Abstract
Autophagy is a highly conserved cellular process responsible for recycling of intracellular material. It is induced by different stress signals, including starvation, cytokines, and pathogens. Type I interferons (IFN) are proteins with pleiotropic functions, such as antiviral, antiproliferative, and immunomodulatory activities. Several recent studies showed type I IFN-induced autophagy in multiple cancer cell lines as evidenced by autophagic markers, for example, the conversion of microtubule-associated protein 1 light chain 3 beta (MAP1LC3B, also known as LC3-I) to LC3-II and the formation of autophagosomes by electron microscopy. In addition, studies suggest the involvement of Janus kinase (JAK)/signal transducer and activator of transcription (STAT) and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/v-akt murine thymoma viral oncogene homolog (AKT) and mechanistic target of rapamycin, serine/threonine kinase (mTOR) pathways in the induction of autophagy. This review highlights a new function of type I IFN as an inducer of autophagy. This new function of type I IFN may play an important role in viral clearance, antigen presentation, inhibition of proliferation, as well as a positive feedback loop for the production of type I IFN.
Introduction
Autophagy is a cellular catabolic process governing the degradation and recycling of intracellular material. This process is evolutionarily conserved in eukaryotic cells. There are 3 distinct autophagic pathways: chaperone-mediated autophagy, microautophagy, and macroautophagy (hereafter referred to as autophagy) (Chen and Klionsky 2011). The process of autophagy includes the formation of pre-autophagosomal structures known as phagophores, which elongate and mature to form double-membraned autophagosomes that contain cytosol as well as organelles. The autophagosome fuses with a lysosome to generate an autolysosome, the internal contents of which are degraded to generate nutrients for the cell (Deretic 2006; Schmid and Munz 2007) (Fig. 1). Autophagy is continuously occurring at a basal level in all cells and increases under stress conditions, such as starvation, the presence of intracellular pathogens, or inflammation (Deretic 2006; Chen and Klionsky 2011). Autophagy is generally believed to have the primary role in the maintenance of cell homeostasis and survival (Kroemer and Levine 2008).
FIG. 1.
The process of autophagy. During autophagy, cytoplasmic and organelle material is sequestered by expanded double membranes to form the autophagosome. The autophagosome then fuses with a lysosome to form an autolysosome, the internal material of which is then degraded.
Induction of Autophagy by Cytokines
Different cytokines can alter autophagy levels in cells (Chang and others 2010; Heaton and Randall 2010; Chen and Klionsky 2011). It was shown that interleukins (IL)-1, IL-2, IL-6, tumor necrosis factor (TNF) alpha, transforming growth factor beta, and interferon (IFN)-γ are autophagy inducers, whereas IL-4, IL-10, and IL-13 can block autophagy (Harris 2011). The proinflammatory cytokine IFN-γ promotes autophagy to eradicate intracellular bacterial pathogens, such as mycobacteria and chlamydia (Gutierrez and others 2004; Al-Zeer and others 2009). Although the mechanism of IFN-γ-induced autophagy is unclear, it was demonstrated that immunity-related GTPases, such as the immunity-related GTPase family M protein (Irgm1) and IFN-inducible member of the immunity-related GTPase family M protein (Irga6), as well as members of the 65-kDa guanylate binding protein family, help to facilitate IFN-γ-induced autophagy (Singh and others 2006; Al Zee and others 2009; Kim and others 2011). In contrast, IFN-γ-induced autophagy has been recently described in Irgm1−/− primary macrophages as signal transducer and activator of transcription (STAT)1 independent, but mitogen-activated protein kinase 14 (MAPK14, also known as p38 MAPK alpha) dependent (Matsuzawa and others 2012). Additionally, a role of interferon regulatory factor-1 (IRF1) and IRF8 in IFN-gamma activated autophagy has also been described (Li and others 2012; Ozato and others 2013). In the study by Li and others (2012), it was shown that induction of autophagy by IFN-γ may contribute to growth inhibition and cell death in human liver cancer cells. Although the primary role of autophagy is to protect cells against death, depending on specific circumstances, it can actually lead to cell death through a process called autophagic or programmed cell death II (PCDII), specifically if cells are defective in an apoptotic pathway (Yu and others 2004; Maiuri and others 2007). The ability of IFN-γ to induce PCDII in HeLa cells was demonstrated for the first time by Inbal and others (2002). Taken together, these studies point to the complexity and importance of autophagy regulation by cytokines in immune responses and inflammation.
Role of Autophagy in Induction of Type I IFN and Viral Replication
Type I IFNs are pleiotropic cytokines that induce antiviral, antiproliferative, and immunomodulatory effects in cells. The major human type I IFNs include the IFN-α subtypes, IFN-β, and IFN-ω. The role of autophagy in viral recognition and induction of type I IFN has been shown (Lee and others 2007). The authors of that study demonstrated the requirement for autophagy in the production of type I IFN by plasmacytoid dendritic cells (pDC) following infection of VSV via TLR7. A similar observation was reported after infection of pDC with HIV-1 (Zhou and others 2012). Viral DNA derived from HSV-1 has also been shown to trigger autophagy and type I IFN production through a mechanism dependent on stimulator of IFN gene protein (STING) (Rasmussen and others 2011). On the other hand, the nucleotide-binding domain leucine-rich repeat containing protein X1 (NLRX1) and Tu translation elongation factor (TUFM) reduced type I IFN production but enhanced autophagy, resulting in an increase of VSV titer in mouse embryonic fibroblast (MEFs) (Lei and others 2012). Importantly, the role of IFN-inducible protein kinase PKR and IFN-regulated 2′,5′-oligoadenylate synthetase (OAS)/RNase pathway in induction of autophagy and suppression of virus replication has been described (Talloczy and others 2006; Chakrabarti and others 2012). Several viruses (e.g. dengue virus, hepatitis C virus, encephalomyocarditis virus, chikungunya virus, poliovirus, and coxsackievirus B3) use autophagy to enhance their replication (Heaton and Randall 2010; Sun and others 2011; Lei and others 2013). Because those viruses can induce type I IFN (Iwasaki 2012), crosstalk between the IFN response and autophagic machinery likely impacts viral replication.
Induction of Autophagy by Type I IFN and Role of JAK/STAT Pathway in Induction of Autophagy by Type I IFN
As discussed in the previous section, a role for autophagy in induction of type I IFN has been reported. Our group first reported that human type I IFN has the ability to induce autophagy in a concentration-dependent manner, starting at 24 to 48 h post-treatment in a number of different cancer cell lines, including Daudi, HeLa, MDA-MB-231, T98G, and A549 (Fey and others 2007; Schmeisser and others 2013). These observations were also confirmed by other groups (Ambjørn and others 2013; Li and others 2013; Zhu and others 2013).
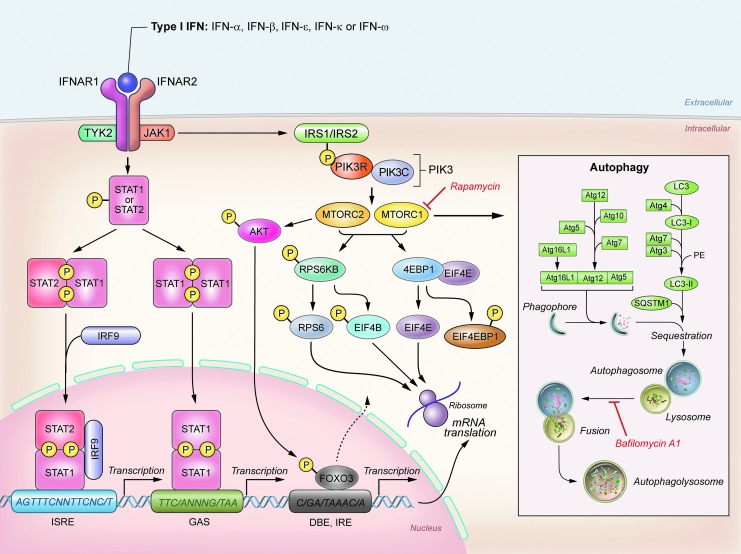
Type I IFN mediates signaling primarily through the Janus kinase (JAK)/STAT pathway (Schindler and others 1992). IFN binds to its receptor IFNAR1/2, which leads to the phosphorylation of STAT1 and STAT2 via tyrosine kinase (TYK) 2 and JAK1. The phosphorylated STATs then associate with IRF9 to form a transcriptional activator complex known as ISGF3 (Fig. 2). The ISGF3 complex translocates to the nucleus, binds to the promoter regions of IFN-stimulated genes (ISGs), and activates their transcription (Kessler and others 1990; Levy and Darnell 1990) (Fig. 2). In addition to STAT1 and STAT2, type I IFN can phosphorylate STAT3, 4, 5, and 6 in a cell type-specific manner (Darnell 1997; Stark and others 1998).
FIG. 2.
Activation of JAK/STAT and PI3K/AKT/mTOR pathways by type I interferons (IFNs). Type I IFNs bind to type I IFN receptor, which comprises 2 subunits on the cell surface, interferon receptor subunit 1 (IFNAR1) and 2 (IFNAR2). Two kinases associate with the IFN receptor: Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2). The binding of type I IFN to its receptor leads to the activation of JAK1 and TYK2, which subsequently mediate phosphorylation of signal transducer and activator of transcription 1 (STAT1) and STAT2, which in turn is necessary for the formation of a complex with IRF9 (ISGF3). This complex translocates to the nucleus, where it binds to IFN-stimulated response elements (ISREs), resulting in the transcription of interferon-stimulated genes (ISGs). Additionally, IFN activates the PI3K/AKT/mTOR pathway. Activation of this pathway is dependent on JAK1/TYK2, which phosphorylate insulin receptor substrate 1 (IRS1) and 2 (IRS2). Phosphorylation of IRS1 and IRS2 leads to the activation of PI3K, which subsequently activates mammalian target of rapamycin complex 1 (mTORC1). MTORC1 activates ribosomal protein S6 kinase (RPS6KB, also known as p70S6 kinase), which then phosphorylates ribosomal protein S6 (RPS6), leading to initiation of mRNA translation. Additionally, mTORC1 also regulates phosphorylation of eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1, also known as translation repressor 4EBP1). Phosphorylation of EIF4EBP1 causes inactivation of EIF4EBP1, which then dissociates from eukaryotic translation–initiation factor E (EIF4E), leading to initiation of cap-dependent mRNA translation. Type I IFN also activates rapamycin-insensitive complex (mTORC2), which is required for phosphorylation of RPS6KB, RPS6, and EIF4BP1. Although upstream signals leading to the activation of mTORC2 complex by type I IFN remain unclear, mTORC2 can activate AKT (Ser437), which plays a role in cell survival and has multiple targets, for example, forkhead box transcription factors (FOXO3). Interestingly, FOXO3 is considered a regulator of the transcription of some IFN-stimulated and autophagy genes. Inhibition of mTORC1 leads to the induction of autophagy. Ubiquitin-like conjugation systems composed of a variety of autophagy-related proteins (Atg) are involved in the process of vesicle expansion and completion. Bafilomycin A1 blocks fusion between autophagosomes and lysosomes.
Results of our study suggested a role for STAT2 in type I IFN-induced autophagy, as IFN-α was unable to induce autophagy in STAT2-deficient Daudi cells (Schmeisser and others 2013). Interestingly, Orvedahl and others (2011) used an image-based genome-wide siRNA screen to identify members of JAK/STAT signaling pathway (e.g. STAT2, TYK2) as candidates important for the destruction of viral components via autophagy (viral autophagy, virophagy) (Orvedahl and others 2011). Moreover, Zhu and others (2013) showed that leukocyte IFN was unable to increase the conversion of microtubule-associated protein 1 light chain 3 beta (MAP1LC3B, also known as LC3-I) to LC3-II after knockdown of JAK1 or STAT1 in K562 cells. Results of this study also highlighted the influence of STAT1 and nuclear factor of kappa light polypeptide gene enhancer in B-cells (NF-κB) on beclin 1 (BECN1) expression and BECN1-PtdIns3K (class II phosphatidylinositol 3-kinase, also known as PI3K-III) complex formation. Although a role of this complex in induction of autophagy had been previously suggested (Kang and others 2011), its role in type I IFN-induced autophagy and a detailed mechanism of involvement of STAT1 and NF-κB in complex formation remain to be explored. In this context, it is important to point out that type I IFN may promote cell survival by activation of NF-κB through the PI3K/AKT pathway (Yang and others 2001). Based on that study, the activation of NF-κB requires STAT3, which acts as an adapter for PI3K. Recently, Vogt and Hart (2011) suggested that there may be a functional link between PI3K/mTOR and STAT3, but the mechanism of this interdependence as well as its role in type I IFN-induced autophagy have yet to be explored. Thus, STAT3 could be an important component that links different signaling pathways. Ambjørn and others (2013) observed an effect of STAT1 silencing on increasing the levels of SQSTM1 (sequestosome 1, also known as p62) but not LC3-II in MCF-7 cells treated with STAT1 siRNA following the treatment with type I IFN. When examined together, the results from the above-mentioned studies provide first evidence that JAK/STAT pathway plays an important role in induction of autophagy by type I IFN.
Role of PI3K-mTOR Pathway in Induction of Autophagy by Type I IFN
In addition to the JAK/STAT pathway, type I IFN also activates the PI3K/AKT/mTORC1 signaling pathway, which is required for transcription and/or mRNA translation of ISGs (Nguyen and others 2001; Platanias 2005; Kaur and others 2008) and promotes cell survival (Ruuth and others 2001; Barca and others 2003). This pathway is also important in regulating autophagy (Chen and Klionsky 2011). A wide range of extracellular signals activate PI3K. For example, PI3K is activated after phosphorylation of insulin receptor substrate 1 (IRS1). Interestingly, type I IFN can induce phosphorylation of IRS1, which provides the docking site for PI3K (Uddin 1995). Active ribosomal protein S6 kinase (RPS6KB, also known as p70S6 kinase) is a negative regulator of IRS1. Thus, feedback inhibition of IRS1 leads to the repression of upstream signaling through PI3K (Tremblay and Marette 2001; Ma and Blenis 2009). A role for PI3K in type I IFN-induced autophagy has been shown by Li and others (2013) in human glioma cells using the chemical inhibitor 3-methyladenine (3-MA), where treatment of glioma cells with 3-MA impaired the induction of autophagy by IFN-β. We showed that nonsaturating concentrations of the PI3K inhibitor LY294002 can augment an increase of LC3-II levels stimulated by IFN-α treatment (Schmeisser and others 2013). The discrepancy between these results could possibly be explained by the activation of RPS6KB-IRS1-PI3K feedback loop in T98G cells, which may lead to the activation of PI3K and thus inhibition of mTORC1. Although the role of RPS6KB-IRS1-PI3K in type I IFN-induced autophagy remains to be explored, results of both studies underscore the importance of PI3K in regulating type I IFN-induced autophagy.
AKT is a downstream substrate of PI3K and had been discovered as a viral proto-oncogene capable of transforming certain cells (Brazil and Hemming 2001). It has been shown that AKT activity is required for the replication of paramyxoviruses (Sun and others 2008). Phosphorylation of AKT (Ser473) occurs shortly after cells are infected with flaviviruses (Lee and others 2005; Das and others 2010). Additionally, influenza viruses also activate the PI3K/AKT pathway (Ehrhardt 2007; Zhirnov and Klenk 2007; Jackson and others 2010). Although the biological significance of this process remains unknown, it has been hypothesized that PI3K/AKT activation may delay virus-induced apoptosis. Thus, these studies suggest that viruses may influence the balance of apoptosis and autophagy to maximize viral replication. The elucidation of this balance is paramount for understanding viral replication and its connection to the cell cycle.
Role of the MAPK Pathway in Induction of Autophagy by Type I IFN
There is increasing evidence that MAPK signaling plays an important role in complementing JAK/STAT signaling pathway for optimal transcription of ISGs (Katsoulidis and others 2005). Matsuzawa and others (2012) suggested a role for MAPK14 (also known as p38) in induction of autophagy by IFN-γ in primary mice macrophages. Our recent study showed that MAPK signaling pathways may also play a role in regulating type I IFN-induced autophagy. Although we observed a correlation between the induction of autophagy and inhibition of phosphorylation at residues Thr202 and Tyr204 of MAPK1 and 3 (also known as ERK1 and 2), the presence of nonsaturating concentrations of PD98059 (inhibitor of MEK1 and 2, upstream regulators of ERK) and IFN-α in Daudi cells did not increase the levels of LC3-II (Schmeisser and others 2013). Interestingly, the use of a different MEK1 and 2 inhibitor (U0126) in both U251 and U87MG cells abrogated the induction of autophagy by IFN-β (Li and others 2013). Taken together, these results emphasize the need to further explore the role of MAP kinases in the induction of autophagy by type I IFN.
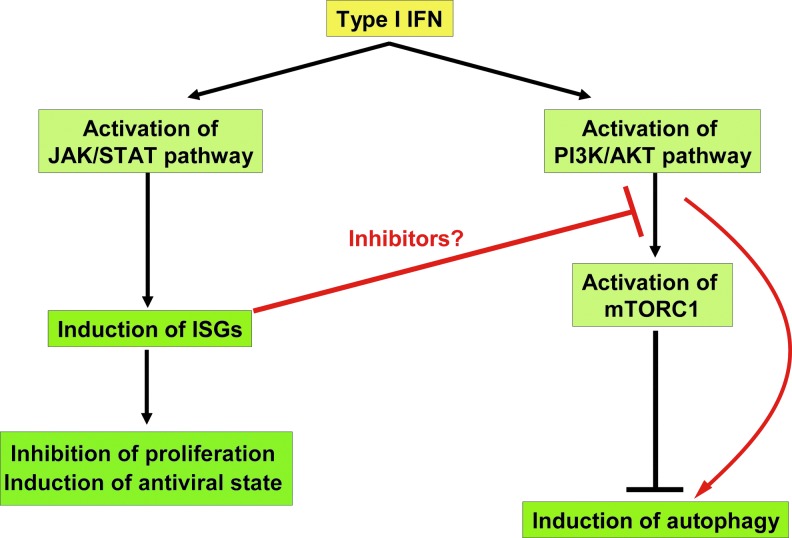
In summary, recent studies of autophagy induction by type I IFN implicate the involvement of JAK/STAT and MAPK signaling, as well as the PI3K/AKT/mTOR signaling axis. Considering that STAT1 and STAT2 are critical factors in autophagy induction and that mTORC1 is inhibited by type I IFN at later time points in different cancer cell lines, these data would suggest the involvement of yet unknown functions of ISGs in the induction of autophagy. Our current model of induction of autophagy by type I IFN is presented in Fig. 3.
FIG. 3.
Proposed model of induction of autophagy by type I IFN. Type I IFNs activate the JAK/STAT pathway, which results in the production of ISGs. Additionally, type I IFNs activate PI3K/AKT/mTOR signaling pathway. We propose that at later time points, negative regulators of PI3K/AKT/mTOR pathway are induced, which inhibit mTORC1 activity and lead to induction of autophagy.
mTORC1 Activity and Induction of Autophagy by Type I IFN
An important component of the PI3K/AKT/mTOR pathway is the mammalian target of rapamycin complex 1 (mTORC1) (Wullschleger and others 2006). Rapamycin inhibits the ability of mTORC1 to phosphorylate downstream substrates, such as RPS6KB and eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1, also known as translation repressor 4EBP1) (Zoncu and others 2011). The mTORC1 complex is composed of serine/threonine protein kinase (mTOR), mLST8 (mTOR-associate protein, LST8 homolog Saccharomyces cerevisiae), Raptor (regulatory associated protein of mTOR, complex 1), and Pras40 (proline-rich AKT1 substrate). mTOR and mLST8 are also part of the mTORC2 complex, which differs from mTORC1 by containing Rictor (rapamycin-insensitive companion of mTOR) and Sin1 (MAPK-associated protein 1) instead of Raptor and Pras40. While mTORC1 governs protein and lipid synthesis as well as mitochondrial metabolism, mTORC2 is a key regulator in cell cycle progression, anabolism, and cell survival. The activity of mTORC1 is regulated by growth factors, nutrients, energy, and stress. One way of inducing mTORC1 is through AKT-mediated phosphorylation of tuberous sclerosis protein 2 (TSC2) (Manning and others 2005). However, AMP-activated kinase (AMPK) can inhibit mTORC1 upon nutrient deprivation (Zoncu and others 2011).
Recently published results by our group (Schmeisser and others 2013) and others (Ambjørn and others 2013; Li and others 2013) suggest that type I IFN may block the function of mTORC1, which leads to the induction of autophagy. To this end, we showed that the treatment of Daudi and T98G cells with nonsaturating concentrations of rapamycin (mTORC1 inhibitor) and IFN-α had an additive effect for autophagy induction, increasing the levels of LC3-II generation. Similarly, the knockdown of mTOR (catalytic subunit of mTORC1) using MTOR siRNA in T98G cells enhanced the ability of IFN-α to increase LC3-II levels. Similar to our results, Ambjørn and others (2013) showed that combinatory treatment of MCF-7 cells with IFN-β and nonsaturating concentrations of rapamycin had an additive effect on the decrease in the level of autophagy marker SQSTM1, supporting the concept that mTORC1 activation opposes pro-autophagy signals conveyed by type I IFN. RPS6KB kinase and translation repressor protein EIF4EBP1 (Fig. 3) are proteins located downstream of mTORC1. We observed that the induction of autophagy by IFN-α correlated with changes in the phosphorylation state of mTORC1 target proteins RPS6KB (Thr389) and EIF4EBP1 (Thr37/Thr46) in Daudi cells starting at 24 h (Schmeisser and others 2013). Both of these proteins play a key role in protein synthesis (Platanias 2005). Ambjørn and others (2013) observed a decrease in phosphorylation of EIF4EBP1 (Thr37/Thr46) in MCF-7 cells starting 12 h after IFN-β treatment, and Li and others (2013) reported that IFN-β may decrease the phosphorylation of AKT (Ser473), mTOR (Ser2448), as well as RPS6KB (Ser371) in U251MG and U87MG cells at 48 h. Taken together, these results suggest that type I IFN-mediated autophagy may proceed with the concomitant inactivation of mTORC1 signaling.
Type I IFN may also regulate the activity of mTORC2. mTORC2-dependent activation of AKT (Ser473) in MEFs by type I IFN has been reported (Kaur and others 2012). The authors of that study showed that mTORC2 plays an important role in driving the expression of ISGs and therefore the generation of the IFN biological response. Interestingly, the transcription factor FOXO3 is a downstream substrate of AKT (Ser473) (Cybulski and Hall 2009). It has been reported that the inactivation of the PI3K/AKT pathway by type I IFN in mice monocytes leads to the degradation of FOXO3 (a repressor of IRF7) and thus feed-forward induction of IFN-β (Litvak and others 2012). Additionally, FOXO3 is also a direct transcription regulator of autophagy genes (Warr and others 2013). Thus, mTORC2/AKT activation by type I IFN may support autophagy induction through FOXO3 regulation (Fig. 2).
Effect of Autophagy on Tumor Growth
An altered level of autophagy has been observed in tumor cells. A rise in the level of autophagy may temporarily enhance the survival of cancer cells but can eventually trigger cell death, especially if cells are apoptosis-defective (Maiuri and others 2007). Increased autophagy was detected in malignant melanoma (Lazova and others 2009) as well as in various gastrointestinal tumors (Yoshioka 2008), human pancreatic cancer cell lines, and tumor specimens (Yang and others 2011). This increase in autophagy may contribute to the resistance of cancer cells to environmental stress or cytotoxic drugs. For example, tumor suppressor phosphatase and tensin homolog (PTEN) is a positive regulator of autophagy, whereas Rat sarcoma (Ras), which activates the catalytic subunit (p110 alpha) of PI3K class I (Rodriguez-Viciana and others 1994), is involved in negative regulation of autophagy. Loss of PTEN or Ras functions could affect the regulation of autophagy and lead to tumor progression (Arico 2001; Furuta and others 2004). Thus, it is possible that type I IFN augments autophagy by interfering with Ras signaling.
Malignant cell development is often associated with abnormalities of the PI3K/AKT/mTOR signaling pathway (Yuan and Cantley 2008). AKT has many downstream targets. One of them is B-cell lymphoma 2 protein (Bcl-2), an anti-apoptotic protein that directly opposes autophagy in MCF-7 cells (Akar and others 2008; Oh and others 2011). The PI3K/AKT/mTOR pathway is a target for breast cancer and malignant glioma therapy (Fan and others 2010; Cidado and Park 2012). In this context, single inhibitors targeting either PI3K (LY294002, wortmannin) or mTOR (rapamycin), as well as dual PI3K/mTOR inhibitors (PI 103), were developed (Fan and others 2010). Rapamycin (sirolimus), the only drug commonly used as an autophagy inducer, and rapamycin analogues, temsirolimus and everolimus, are mTOR inhibitors approved by the U.S. Food and Drug Administration for the treatment of renal cell carcinoma, and are currently evaluated in Phase III studies in combination with type I IFN (Fasolo and others 2012). Although their mechanism of action is not known, it would be interesting to explore the role of PCDII in this treatment. Despite the extensive study of autophagy in cancer treatment, further studies will be required to find the mechanism(s) that can regulate autophagy and induction of apoptosis or PCDII. Such mechanism(s) would have a significant impact on the treatment of cancer.
It has been reported that autophagy correlates with cell cycle regulation, possibly through cyclin-dependent kinases, as a higher level of autophagy was detected in G1 and S phases of the cell cycle (Tasdemir and others 2007; Filippi-Chiela and others 2011). We have shown that type I IFN-induced autophagy correlates with cell cycle perturbation and inhibition of cellular proliferation in several cancer cell lines (Schmeisser and others 2013). Results of our study are in agreement with results published by Ambjørn and others (2013). Additionally, recent studies of type I IFN-induced autophagy indicated that blocking autophagy increased the pro-apoptotic effect of type I IFN (Ambjørn and others 2013; Li and others 2013; Zhu and others 2013). Zhu and others (2013) showed that the knockdown of STAT1 and NF-κB increased type I IFN-induced activation of Caspase 3 (CASP3) in K562 cells. They also showed that the knockdown of BECN1 did not influence the induction of TNF ligand (TNFSF10, also known as TRAIL) by type I IFN but increased the activation of CASP8 and 9 and cleavage of BH3 interaction domain death agonist (BID), as well as a decrease in mitochondrial membrane potential. In agreement with this observation, we previously reported type I IFN-induced CASP8-dependent apoptosis in OVCAR-3 cells (Miyake and others 2012; Tsuno and others 2012). Additionally, Li and others (2013) showed that the inhibition of type I IFN-induced autophagy by 3-MA or hydroxychloroquine can cause an increase in CASP3 cleavage in U251MG and U87MG glioma cells. Interestingly, treatment of glioma cells with pan-caspase inhibitor z-VAD-fmk not only inhibited caspase-dependent apoptosis but also impaired the levels of LC3-II and decreased the levels of double-membrane structures 48 h after IFN-β treatment, suggesting a dual function for caspases in regulating autophagy. An increase in these apoptotic markers was also observed by Ambjørn and others (2013) in MCF-7 cells after IFN-β treatment. They observed results similar to those published by Li and others (2013) in that the treatment of MCF-7 cells with pan-caspase inhibitor z-VAD-fmk prevented the induction of TUNEL-positive cells by IFN-β. However, the effect of z-VAD-fmk inhibitor on type I IFN-induced autophagy was not explored in this study. Silencing of core autophagy-related proteins (ATG) 5, ATG7, and unc-51-like autophagy activating kinase (ULK) 1/2 leads to robust induction of TUNEL-positive cells after IFN-β treatment (Ambjørn and others 2013). All the above-described observations are in contrast to data obtained by Buchser and others (2012), which showed that depending on cell type, IFN-α had either no significant effect (in pancreatic cell lines Panc 2.03 and T-24) or was able to inhibit autophagy (colon cancer cell line HCT116). This inhibition was even more pronounced when HCT116 cells were cocultured with lymphocytes in the presence of IFN-α. Thus, the authors of this study showed that type I IFN may play a role in regulation of lymphocyte-induced cell-mediated autophagy. The complexity of experimental approaches in this study could explain the discrepancies between observations made by Buchser and others (2012) and other groups (Ambjørn and others 2013; Li and others 2013; Schmeisser and others 2013; Zhu and others 2013). In summary, we would stress that the induction of autophagy by type I IFN may not be an unintended consequence of IFN signaling, or an attempt by transformed cells to withstand the pro-apoptotic function of IFN, but rather a new function for type I IFN, the biological significance of which has to be explored. Considering that type I IFN is clinically used for the treatment of viral infections, autoimmune disorders and certain cancers, the newly established connection between type I IFN and induction of autophagy is of substantial clinical importance.
Conclusions
Several recent studies reported the induction of autophagy by type I IFN in multiple cancer cell lines (Ambjørn and others 2013; Li and others 2013; Schmeisser and others 2013; Zhu and others 2013). An important conclusion from these studies is that autophagy may counteract the pro-apoptotic functions of type I IFN in tumor cells. The observation of this phenomenon opens many questions: How does type I IFN induce autophagy? What is the biological significance of type I IFN-induced autophagy? Which ISGs are involved in type I IFN-induced autophagy? What are the key factors that control the switch between autophagy and apoptosis? Does type I IFN-induced autophagy play a role in the immunomodulatory function of IFN? Future studies will be necessary to find the answers to those questions.
Acknowledgments
We thank Drs. A.L. Snow (USUHS), M. Lenardo (NIAID), F. Schmeisser (FDA), C. Balinsky (NIAID), and C. Johnson (NIAID) for reviewing the article and valuable discussions. This research was supported by the Intramural Research Program of the NIH (NIAID). We also thank Ms. Lydia Kibiuk (NIH, Medical Arts Branch) for her help in generating Figs. 1 and 2.
Author Disclosure Statement
The authors have no financial conflicts of interest.
References
- Akar U, Chaves-Reyez A, Barria M, Tari A, Sanguino A, Kondo Y, Kondo S, Arun B, Lopez-Berestein G, Ozpolat B. 2008. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy 4(5):669–679 [DOI] [PubMed] [Google Scholar]
- Al-Zeer MA, Al-Younes AI, Braun PR, Zerrahn J, Meyer TF. 2009. IFN-gamma-inducible Irga6 mediate host resistance against Chlamydia trachomatis via autophagy. PLoS One 4(2):e4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambjørn M, Ejlerskov P, Liu Y, Lees M, Jäätelä M, Issazadeh-Navikas S. 2013. IFNbeta1/interferon-beta-induced autophagy in MCF-7 breast cancer cells counteracts its proapoptotic function. Autophagy 9(3):287–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico S. 2001. Tumor suppressor PTEN positively regulate macrophagy by inhibiting the phosphatidylinositol 3-kinase/protein B pathway. J Biol Chem 276(38):35243–35246 [DOI] [PubMed] [Google Scholar]
- Barca O, Ferre S, Seoane M, Prieto J M, Lema M, Senaris R, Arce VM. 2003. Interferon beta promotes survival in primary astrocytes through phosphatidylinositol 3-kinase. J Neuroimmunol 139(1–2):155–159 [DOI] [PubMed] [Google Scholar]
- Brazil DP, Hemming BA. 2001. Ten years of protein kinase B signaling: a hard Akt to follow. Trends Biochem Sci 26(11):657–664 [DOI] [PubMed] [Google Scholar]
- Buchser W-J, Laskow T-C, Pavlik P-J, Lin H-M, Lotze M-T. 2012. Cell-mediated autophagy promotes cancer cell survival. Cancer Res 72(12):2978–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Ghosh PK, Banerjee S, Gaughan Ch, Silverman RH. 2012. RNase L triggers autophagy in response to viral infections. J Virol 86(20):11311–11321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YP, Tsai ChCh, Huang WCh, Wang ChY, Chen ChL, Lin YS, Kai JI, Hsieh ChY, Cheng YL, Choi PCh, Chen SH, Chang SP, Liu HS, Lin ChF. 2010. Autophagy facilitates IFN-gamma-induced Jak2-STAT1 activation and cellular inflammation J Biol Chem 285(37):28715–28721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Klionsky DJ. 2011. The regulation of autophagy–unanswered questions. J Cell Sci 124(Pt2):161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cidado J, Park B-H. 2012. Targeting the PI3K/Akt/mTOR pathway for breast cancer therapy. J Mammary Gland Biol Neoplasia 17(3–4):205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski N, Hall MN. 2009. TOR complex 2: a signaling pathway of its own. Trends Biochem Sci 34(12):620–627 [DOI] [PubMed] [Google Scholar]
- Darnell JE. 1997. Stats and gene regulation. Science 277(5332):1630–1635 [DOI] [PubMed] [Google Scholar]
- Das S, Charkraborty S, Basu A. 2010. Critical role of lipid rafts in virus entry and activation of phosphoinositide 3′ kinase/Akt signaling during early stages of Japanese encephalitis virus infection in neural stem/progenitor cells. J. Neurochem 115(2):537–549 [DOI] [PubMed] [Google Scholar]
- Deretic V. 2006. Autophagy as an immune defense mechanism. Curr Opin Immunol 18:375–382 [DOI] [PubMed] [Google Scholar]
- Ehrhardt C. 2006. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell Microbiol 8(8):1336–1348 [DOI] [PubMed] [Google Scholar]
- Fan QW, Cheng C, Hackett C. 2010. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal 3(147):ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasolo A, Sessa C, Targetin M. 2012. TOR pathways in human malignancies. Curr Pharm Des 18(19):2766–2777 [DOI] [PubMed] [Google Scholar]
- Fey SB, Schmeisser H, Mejido J, Nie H, Tsuno T, Zoon KC. 2007. Influence of IFN-α2c on growth arrest, apoptosis, and autophagy in Daudi cells. J Interferon Cytokine Res 98:738.(published abstract) [Google Scholar]
- Filippi-Chiela EC, Villodre ES, Zamin LL, Lenz G. 2011. Autophagy interplay with apoptosis and cell cycle regulation in the growth inhibition effect of resveratrol in glioma cells. PLoS One 6(6):e20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. 2004. Ras is involved in negative control of autophagy through the class I PI3-kinase. Oncogene 23(22):3898–3904 [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119(6):753–766 [DOI] [PubMed] [Google Scholar]
- Harris J. 2011. Autophagy and cytokines. Cytokine 56(2):140–144 [DOI] [PubMed] [Google Scholar]
- Heaton NS. and Randall G. 2010. Dengue virus–induced autophagy regulates lipid metabolism. Cell Host and Microbe 8(5):422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. 2002. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol 157(3):455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A. 2012. A virological view of innate immune recognition. Annu Rev Microbiol 66:177–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Killip MJ, Galloway CS, Russell RJ, Randall RE. 2010. Loss of function of the influenza A virus NS1 protein promotes apoptosis but this is not due to a failure to activate phosphatidilinositol 3-kinase (PI3K). Virology 396(1):94–105 [DOI] [PubMed] [Google Scholar]
- Kang R, Zeh HJ, Lotze MT, Tang D. 2011. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 18(4):571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsoulidis E, Li Y, Mears H, Platanias LC. 2005. The p38 mitogen activated protein kinase pathway in interferon signal transduction. J Interferon Cytokine Res 25(12):749–756 [DOI] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, Baker DP, Hay N, Fish EN, Platanias LC. 2008. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci U S A 105(12):4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Majchrzak-Kita B, Baker DP, Su B, Fish EN, Platanias L. 2012. Regulatory effects of mTORC2 complex in type I IFN signaling and in the generation of IFN responses. Proc Natl Acad Sci U S A 109(20):7723–7728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler DS, Veals SA, Fu XY, Levy DE. 1990. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev 4(10):1753–1765 [DOI] [PubMed] [Google Scholar]
- Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. 2011. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science 332(6030):717–721 [DOI] [PubMed] [Google Scholar]
- Kroemer G, Levine B. 2008. Autophagy cell death: the story of a misnomer. Nat Rev Mol Cell Biol 9(12):1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazova R, Klump V, Pawelek J. 2009. Autophagy in cutaneous malignant melanoma. J Cutan Pathol 37(2):256–268 [DOI] [PubMed] [Google Scholar]
- Lee CJ, Liao CL, Lin YL. 2005. Flavivirus activates phosphatidylinositol 3-kinase signaling to block caspase–dependent apoptotic cell death at the early stages of virus infection. J Virol 79(13):8388–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. 2007. Autophagy dependent viral recognition by plasmocytoid dendritic cells. Science 315(5817):1398–1400 [DOI] [PubMed] [Google Scholar]
- Lei Y, Wen H, Ting JPY. 2013. The NLR protein, NLRX1 and its partner TUFM reduce type I interferon, and enhance autophagy. Autophagy 9(3):1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG, Swanson KV, Wen K-W, Damania B, Moore ChB, Giguere PM, Siderovski DP, Hiscott J, Razani B, Semenkovich CF, Chen X, Ting JPY. 2012. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity 36(6):933–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Darnell JE, Jr., 1990. Interferon-dependent transcriptional activation: signal transduction without second messenger involvement? New Biol 2(10):923–928 [PubMed] [Google Scholar]
- Li P, Du Q, Cao Z, Guo Z, Evankovich J, Yan W, Chang Y, Shao L, Stolz DB, Tsung A, Geller DA. 2012. Interferon-gamma induces autophagy with growth inhibition and cell death in human hepatocellular carcinoma (HCC) cells through interferon regulatory factor-1 (IRF-1). Cancer Lett 314(2):213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhu H, Zeng X, Fan J, Qian X, Wang S, Wang Z, Sun Y, Wang X, Wang W, Ju D. 2013. Suppression of autophagy enhanced growth inhibition and apoptosis of interferon-beta in human glioma cells. Mol Neurobiol 47(3):1000–1010 [DOI] [PubMed] [Google Scholar]
- Litvak V, Ratushny AV, Lampano AE, Schmitz F, Huang AC, Raman A, Rust AG, Bergthaler A, Aitchison JD, Aderem A. 2012. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 490(7420):421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. 2009. Molecular mechanisms of mTOR-mediated translation control. Nat Rev Mol Cell Biol 10(5):307–318 [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. 2007. Self-eating and self killing: crosstalk between autophagy and apoptosis. Nat Rev 8(9):741–752 [DOI] [PubMed] [Google Scholar]
- Manning B-D, Logsdon MN, Lipovsky AI, Abbott D, Kwiatkowski DJ, Cantley LC. 2005. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev 19(15):1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa T, Kim BH, Shenoy AR, Kamitani S, Miyake M, Macmicking JD. 2012. IFN-gamma elicits macrophage autophagy via the p38 MAPK signaling pathway. J Immunol 189(2):813–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K, Bekisz J, Zhao T, Clark CR, Zoon K. 2012. Apoptosis-inducing factor (AIF) is targeted in IFN-alpha2a-induced Bid-mediated apoptosis through Bak activation in ovarian cancer cells. Biochim Biophys Acta 1823(2):1378–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Ramana CV, Bayes J, Stark GR. 2001. Roles of phosphatidylinositol 3-kinase in interferon-gamma-dependent phosphorylation of STAT1 on serine 727 and activation of gene expression. J Biol Chem 276(36):33361–33368 [DOI] [PubMed] [Google Scholar]
- Oh S, Xiaofei E, Ni D, Pirooz SD, Lee JY, Lee D, Zhao Z, Lee S, Lee H, Ku B, Kowalik T, Martin SE, Oh BH, Jung JU, Liang C. 2011. Downregulation of autophagy by Bcl-2 promotes MCF7 breast cancer cell growth independent of its inhibition of apoptosis. Cell Death Differ 18(3):452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin Ch, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier RJ, Xie Y, Levine B. 2011. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480(7375):113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozato K, Yoshimi R, Yoshida Y, Gupta M, Yoshii H, Munasinghe J, Maximova O, Xiong H, Wang H, Morse HC. 2013. IRF-8 regulates autophagy and activates microglia to exacerbate neuroinflammation. Cytokine 63:288.(published abstract) [Google Scholar]
- Platanias LC. 2005. Mechanisms of Type I and Type II interferon mediated signaling. Nature Rev Immunol 5(5):375–386 [DOI] [PubMed] [Google Scholar]
- Rasmussen SB, Horan KA, Holm ChK, Stranks AJ, Mettenleiter TC, Simon AK, Jensen SB, Rixon FJ, He B, Paludan SR. 2011. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J Immunol 187(10):5268–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. 1994. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370(6490):527–532 [DOI] [PubMed] [Google Scholar]
- Ruuth K, Carlsson L, Hallberg B, Lundgren E. 2001. Interferon-α promotes survival of human primary B-lymphocytes via phosphatidylinositol 3-kinase. Biochem Biophys Res Commun 284(3):583–586 [DOI] [PubMed] [Google Scholar]
- Schindler C, Shuai K, Prezioso VR, Darnell JE, Jr., 1992. Interferon–dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257(5071):809–813 [DOI] [PubMed] [Google Scholar]
- Schmeisser H, Fey SB, Horowitz J, Fischer ER, Balinsky CA, Miyake K, Bekisz J, Snow AL, Zoon KC. 2013. Type I interferons induce autophagy in certain human cancer cell lines. Autophagy 9(5):683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Munz C. 2007. Innate and adoptive immunity through autophagy. Immunity 27(1):11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Deretic V. 2006. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313(5792):1438–1441 [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu Rev Biochem 67:227–264 [DOI] [PubMed] [Google Scholar]
- Sun J, Mayura MD, Soong L, Ou JHJ. 2011. Tug-of-war between HCV and the host. Autophagy 7(11):1394–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Fuentes SM, Timani K, Sun D, Murphy Ch, Lin Y, August A, Teng MN, He B. 2008. Akt plays a critical role in replication of non-segmented negative-stranded RNA viruses. J Virol 82(1):105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z, Virgin HW, Levine B. 2006. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2(1):24–29 [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Tajeddine N, Vitale I, Criollo A, Vicencio JM, Hickman JA, Geneste O, Kroemer G. 2007. Cell cycle-dependent induction of autophagy, mitophagy and reticulophagy. Cell Cycle 6(18):2263–2267 [DOI] [PubMed] [Google Scholar]
- Tremblay F, Marette A. 2001. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem 276(21):38052–38060 [DOI] [PubMed] [Google Scholar]
- Tsuno T, Mejido J, Zhao T, Phillips T, Myers TG, Bekisz J, Zoon KC. 2012. BID is a critical factor controlling cell viability regulated by IFN-alpha. J Immunother 35(1):23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin S. 1995. IFN-alpha engages the insulin receptor substrate-1 to associate with the phosphatidylinositol 3′-kinase. J Biol Chem 270(27):15938–159341 [DOI] [PubMed] [Google Scholar]
- Vogt PK, Hart JR. 2011. PI3K and STAT3: A new alliance. Cancer Discov 1(6):481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J, Passegue E. 2013. FOXO3A directs a protective autophagy program in hematopoietic stem cells. Nature 494(7437):33–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124(3):5–19 [DOI] [PubMed] [Google Scholar]
- Yang CH, Murti A, Pfeffer SR, Kim JG, Donner DB, Pfeffer LM. 2001. Interferon α/β promotes cell survival by activating nuclear factor κB through phosphatidylinositol 3-kinase Akt. J Biol Chem 276(17):13756–13761 [DOI] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H. 2011. Pancreatic cancer requires autophagy for tumor growth. Genes Dev 25(7):717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka A. 2008. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int. J Oncol 33(3):461–468 [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. 2004. Regulation of an ATG-7 beclin program of autophagic cell death by caspase −8. Science 304(5676):1500–1502 [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. 2008. PI3K pathway alterations in cancer: variations in a theme. Oncogene 27(41):5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhirnov OP, Klenk HD. 2007. Control of apoptosis in influenza virus-infected cells by up-regulation of Akt and p53 signaling. Apoptosis 12(8):1419–1432 [DOI] [PubMed] [Google Scholar]
- Zhou D, Kang KH, Spector SA. 2012. Production of interferon alpha by human immunodeficiency virus type 1 in human plasmocytoid dendritic cells is dependent on induction of autophagy. J Infect Dis 205(8):1258–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Cao L, Yu Y, Yang L, Yang M, Liu K, Huang J, Kang R, Livesey KM, Tang D. 2013. Inhibition autophagy potentiates the anticancer activity of IFN1@/IFNalpha in chronic myeloid leukemia cells. Autophagy 9(3):317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12(1):21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]