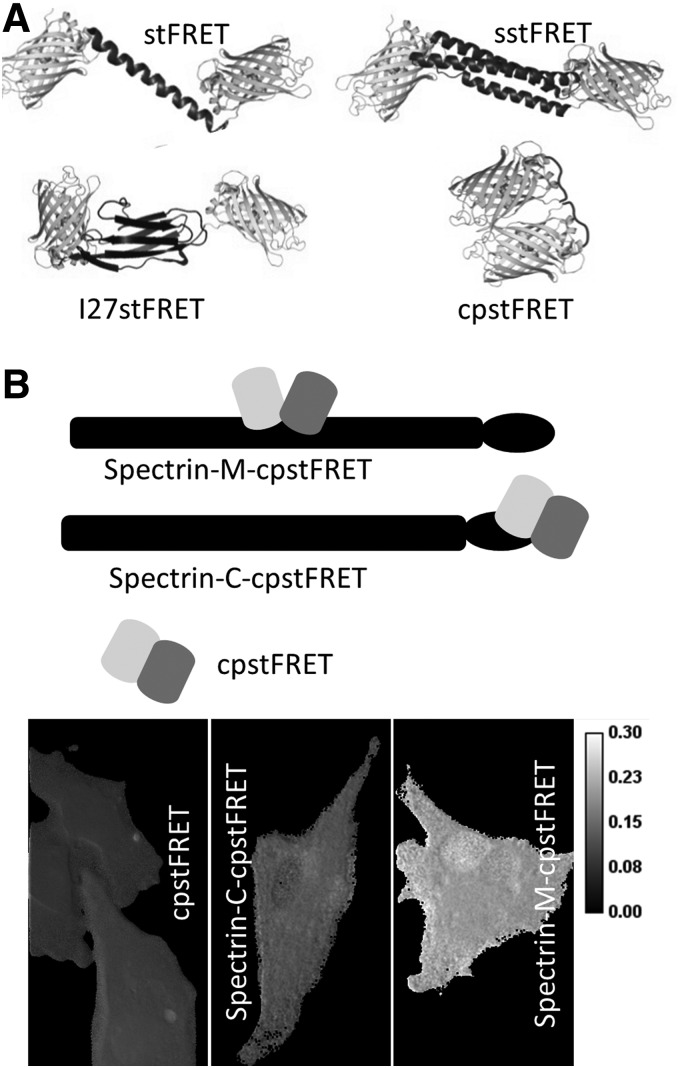
stFRET sensor variants. (A) Diagram of stFRET, sstFRET, cpstFRET, and I27stFRET that uses the I27 domain beta sheet as the linker; (B) spectrin-M-cpstFRET, with cpstFRET inserted in the middle of spectrin, Spectrin-C-cpstFRET, with spectrin tagged by cpstFRET at the C terminal as a stress free configuration. Lower panel shows BAEC cells expressing the three constructs. Spectrin-M-cpstFRET displays a higher FRET ratio reflecting higher constitutional stress. BAEC, bovine aortic endothelial cells; cpstFRET, circularly permutated stFRET; sstFRET, spectrin repeat stFRET.
We conducted the same sets of in vitro characterization on sstFRET as we did with the stFRET. We inserted sstFRET into alpha-actinin and expressed the chimera in human embryonic kidney (HEK) and BAEC cells. The actinin distribution and functions appeared normal compared with terminal GFP-tagged versions. We found similar stress patterns in actinin using either stFRET or sstFRET. Actinin-sstFRET displays significant constitutive stress and force modulation during cell contraction, extension and migration (63). Time lapse imaging showed rapid modulation of the stress in actinin when we challenged the cell with cyclic osmotic pressure. Hypotonic swelling increased actinin tension, even though it was deep in the cell as predicted (90), and tension was rapidly released when the bath was returned to isotonic saline.
The accessibility of different cell types for labeling may be restrained due to the limitations of the transient transfection methods for gene delivery and the arcane environment of a culture dish relative to tissues. To make the most cell types available, we created a transgenic mouse line with actinin-sstFRET by randomly incorporating the construct into the genome using the pronuclear injection method. PCR confirmed the presence of the correct genes in adult mice without showing morphological abnormality or abnormal behavior. However, we observed little fluorescence in the mouse and are now working on other transgenic methods to create new strains. These transgenic animals will open vast new perspectives in biology.
Circularly permutated stFRET
The force sensors discussed earlier rely on stress-induced strain in the linkers. However, the molecular dimensions of the linker limit sensitivity and dynamic range. The relative orientation of the dipoles modulates FRET with a cos2 dependence. When the dipoles are parallel and close together, the efficiency can be nearly 100% and when they are orthogonal, it can be nearly 0%. Hence, we designed a new FRET sensor from circularly permutated stFRET named cpstFRET (64). The sensor consists of tandemly connected circularly permutated Cerulean and Venus. The FRET pair were oriented so that at zero stress, the donor and acceptor are close and parallel (Fig. 8A). The parallel dipoles have κ2=4, which yields the highest FRET at a fixed distance. Torsion exerted at the ends of cpstFRET will twist the dipoles toward a perpendicular position, where κ2=0. Theoretically, no FRET occurs at the perpendicular conformation regardless of how close they are. We demonstrated this angular dependence of FRET efficiency in our original work by sequentially deleting or adding amino acids to the helical linker (66). Using the same in vitro characterization methods for stFRET, we confirmed the normal expression of cpstFRET in eukaryotic cells and the efficiency of cpstFRET is approximately 80% when stress free in cells. Stress calibration with DNA springs showed a dramatic decrease with 5–7 pN forces (64). With efficiency E ranging from 80% to 0, this sensor has a much wider dynamic range than the previous ones.
cpstFRET is also smaller than stFRET and less likely to interfere with host function. cpstFRET, at 54 kDa, is about ∼30% smaller than stFRET. We incorporated cpstFRET into spectrin and expressed the chimeras in HEK293, Madin-Darby canine kidney cells (MDCK), and BAECs and then examined the constitutive stress in spectrin at rest and during cell migration. The negative control was spectrin tagged at the C terminal with cpstFRET (Fig. 8B). Spectrin was under significant constitutive stress in MDCK and BAEC cells but not in HEK cells (64) with obvious differences in the cell physiology, but without the probe, we never would know that differences existed. The stress in spectrin appears to originate from F-actin and microtubules, as cytochalasin D or colchicine can rapidly release the stress in spectrin. Even though spectrin is anchored to transmembrane protein complexes (3), the bilayer itself does not appear to exert detectable forces on spectrin, because in HEK cells, the spectrin was largely unstressed. The variations of constitutive stress in spectrin are just one example of what we believe will be a wide span of variability in different cells, and that is just a token of the influence of mechanical stress on cell physiology.
Tension sensor module
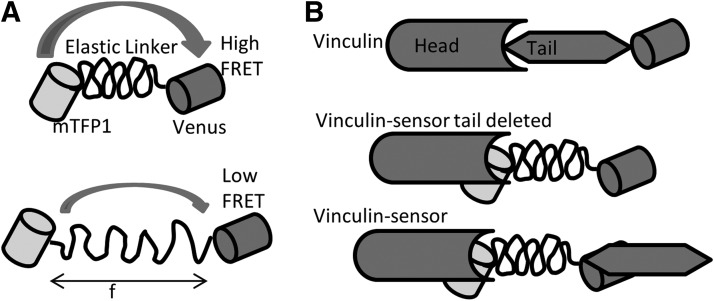
Using a similar linear FRET sensor concept, Grashoff et al. later developed a variant sensor called the “tension sensor module” (TSMod). The module consists of a FRET pair Venus and mTFP1 (1) connected by a 40-amino-acid elastic domain derived from silk protein flagelliform (Fig. 9) (29). They integrated the sensor module into vinculin between the head and tail domain after amino acid 883. As we did with stFRET, they constructed vinculin C terminal tagged with Venus as a standard for normal protein distribution and a reference for the normal function of chimeric constructs. Their force-free control is a tail-truncated mutant that cannot bind to F-actin or paxillin. The expression and compatibility of chimeric vinculin was tested in murine embryonic fibroblasts and BAECs. Using wild-type vinculin null cells, they transiently transfected vinculin-TSMod, which displayed the proper distribution of the protein to focal adhesions. The FA and F-actin phenotype were indistinguishable from cells expressing the control vinculin-Venus. Surprisingly, TSMod also localized to the nucleus as we saw with free stFRET (65, 66). This sensor was calibrated in vitro using single-molecule spectroscopy, where a confocal microscope was combined with optical tweezers. Due to the low photo stability of FPs at the single-molecule scale, they replaced the FPs by Cy3 and Cy5 for the calibration. Under cyclic stress applied by laser tweezers, the tension module showed reversibility without hysteresis and responded to stress as low as 1–6 pN. These data agree with those acquired from single and dsDNA stretching.
FIG. 9.
TSMod and tension studies in Vinculin. (A) Structure of TSMod. Upper panel shows resting TSMod with high FRET, the lower panel shows TSMod with force loading and low FRET; (B) three constructs of vinculin. Vinculin tagged by Venus at C terminal, vinculin head domain tagged by TSMod as a force free control, and vinculin with TSMod inserted between head and tail domain for force measurements. TSMod, tension sensor module.
For live cell measurements, they mapped the mechanical force in vinculin during cell migration and found it experienced higher force in the FAs at the leading edge and less at the trailing edge that possessed larger FAs. The tail truncated controls displayed uniformly higher FRET, indicating lower force, similar to what we reported for the force distribution in actinin and filamin in BAECs (63, 65). TSMod has been optimized and applied to the detection of tension in cadherin and PECAM-1 in epithelial and endothelial cells (9, 17). These genetically encoded force sensors for the first time open up cell mechanics and permit the exploration of its coupling to the other sources of free energy
Conclusions and Future Directions
FRET-based force sensors enabled us to measure the forces in many structural proteins in living cells in real time (video rates). They enable us to introduce the sensors into embryos and transgenic animals and map in four dimensions the mechanics of specific proteins. Transgenic animals can be created to express the protein in particular cell types or specific organs, or with a general promoter, into nearly all cells. Transgenic animals universally expressing prelabeled structural proteins can provide a vast spectrum of cell types and organs and even permit imaging of the moving animal, facilitating unlimited biomechanics studies. Zebrafish are the latest transgenic animals to be created (unpublished). It would be fascinating to examine how embryo development and stem cell differentiation are influenced by forces in specific proteins (21). Force profiles may be important in understanding cancer stem cells and provide critical hints and possible drug targets for cancer metastasis (52). Since malignant transformation and metastasis involve rapid changes in the ECM and cytoskeleton stress (41), profiling mechanical stress in metastatic and benign cancer cells may set important fiducial parameters for early diagnosis, optimizing therapy, and improving drug screening. Obviously there is no limit of the applications of these sensors in biological systems, and there is a vast promise of insights into new therapies. The availability of the stress sensors promises to have an equivalent effect of the introduction of Ca2+ probes and GFP.
Abbreviations Used
- AFM
atomic force microscopy
- BAEC
bovine aortic endothelial cells
- cpstFRET
circularly permutated stFRET
- dsDNA
double-stranded DNA
- ECM
extracellular matrix
- FA
focal adhesion
- FP
fluorescence protein
- FRET
Förster resonance energy transfer
- GFP
green fluorescence protein
- HEK
human embryonic kidney
- MDCK
Madin-Darby canine kidney cells
- ssDNA
single-stranded DNA
- sstFRET
spectrin repeat stFRET
- stFRET
stretch sensitive FRET
- TSMod
tension sensor module
Acknowledgment
This work was supported by grants from NIH and the Children's Guild of Buffalo.
References
- 1.Ai HW, Henderson JN, Remington SJ, and Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J 400: 531–540, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashkin A. and Dziedzic JM. Optical trapping and manipulation of viruses and bacteria. Science 235: 1517–1520, 1987 [DOI] [PubMed] [Google Scholar]
- 3.Baines AJ. Evolution of spectrin function in cytoskeletal and membrane networks. Biochem Soc Trans 37: 796–803, 2009 [DOI] [PubMed] [Google Scholar]
- 4.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, and Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol 3: 466–472, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Barry SP, Davidson SM, and Townsend PA. Molecular regulation of cardiac hypertrophy. Int J Biochem Cell Biol 40: 2023–2039, 2008 [DOI] [PubMed] [Google Scholar]
- 6.Besch S, Snyder KV, Zhang RC, and Sachs F. Adapting the Quesant (c) Nomad (TM) atomic force microscope for biology and patch-clamp atomic force microscopy. Cell Biochem Biophys 39: 195–210, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Beyder A. and Sachsb F. Electromechanical coupling in the membranes of Shaker-transfected HEK cells. Proc Natl Acad Sci USA 106: 6626–6631, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Block SM, Goldstein LS, and Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature 348: 348–352, 1990 [DOI] [PubMed] [Google Scholar]
- 9.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, and Dunn AR. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci USA 109: 12568–12573, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton K. and Taylor DL. Traction forces of cytokinesis measured with optically modified elastic substrata. Nature 385: 450–454, 1997 [DOI] [PubMed] [Google Scholar]
- 11.Charvin G, Strick TR, Bensimon D, and Croquette V. Tracking topoisomerase activity at the single-molecule level. Annu Rev Biophys Biomol Struct 34: 201–219, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Brock R, Luh F, Chou PJ, Larrick JW, Huang RF, and Huang TH. The solution structure of the active domain of CAP18—a lipopolysaccharide binding protein from rabbit leukocytes. FEBS Lett 370: 46–52, 1995 [DOI] [PubMed] [Google Scholar]
- 13.Cheng C, Tempel D, van Haperen R, van der Baan A, Grosveld F, Daemen MJ, Krams R, and de Crom R. Atherosclerotic lesion size and vulnerability are determined by patterns of fluid shear stress. Circulation 113: 2744–2753, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Cherney DP, Bridges TE, and Harris JM. Optical trapping of unilamellar phospholipid vesicles: investigation of the effect of optical forces on the lipid membrane shape by confocal-Raman microscopy. Anal Chem 76: 4920–4928, 2004 [DOI] [PubMed] [Google Scholar]
- 15.Chien KR, Knowlton KU, Zhu H, and Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive physiologic response. FASEB J 5: 3037–3046, 1991 [DOI] [PubMed] [Google Scholar]
- 16.Claridge SA, Schwartz JJ, and Weiss PS. Electrons, photons, and force: quantitative single-molecule measurements from physics to biology. ACS Nano 5: 693–729, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conway DE, Breckenridge MT, Hinde E, Gratton E, Chen CS, and Schwartz MA. Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr Biol 23: 1024–1030, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies PF, Spaan JA, and Krams R. Shear stress biology of the endothelium. Ann Biomed Eng 33: 1714–1718, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Depry C. and Zhang J. Visualization of kinase activity with FRET-based activity biosensors. Curr Protoc Mol Biol Chapter 18: Unit 18 15, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Dietz H. and Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc Natl Acad Sci USA 101: 16192–16197, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Discher DE, Mooney DJ, and Zandstra PW. Growth Factors, Matrices, and Forces Combine and Control Stem Cells. Science 324: 1673–1677, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djinovic-Carugo K, Gautel M, Ylanne J, and Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett 513: 119–123, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Egginton S. Angiogenesis - may the force be with you! J Physiol 588: 4615–4616, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, and Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol 166: 877–887, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engler AJ, Sen S, Sweeney HL, and Discher DE. Matrix elasticity directs stem cell lineage specification. Cell 126: 677–689, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Förster T. Experimentelle und theoretische Untersuchung des zwischenmolekularen Ubergangs von Elektronenanregungsenergie. Z Naturforsch A 4: 321–327, 1949 [Google Scholar]
- 27.Forster VT. Zwischenmolekulare energiewanderung und fluoreszenz. Ann Phys 6: 54–75, 1948 [Google Scholar]
- 28.Galbraith CG. and Sheetz MP. A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci USA 94: 9114–9118, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, and Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466: 263–266, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, Romeyke M, Lenz D, Erickson HM, Ananthakrishnan R, Mitchell D, Kas J, Ulvick S, and Bilby C. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J 88: 3689–3698, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hampton T. Lessening of cell stiffness might serve as new biomarker for malignancy. JAMA 299: 276, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Hansma PK, Elings VB, Marti O, and Bracker CE. Scanning tunneling microscopy and atomic force microscopy: application to biology and technology. Science 242: 209–216, 1988 [DOI] [PubMed] [Google Scholar]
- 33.Harris AK, Wild P, and Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208: 177–179, 1980 [DOI] [PubMed] [Google Scholar]
- 34.Haswell ES. Gravity perception: how plants stand up for themselves. Curr Biol 13: R76 1–R763, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Heydemann A. and McNally EM. Consequences of disrupting the dystrophin-sarcoglycan complex in cardiac and skeletal myopathy. Trends Cardiovasc Med 17: 55–59, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Hoffman BD. and Crocker JC. Cell mechanics: dissecting the physical responses of cells to force. Annu Rev Biomed Eng 11: 259–288, 2009 [DOI] [PubMed] [Google Scholar]
- 37.Hollander W. Role of hypertension in atherosclerosis and cardiovascular disease. Am J Cardiol 38: 786–800, 1976 [DOI] [PubMed] [Google Scholar]
- 38.Horner VL. and Wolfner MF. Mechanical stimulation by osmotic and hydrostatic pressure activates Drosophila oocytes in vitro in a calcium-dependent manner. Dev Biol 316: 100–109, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horner VL. and Wolfner MF. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev Dyn 237: 527–544, 2008 [DOI] [PubMed] [Google Scholar]
- 40.Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, and Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature 421: 172–177, 2003 [DOI] [PubMed] [Google Scholar]
- 41.Huang S. and Ingber DE. Cell tension, matrix mechanics, and cancer development. Cancer Cell 8: 175–176, 2005 [DOI] [PubMed] [Google Scholar]
- 42.Indra I, Undyala V, Kandow C, Thirumurthi U, Dembo M, and Beningo KA. An in vitro correlation of mechanical forces and metastatic capacity. Phys Biol 8: 015015, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, and Kinosita K. Mechanically driven ATP synthesis by F1-ATPase. Nature 427: 465–468, 2004 [DOI] [PubMed] [Google Scholar]
- 44.Jaalouk DE. and Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol 10: 63–73, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang G, Giannone G, Critchley DR, Fukumoto E, and Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424: 334–337, 2003 [DOI] [PubMed] [Google Scholar]
- 46.Johnson CP, Tang HY, Carag C, Speicher DW, and Discher DE. Forced unfolding of proteins within cells. Science 317: 663–666, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson DB. and Dell'Italia LJ. Cardiac hypertrophy and failure in hypertension. Curr Opin Nephrol Hypertens 5: 186–191, 1996 [DOI] [PubMed] [Google Scholar]
- 48.Kamgoue A, Ohayon J, and Tracqui P. Estimation of cell Young's modulus of adherent cells probed by optical and magnetic tweezers: influence of cell thickness and bead immersion. J Biomech Eng 129: 523–530, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Kirmizis D. and Logothetidis S. Atomic force microscopy probing in the measurement of cell mechanics. Int J Nanomed 5: 137–145, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y, and Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell 22: 4647–4656, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar A, Khandelwal N, Malya R, Reid MB, and Boriek AM. Loss of dystrophin causes aberrant mechanotransduction in skeletal muscle fibers. FASEB J 18: 102–113, 2004 [DOI] [PubMed] [Google Scholar]
- 52.Kumar S. and Weaver VM. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev 28: 113–127, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, and Weaver VM. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139: 891–906, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li IT, Pham E, Chiang JJ, and Truong K. FRET evidence that an isoform of caspase-7 binds but does not cleave its substrate. Biochem Biophys Res Commun 373: 325–329, 2008 [DOI] [PubMed] [Google Scholar]
- 55.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, and Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem 280: 19883–19887, 2005 [DOI] [PubMed] [Google Scholar]
- 56.Loufrani L, Dubroca C, You D, Li Z, Levy B, Paulin D, and Henrion D. Absence of dystrophin in mice reduces NO-dependent vascular function and vascular density: total recovery after a treatment with the aminoglycoside gentamicin. Arterioscler Thromb Vasc Biol 24: 671–676, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lovelock JE. Affinity of organic compounds for free electrons with thermal energy: its possible significance in biology. Nature 189: 729–732, 1961 [DOI] [PubMed] [Google Scholar]
- 58.Mammoto T. and Ingber DE. Mechanical control of tissue and organ development. Development 137: 1407–1420, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marti O, Ribi HO, Drake B, Albrecht TR, Quate CF, and Hansma PK. Atomic force microscopy of an organic monolayer. Science 239: 50–52, 1988 [DOI] [PubMed] [Google Scholar]
- 60.Martinac B. Mechanosensitive ion channels: molecules of mechanotransduction. J Cell Sci 117: 2449–2460, 2004 [DOI] [PubMed] [Google Scholar]
- 61.Mathieu F, Liao S, Kopatsch J, Wang T, Mao C, and Seeman NC. Six-helix bundles designed from DNA. Nano Lett 5: 661–665, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maul TM, Chew DW, Nieponice A, and Vorp DA. Mechanical stimuli differentially control stem cell behavior: morphology, proliferation, and differentiation. Biomech Model Mechanobiol 10: 939–953, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng F. and Sachs F. Visualizing dynamic cytoplasmic forces with a compliance-matched FRET sensor. J Cell Sci 124: 261–269, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meng F. and Sachs F. Orientation-based FRET sensor for real-time imaging of cellular forces. J Cell Sci 125: 743–750, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meng F, Suchyna TM, Lazakovitch E, Gronostajski RM, and Sachs F. Real time FRET based detection of mechanical stress in cytoskeletal and extracellular matrix proteins. Cell Mol Bioeng 4: 148–159, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meng F, Suchyna TM, and Sachs F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J 275: 3072–3087, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milinkovitch MC, Manukyan L, Debry A, Di-Poi N, Martin S, Singh D, Lambert D, and Zwicker M. Crocodile head scales are not developmental units but emerge from physical cracking. Science 339: 78–81, 2013 [DOI] [PubMed] [Google Scholar]
- 68.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, and Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388: 882–887, 1997 [DOI] [PubMed] [Google Scholar]
- 69.Morgan A. and Weiss Jarrett J. Markers of bone turnover across a competitive season in female athletes: a preliminary investigation. J Sports Med Phys Fitness 51: 515–524, 2011 [PubMed] [Google Scholar]
- 70.Munevar S, Wang Y, and Dembo M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J 80: 1744–1757, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, and Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90, 2002 [DOI] [PubMed] [Google Scholar]
- 72.Neuman KC, Chadd EH, Liou GF, Bergman K, and Block SM. Characterization of photodamage to Escherichia coli in optical traps. Biophys J 77: 2856–2863, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park SH, Barish R, Li H, Reif JH, Finkelstein G, Yan H, and Labean TH. Three-helix bundle DNA tiles self-assemble into 2D lattice or 1D templates for silver nanowires. Nano Lett 5: 693–696, 2005 [DOI] [PubMed] [Google Scholar]
- 74.Park SH, Yin P, Liu Y, Reif JH, LaBean TH, and Yan H. Programmable DNA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett 5: 729–733, 2005 [DOI] [PubMed] [Google Scholar]
- 75.Pelham RJ, Jr., and Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA 94: 13661–13665, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Plodinec M. and Schoenenberger CA. Spatial organization acts on cell signaling: how physical force contributes to the development of cancer. Breast Cancer Res 12: 308, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poelmann RE, Gittenberger-de Groot AC, and Hierck BP. The development of the heart and microcirculation: role of shear stress. Med Biol Eng Comput 46: 479–484, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rizzo MA, Springer GH, Granada B, and Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol 22: 445–449, 2004 [DOI] [PubMed] [Google Scholar]
- 79.Robling AG, Niziolek PJ, Baldridge LA, Condon KW, Allen MR, Alam I, Mantila SM, Gluhak-Heinrich J, Bellido TM, Harris SE, and Turner CH. Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J Biol Chem 283: 5866–5875, 2008 [DOI] [PubMed] [Google Scholar]
- 80.Sachs F. and Morris CE. Mechanosensitive ion channels in nonspecialized cells. Rev Physiol Biochem Pharmacol 132: 1–77, 1998 [DOI] [PubMed] [Google Scholar]
- 81.Saeger J, Hytonen VP, Klotzsch E, and Vogel V. GFP's mechanical intermediate states. PLoS One 7: e46962, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saif MT, Sager CR, and Coyer S. Functionalized biomicroelectromechanical systems sensors for force response study at local adhesion sites of single living cells on substrates. Ann Biomed Eng 31: 950–961, 2003 [DOI] [PubMed] [Google Scholar]
- 83.Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 23: 160–170, 2008 [DOI] [PubMed] [Google Scholar]
- 84.Schwarz US, Balaban NQ, Riveline D, Bershadsky A, Geiger B, and Safran SA. Calculation of forces at focal adhesions from elastic substrate data: the effect of localized force and the need for regularization. Biophys J 83: 1380–1394, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shin JH, Tam BK, Brau RR, Lang MJ, Mahadevan L, and Matsudaira P. Force of an actin spring. Biophys J 92: 3729–3733, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shroff H, Reinhard BM, Siu M, Agarwal H, Spakowitz A, and Liphardt J. Biocompatible force sensor with optical readout and dimensions of 6 nm3. Nano Lett 5: 1509–1514, 2005 [DOI] [PubMed] [Google Scholar]
- 87.Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, and Vogel V. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol 5: e268, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith SB, Cui Y, and Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 271: 795–799, 1996 [DOI] [PubMed] [Google Scholar]
- 89.Smith SB, Finzi L, and Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science 258: 1122–1126, 1992 [DOI] [PubMed] [Google Scholar]
- 90.Spagnoli C, Beyder A, Besch S, and Sachs F. Atomic force microscopy analysis of cell volume regulation. Phys Rev E Stat Nonlin Soft Matter Phys 78: 031916, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strick TR, Allemand JF, Bensimon D, Bensimon A, and Croquette V. The elasticity of a single supercoiled DNA molecule. Science 271: 1835–1837, 1996 [DOI] [PubMed] [Google Scholar]
- 92.Suchyna TM. and Sachs F. Mechanosensitive channel properties and membrane mechanics in mouse dystrophic myotubes. J Physiol 581: 369–387, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suresh S. Biomechanics and biophysics of cancer cells. Acta Biomater 3: 413–438, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Szent-Gyorgyi A. Molecules, electrons and biology. Trans NY Acad Sci 31: 334–340, 1969 [DOI] [PubMed] [Google Scholar]
- 95.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, and Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci USA 100: 1484–1489, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Loenhout MT, De Vlaminck I, Flebus B, den Blanken JF, Zweifel LP, Hooning KM, Kerssemakers JW, and Dekker C. Scanning a DNA molecule for bound proteins using hybrid magnetic and optical tweezers. PLoS One 8: e65329, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang A. and Zocchi G. Elastic energy driven polymerization. Biophys J 96: 2344–2352, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, and Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol 137: 231–245, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K, Appleby M, Brunkow ME, and Latham JA. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J 22: 6267–6276, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wirtz D, Konstantopoulos K, and Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer 11: 512–522, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wirtz HR. and Dobbs LG. The effects of mechanical forces on lung functions. Respir Physiol 119: 1–17, 2000 [DOI] [PubMed] [Google Scholar]
- 102.Wozniak MA. and Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol 10: 34–43, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu W, Mezencev R, Kim B, Wang L, McDonald J, and Sulchek T. Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PLoS One 7: e46609, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang J, Li CW, and Yang M. Hydrodynamic simulation of cell docking in microfluidic channels with different dam structures. Lab Chip 4: 53–59, 2004 [DOI] [PubMed] [Google Scholar]
- 105.Yang MT, Fu J, Wang YK, Desai RA, and Chen CS. Assaying stem cell mechanobiology on microfabricated elastomeric substrates with geometrically modulated rigidity. Nat Protoc 6: 187–213, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yano T, Oku M, Akeyama N, Itoyama A, Yurimoto H, Kuge S, Fujiki Y, and Sakai Y. A novel fluorescent sensor protein for visualization of redox states in the cytoplasm and in peroxisomes. Mol Cell Biol 30: 3758–3766, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu H, Mouw JK, and Weaver VM. Forcing form and function: biomechanical regulation of tumor evolution. Trends Cell Biol 21: 47–56, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]