Abstract
The autoinhibition/activation of the PMCA (plasma membrane Ca2+ -ATPase) involves conformational changes in the membrane region of the protein that affect the amount of lipids directly associated with the transmembrane domain. The lipid–protein-dependence of PMCA isoforms 2 and 4 expressed and obtained in purified form from Saccharomyces cerevisiae was investigated using the phosphatidylcholine analogue [125I]TID-PC/16 {l-O-hexadecanoyl-2-O-[9-[[[2-[125I]iodo-4-(trifluoromemyl-3H-diazirin-3-yl)benzyl]oxy]carbonyl]nonanoyl]-sn-glycero-3-phosphocholine}, which was incorporated into mixtures of dimyristoylphosphatidylcholine and the non-ionic detergent C12E10 [deca(ethylene glycol) dodecyl ether]. We found no differences between the recombinant PMCA4 and PMCA purified from erythrocytes (ePMCA). However, titration of the half-maximal activation by Ca2+/calmodulin of PMCA2 showed 30-fold higher affinity than PMCA4. PMCA2 exhibited a lower level of labelling in the autoinhibited conformation relative to PMCA4, indicating that the lower autoinhibition was correlated with a lower exposure to lipids in the autoinhibited state. Analysis of the lipid–protein stoichiometry showed that the lipid annulus of PMCA varies: (i) in accordance to the conformational state of the enzyme; and (ii) depending on the different isoforms of PMCA. PMCA2 during Ca2+ transport changes its conformation to a lesser extent than PMCA4, an isoform more sensitive to modulation by calmodulin and acidic phospholipids. This is the first demonstration of a dynamic behaviour of annular lipids and PMCA.
Keywords: autoinhibition, calmodulin, lipid–protein interaction, phospholipid, plasma membrane calcium-ATPase (PMCA)
INTRODUCTION
PMCA (plasma membrane Ca2+ -ATPase) is a P2-type ATPase that controls intracellular Ca2+ and the shape of the Ca2+ signal by actively extruding Ca2+ to the extracellular space [1]. It is a single polypeptide of approximately 135 kDa and ten transmembrane segments. Following solvation of the crystal structure of SERCA (sarcoplasmic/endoplsmic reticulum Ca2+ -ATPase), the soluble portion of the PMCA can be divided into phosphorylation (P), nucleotide-binding (N) and action (A) domains [2]. The PMCA is an autoinhibited P-ATPase that contains regulatory elements in the AL region in the A-M3 linker and at the C-terminal region. The interaction with CaM (calmodulin) or acidic lipids, or the removal of the autoinhibitory regions, activates the PMCA [3,4].
In humans, there are four genes that code for PMCA isoforms 1–4, and alternative splicing of mRNA occurring in each of the two regulatory regions adds more isoform diversity. PMCA4 is widely distributed in different tissues, whereas PMCA2 shows a more restricted expression pattern with predominance in excitable cells [5]. Previous studies using isolated membranes have shown that the recombinant PMCA4 and PMCA2 also differ in its degree of autoinhibition [6–8].
Lipids interact with membrane proteins in three different manners: (i) the annular shell of lipids bound to the protein surface which mediates between the protein and the bilayer; (ii) lipid molecules that are immersed in cavities and clefts of the protein surface that are more important in multimeric assemblies; and (iii) lipids which reside within a membrane protein or a membrane protein complex and are in unusual positions [9].
Although the bulk of the lipid molecules in contact with an intrinsic membrane protein act as a solvent for the protein, interacting with the protein relatively non-specifically, some proteins interact with much greater specificity with a small number of lipid molecules, these lipid molecules often being essential for activity and acting like a traditional cofactor [10]. The crystal structures of SERCA show that extensive conformational changes occur during the transport cycle and are transmitted to the membrane portion of the protein where the ion is translocated [2,11–14] The structure of the solvent lipids could be important in determining the conformation of the protein and hence its activity [10].
In previous studies, we assessed the overall exposure of a purified PMCA obtained from human erythrocytes (ePMCA) to surrounding lipids by quantifying the extent of protein labelling by the photoactivatable phosphatidylcholine analogue [125I]TID-PC/16 {l-O-hexadecanoyl-2-O-[9-[[[2-[125I]-iodo-4-(trifluoromemyl-3H-diazirin-3-yl)benzyl]oxy]carbonyl]-nonanoyl]-sn-glycero-3-phosphocholine} [15–18]. This probe locates in the phospholipid milieu and, upon photolysis, reacts indiscriminately with its molecular cage [19–21]. It is thus possible to analyse directly the interaction between a membrane protein and lipids belonging to its immediate environment [22]. Moreover, we showed that the incorporation of [125I]TID-PC/16 is a useful tool to evaluate the conformation of PMCA by the extent of labelling of the PMCA in its major known conformational states [17,18].
Recently, we investigated the annular lipid–protein stoichiometry in native pig kidney Na+/K+ -ATPase preparation [23]. In that study, we showed that the transmembrane domain of the Na+/K+ -ATPase in the E1 state is less exposed to the lipids than in E2, i.e. the conformational transitions are accompanied by changes in the number of annular lipids, but not in the affinity of these lipids for the protein [23].
In the present study, we have used the hydrophobic photolabelling method described previously [22] to study the non-covalent interactions between the membrane domain of the recombinant PMCA purified from Saccharomyces cerevisiae and surrounding phospholipids under different experimental conditions that lead to different degrees of autoinhibition. Furthermore, we compare the behaviour of expressed PMCA4 and ePMCA with that of the naturally activated PMCA2 showing a dynamic interaction of annular phospholipids and PMCA depending on the degree of autoinhibition.
EXPERIMENTAL
Reagents
All chemicals used were of analytical grade and purchased from Sigma Chemical Co. The reagents used in DNA manipulations were obtained from New England Biolabs and Qiagen.
Expression and purification of PMCA from yeast
PMCA4 (human isoform 4xb) was expressed and purified from S. cerevisiae strain DBY 2062 (MATa his4–619 leu2–3,112) as described previously [24–26]. The cDNA coding for PMCA2 (rat isoform 2zb) was a gift from Dr Gary Shull (Department of Molecular Genetics, Biochemistry and Microbiology, University of Cincinnati, Cincinnati, OH, U.S.A.) and was the same as in previous studies of PMCA4 and PMCA2 [7,8]. Yeast cells were transformed with the pMP625 vector containing a Leu+ marker and the PMAI promoter and transformants were selected for their ability to grow in the absence of leucine on plates containing 6.7 % YNB (yeast nitrogen base) without amino acids, 0.67 % complete supplemented medium lacking leucine (Leu− ), 2.2 % dextrose and 1.5 % agar.
Yeast culture (4 litres) expressing the protein was grown in YNB Leu− medium at 28°C in glass flasks with agitation and, after the culture reached an approximate D600 of 1.5, 14 litres of complete medium was added and the incubation was continued for 6 h (to a D600 of 4.0–5.0). Total yeast membranes were solubilized with C12E10 [deca(ethylene glycol) dodecyl ether] and the PMCA was purified using CaM-affinity chromatography as described previously [27], as modified by Filomatori and Rega [28]. Approximately 400 μg of purified PMCA was obtained from one batch of yeast.
Purification of ePMCA
ePMCA was isolated from CaM-depleted erythrocyte membranes using the CaM-affinity chromatography procedure [27], as modified by Filomatori and Rega [28]. The protein concentration after purification was approximately 10 μg/ml. No phospholipids were added at any step in the purification procedure. The purification procedure described preserves transport activity and maintains the kinetic properties and regulatory characteristics of the enzyme in its native milieu [27,28].
Measurement of Ca2+ -ATPase activity
ATPase activity was measured at 37°C by following the release of Pi from ATP as described previously [28]. The incubation medium was 120 mM KCl, 30 mM potassium Mops (pH 7.4), 3.75 mM MgCl2, 1mM EGTA, 80 μM C12E10, enough CaCl2 to give the desired final free Ca2+ concentration and DMPC (dimyristoyl phosphatidylcholine) at the concentration indicated in each Figure. When necessary, CaM and/or PS (L-α-phosphatidyl-L-serine) were added and their concentration is indicated in each Figure. The reaction was started by the addition of ATP (final concentration 2 mM). Release of Pi was estimated using the procedure of Fiske and Subbarow [29]. Measurements were performed in a Jasco V-630 Bio spectrophotometer.
Preparation of [125I]TID-PC/16
TTD-PC/16 {l-O-hexadecanoyl-2-O-[9-[[[2-(tributylstannyl)-4-(trifluoromethyl-3H-diazirin-3-yl)benzyl]oxy]carbonyl]nonanoyl]-sn-glycero-3-phosphocholine, tin precursor) was a gift from Dr J. Brunner (Department of Biochemistry, ETH Zurich, Zurich, Switzerland). [125I]TID-PC/16 was prepared by radioiodination of its tin precursor using the method of Weber and Brunner [21]. After the reaction was completed, the mixture was extracted with chloroform/methanol (2:1, v/v) and [125I]TID-PC/16 was purified by passage through a silica gel column (2.5 ml) using chloroform/methanol/water/acetic acid (65:25:4:1, by vol.) as solvent. The elution was monitored by TLC/autoradiography and the fractions containing the product were dried and stored at −20°C.
Labelling procedure
For labelling of PMCA, a dried film of the photoactivatable reagent was suspended in C12E10/DMPC or C12E10/PS mixed micelles containing 10 μg/ml membrane protein, 120 mM KCl, 30 mM potassium Mops (pH 7.4), 3.75 mM MgCl2, 1mMEGTA and, where indicated, CaCl2 to give the desired final free Ca2+ concentration. The CaM, DMPC and PS concentrations depend on the experimental strategy and are indicated in each Figure. The samples were incubated for 20 min at 37°Cbefore being irradiated for 15 min with light from a filtered UV source (λ≈360 nm).
Radioactivity and protein determination
Electrophoresis was performed according to the Tris/Tricine SDS/PAGE method [30]. Polypeptides were stained with Coomassie Blue R, the isolated bands were excised from the gel, and the incorporation of radioactivity was measured directly on a γ -counter. The amount of protein was quantified by eluting each stained band as described previously [31], including BSA in each gel as a standard for protein quantification. Specific incorporation was calculated as the ratio between measured radioactivity and amount of protein determined for each band.
Derivation of the lipid–protein stoichiometry from the photolabelling data
Displacement experiments were carried out as follows. A dried film of the photoactivatable reagent was suspended in C12E10 micelles containing the membrane protein (10 μg/ml), to which increasing amounts of DMPC suspended in the same detergent were added. Mixtures of DMPC/C12E10 were prepared by sonication for 15 min. The final C12E10 concentration was adjusted to 80 μM. Incorporation of the probe to the membrane protein was calculated as the ratio between the amount of radioactivity associated with the band and the amount of protein. The number of lipid molecules in direct contact with the protein (α) was calculated as the ratio between the incorporation of [125I]TID-PC/16 to the protein (c.p.m./mol of protein) and the specific radioactivity of the reagent for each amount of DMPC added (c.p.m./mol of PC). This value was corrected taking into account the yield of the labelling reaction (14 %, see [16] for an explanation).
A hyperbolic function (eqn 1):
| (1) |
was fitted to the experimental data (see Figure 4B) and its graphical representation is shown by continuous lines. Each plot includes data from three or four different experiments. Parameter a represents the maximum number of PC molecules in direct contact with the protein, and parameter b is the concentration of PC required for reaching half coverage of the membrane-embedded surface of the protein.
Figure 4. Lipid–protein stoichiometry of PMCA isoforms.
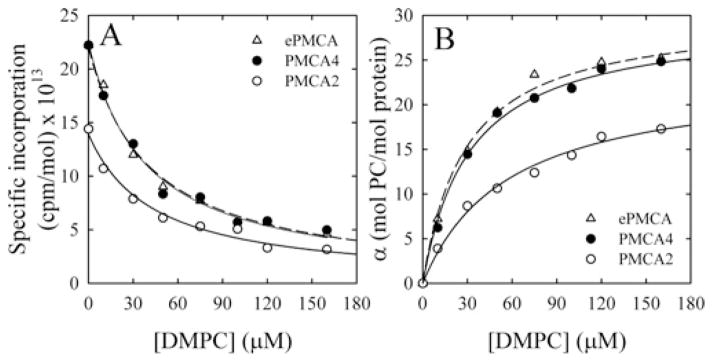
(A) [125I]TID-PC/16 specific incorporation into ePMCA, PMCA4 and PMCA2 in the presence of 100 μM for different DMPC concentrations. The continuous line represents the fit of hyperbolic decay to the experimental data. (B) The number of lipid molecules in direct contact with the pump, α, as a function of DMPC concentration under the conditions mentioned was calculated as the ratio between the incorporation of the probe to the protein (c.p.m./mol of protein) and the specific radioactivity of the photoactivatable reagent (c.p.m./mol of DMPC), corrected to the yield of the labelling reaction. The continuous lines are the graphical representation of eqn (1) fitted to the experimental data. The best fit values for parameter a for all conditions tested are shown in Table 1.
RESULTS
Activity of recombinant PMCA4 and PMCA2 expressed in yeast
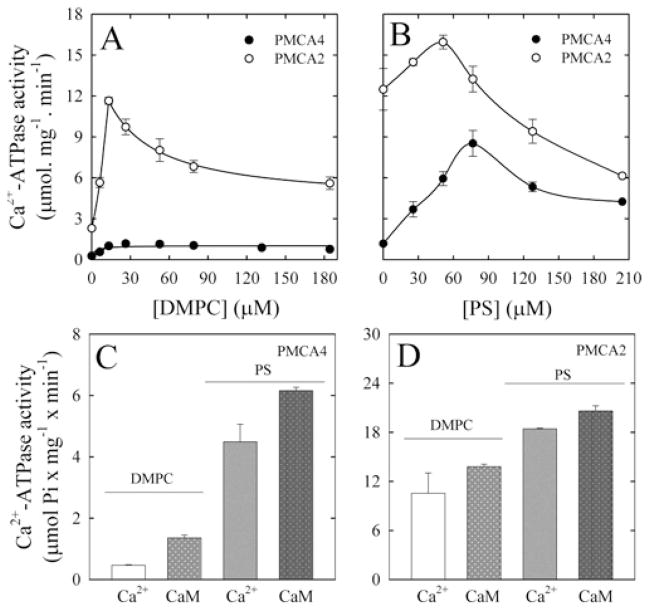
PMCA4 and PMCA2 were expressed in S. cerevisiae and purified by CaM-affinity chromatography. No exogenous lipids were added during the purification procedure. Figures 1(A) and 1(B) show the Ca2+ -ATPase activity of the recombinant proteins reconstituted with increasing concentrations of DMPC and/or PS, assayed in the presence of 100 μM Ca2+. The activity increased with the concentration of phospholipids to a maximum and then decreased. Under all conditions, PMCA2 activity was higher than that of PMCA4, but this latter isoform showed a larger activating effect in the presence of acidic phospholipids.
Figure 1. Effect of DMPC and PS concentration on PMCA.
Ca2+ -ATPase activity of purified recombinant PMCA4 and PMCA2 in the presence of 100 μM Ca2+ and as a function of DPMC (A) and PS (B) concentration. CaM activation was determined in the presence of 3 μM Ca2+ and 3 μM Ca2+ plus 100 nM CaM. PMCA4 was reconstituted with 30 μM DMPC or 30 μM DMPC/70 μM PS (C), and PMCA2 was reconstituted with 20 μM DMPC or 20 μM DMPC/50 μM PS (D).
Figures 1(C) and 1(D) show the activity of both PMCA4 and PMCA2 in the presence or absence of 100 nM CaM and at a concentration of Ca2+ of 3 μMto enhance the effect of PS or CaM activation of PMCA. However, the Ca2+ -ATPase activity in the absence of CaM is lower at this concentration of Ca2+ (compare Figures 1A and 1B with Figures 1C and 1D). Each isoform was reconstituted with the optimal phospholipid concentration as described in Figures 1(A) and 1(B).
Activation of PMCA2 by CaM was approximately 33 %, whereas activation of PMCA4bwas nearly 250 %. When PMCA4 was reconstituted in the presence of PS, activity increased nearly 10-fold; under those conditions, the enzyme is fully activated and CaM increased the activity barely 45 %. PMCA2 activity was higher than that of PMCA4 under all conditions tested; however, the presence of acidic phospholipids and CaM have a minimal activating effect, consistent with the fact that PMCA2 is a naturally activated isoform.
Relative specific incorporation of [125I]TID-PC/16 to PMCA under different conditions
We determined the extent of [125I]TID-PC/16 labelling of PMCA2 and PMCA4 in their major known conformational states. The E2 state was attained by incubating PMCAs in the absence of Ca2+ (1 mM EGTA). The absolute value of incorporation of [125I]TID-PC/16 to PMCA4 and PMCA2 (expressed as c.p.m. per mol of protein) were not significantly different between PMCA isoforms in the E2 conformation, thus the incorporation of [125I]TID-PC/16 under these conditions was considered as the control (100 %). Figure 2 also shows the specific [125I]TID-PC/16 incorporation to purified human ePMCA (the majority isoform is PMCA4b) under the same conditions as a control. The specific incorporation of the probe was similar to that obtained for the same isoform overexpressed in yeast, suggesting that the study of the transmembrane domain structure with [125I]TID-PC/16 in this system is representative of the isoforms obtained from their biological sources.
Figure 2. Specific incorporation of [125I]TID/PC16 to PMCA isoforms under different conditions.
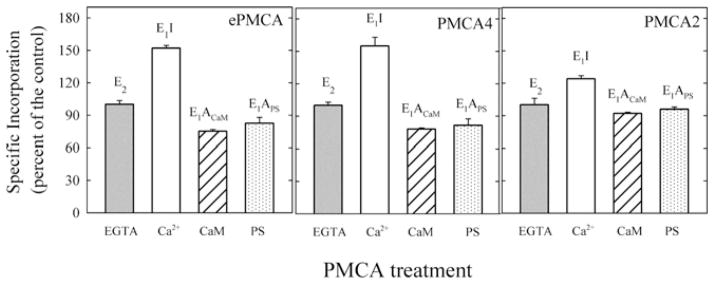
Relative specific incorporation of [125I]TID-PC/16 into ePMCA, PMCA4 and PMCA2 under different conditions: 1 mM EGTA (E2, EGTA); 100 μMCa2+ (E1I, Ca2+ ); 100 μMCa2+ and 200 nM CaM (E1ACaM, CaM); and 100 μM Ca2+ and 70–50 μM PS (E1APS, PS) for PMCA4, ePMCA and PMCA2. Incorporation in the presence of 1 mM EGTA for each isoform was taken as 100 % (control).
In the presence of Ca2+ and in the absence of activators, the PMCA enzymes are in the autoinhibited state E1I. As reported previously [17] for the purified ePMCA, under these conditions, the binding of [125I]TID-PC/16 was the highest. However, whereas the level of labelling of PMCA4 reached 155 ± 8 %, that of PMCA2 was 124 ± 2 %. In contrast, in the activated conformation E1A (E1ACaM), attained in the presence of Ca2+ and CaM, the labelling of PMCA4 and PMCA2 were reduced to 78 ± 1 %and 92 ± 1 %respectively. To determine the specific [125I]TID-PC/16 incorporation when the pump is activated with PS (E1APS), each isoform was reconstituted in the presence of its optimal phospholipid concentration. The specific incorporation to PMCA2 in the presence of PS was 96 ± 2 % and for PMCA4 was 82 ± 6 %. This value was not significantly different from the value obtained for ePMCA (81 ± 2 %).
These results indicate that autoinhibition in the presence of Ca2+ and activation by Ca2+/CaM or Ca2+/PS involve a small change in phospholipid exposure in the PMCA2 isoform compared with PMCA4.
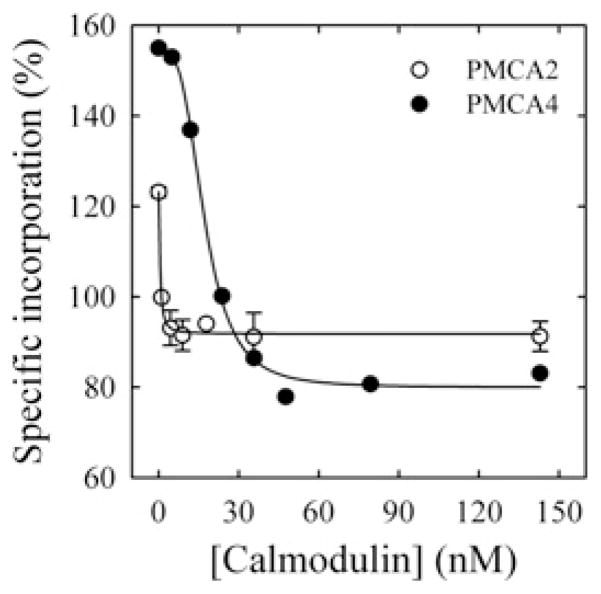
Titration of the E1I to E1A conformer shift with CaM
The dependence of the amount of bound [125I]TID-PC/16 to PMCA with the degree of activation of the PMCA was followed by quantifying the reduction in the labelling as a function of CaM concentration. In this case, each isoform was reconstituted in the present of DMPC, because the CaM activation is higher under these conditions than when the pump is pre-activated by acidic phospholipids (Figures 1C and 1D). Results in Figure 3 show that the concentration of CaM required for half-maximal reduction of the labelling was approximately 30-fold lower for PMCA2 than for PMCA4.
Figure 3. CaM-dependence of the incorporation of [125I]TID-PC/16 to PMCA4 (●) and PMCA2 (○).
Incorporation of [125I]TID-PC/16 was determined in a medium containing 100 μM Ca2+ at 37°C. Eqn (2) was fitted to the experimental points, where [PCB] is the percentage relative to EGTA condition of [125I]TID-PC/16 bound to PMCA at a given concentration of CaM; [PCmin] is the minimal amount (%) of [125I]TID-PC/16 bound to PMCA (at saturating concentration of CaM), [PC0] is the maximal amount (%) of PC bound to PMCA (at zero [CaM]) and K d(CaM) is the concentration of CaM for half-maximal binding to PMCA. This resulted in the following values: [PCmin] = 79.9 ± 1.9 %, [PC0] = 154.5 ± 2.2 %, n = 3.3 ± 0.4 and K d(CaM) = 17.6 ± 0.9 nM for PMCA4; and [PCmin] = 91.8 ± 0.9 %, [PC0] = 123.2 ± 1.4 %, n = 1.4 ± 0.6 and K d(CaM) = 0.52 ± 0.08 nM for PMCA2.
Lipid–protein stoichiometry
In order to determine the number of lipid molecules in direct contact with the protein (α), the displacement of the [125I]TID-PC/16 probe bound to ePMCA, PMCA4 and PMCA2 by non-radiolabelled DMPC was estimated. Figure 4 shows that the α parameter increases with the concentration of DMPC along a hyperbolic function. For best clarity, we show in Figure 4 only the results obtained in the presence of Ca2+ for PMCA2, PMCA4 and ePMCA as an example of all conditions tested and the maximum number of annular lipids determined in the same way are shown in Table 1.
Table 1.
Best fit values (mean ± S.E.M.) of the parameter a of the curves of α as a function of DMPC concentration for PMCA isoforms tested in different conformations
| Conformation | Lipid/protein stoichiometry a (mol of DMPC/mol of protein)
|
||
|---|---|---|---|
| PMCA4 | PMCA2 | ePMCA | |
| E2 | 19 ± 1 | 18 ± 1 | 19 ± 2 |
| E1I | 29 ± 1 | 23 ± 1 | 31 ± 1 |
| E1ACaM | 14 ± 1 | 17 ± 1 | 14 ± 1 |
| E1APS | 14 ± 2 | 17 ± 1 | 15 ± 2 |
In the presence of Ca2+ (E1I state), the number of DMPC molecules surrounding PMCA4 and ePMCA were similar. However, the number of annular DMPC molecules changed with the conformation of the protein, most notably from the Ca2+ -autoinhibited state E1I which showed approximately 29 lipid molecules/PMCA to the activated state E1A obtained in the presence of CaM or PS, which show only 14 lipid molecules/PMCA. On the other hand, PMCA2 is surrounded by 23 lipid molecules in the autoinhibited state and 17 in the fully activated state E1A, with either CaM or PS. The amounts of DMPC needed for half-maximal labelling were similar for all conditions of PMCA4, but lower than those for PMCA2 (results not shown).
DISCUSSION
Properties of PMCA2 purified from yeasts
PMCA4, the most abundant isoform in human erythrocytes, is the best known PMCA. Previous studies of the recombinant PMCA4 used isolated membranes and purified preparations of mammalian and insect PMCA-expressing cells [6–8]. Previously, we obtained PMCA4 in purified form from overexpressing yeasts [24–26]. In the present study, we show that the yeast expression system can be used successfully to obtain purified PMCA2, which is difficult to obtain from natural sources.
The specific Ca2+ -ATPase activity of the purified recombinant PMCAs is lower than the specific activity of ePMCA. The reason for this difference is still not known, but we have suggested previously that the lipid environment of the recombinant proteins may not be optimal for the preservation of the activity when faced with the detergents used in the purification [26]. This was particularly apparent when phosphatidylcholine was used to reconstitute the recombinant proteins. On the other hand, if acidic lipids were used for reconstitution, the activity of the recombinant proteins nearly reach that of ePMCA. Thus, despite these differences, the results of the present study show that the natural ePMCA and the recombinant PMCA4 have a similar number of annular lipids. Furthermore, the changes in the surrounding lipids associated with the activation/autoinhibition seem to not be dependent on the activity of the enzyme since they are preserved even when the full cycling of the pump is inhibited by EITC-PMCA (I.C. Mangialavori, M. Dalghi and J.P.F.C. Rossi, unpublished work).
In agreement with previous studies performed using cell membranes [6–8], we found that PMCA2 has a higher basal activity in the absence of CaM than PMCA4. Because PMCA2 was purified by CaM-affinity chromatography, no endogenous CaM should be present in the final preparation, thus the higher activity of PMCA2 cannot be due to residual CaM bound to the enzyme. Therefore our results confirm that PMCA2 is naturally less autoinhibited than PMCA4.
On the other hand, Cura et al. [26] reported that the activity of PMCA overexpressed in yeasts is restored in the presence of brain extract, a mixture rich in PS. In the present study, we used pure PS to rule out the possibility that the interaction between [125I]TID-PC/16 and the enzyme may be affected by the presence of other components in the brain extract. In the presence of PS, PMCA4 activity is increased 10-fold over the activity in the presence of DMPC, whereas PMCA2 activity is increased only twice. This result agrees with the fact that PMCA2 is partially activated, as observed after CaM activation as described above.
Autoinhibited state and activation of PMCA4 and PMCA2 as revealed by [125I]TID-PC/16 incorporation
The results of [125I]TID-PC/16 labelling of PMCA4 from S. cerevisiae are very similar to those reported for PMCA purified from membrane erythrocytes [17]; i.e. E1I for PMCA4 incorporates 155 ± 8 %, and E1A obtained in the presence of CaM or PS is 78 ± 1 % and 82 ± 6 % respectively. In contrast, in the presence of Ca2+, the incorporation of [125I]TID-PC/16 to PMCA2 was lower than in PMCA4, consistent with a less autoinhibited conformation. Interestingly, the level of labelling of PMCA2 in the E1A conformation at saturating concentrations of CaM (92 ± 1 %), or optimal PS concentration (96 ± 2 %), was significantly higher than that of PMCA4. Thus the active conformation of PMCA4 is less exposed to the surrounding lipids than is PMCA2. This fact also points out that differences between PMCA4 and PMCA2 still persist in the fully CaM-activated state of the proteins.
The titration of the E1I to E1A transition induced by Ca2+/CaM showed that PMCA2 has a higher apparent affinity for Ca2+/CaM. Because the binding affinity of Ca2+/CaM to the isolated binding site of PMCA2 is thought to be similar or higher than of the PMCA4, these results indicate that in PMCA4 either the CaM-binding domain is more occluded than in PMCA2 or that Ca2+/CaM dissociates faster from PMCA4 than from PMCA2. This is in agreement with previous studies by Caride et al. [32].
Lipid–protein stoichiometry
The displacement experiments showed that the levels of [125I]TIDPC/16 incorporation attained under different conditions do in fact reflect different numbers of maximal DMPC molecules in contact with the membrane-embedded surface of the proteins, as indicated by the different values of the parameter a. In the absence of Ca2+ (E2), both PMCA4 and PMCA2 have similar a values, a result which suggests that, in the E2 conformation, there is no significant difference in the lipid-covered surface, despite the different sequences among the proteins. However, a major difference was detected in the lipid–protein stoichiometry in the presence of Ca2+ between the autoinhibited and the activated forms. Results of the present study indicate that the activation involves a reorganization of the transmembrane region with the removal of lipid molecules from the protein annulus. Although in PMCA4, activation involved the loss of approximately 15 phospholipid molecules per protein, in PMCA2, the loss was of only six. This fact probably reflects that, during transport of Ca2+, PMCA2 changes its conformation to a lesser extent than PMCA4, an isoform more sensitive to modulation by CaM and acidic phospholipids.
The number of lipid molecules surrounding the membrane-spanning domain has been deduced for several proteins from EPR spin labelling studies [33], which quantify the association of motional restricted lipids with membrane proteins. In the present study, we have used the hydrophobic photolabelling method described previously [22] to study the non-covalent interactions between the membrane domain of the recombinant PMCA purified from S. cerevisiae and surrounding phospholipids under different experimental conditions that lead to different degrees of autoinhibition.
Considering that the average thickness of the transmembrane domain of the protein is approximately 30 Å (1 Å = 0.1 nm), the match between the lipidic domain and the protein should include tilting of the helices and rotation of side chains at the end of the helices, a fact demonstrated for SERCA by the analysis of crystals obtained at different conformations. We have calculated the accessible surface area of the SERCA transmembrane region using crystallographic data for several E1 and E2 conformers and found an approximately 20 % difference of exposure to annular lipids. This result agrees well with our experimental results using [125I]TID-PC/16 labelling [17].
Because the rate of exchange of lipid molecules between the annular shell around a membrane protein and the bulk phase is fast, the lipid–protein interaction has been described as ‘a non-sticky one’ [34]. In agreement with this, results of the present study show, for the first time, a dynamic interaction of annular phospholipids and PMCA under the different distinct conformations attained during the pumping cycle.
The transmembrane domain of PMCA (ten helices) can be represented by a cylinder with a diameter of 45 Å. Assuming that the lipid chains are circles 4.8 Å in diameter, we can calculate a mean of 29–30 annular lipids per molecule of PMCA4 in its loose conformation (E1I), a result that agrees well with the 29 phospholipid molecules found in our experiments. A relative small conformational change in the compaction or tilt of the transmembrane domain would be sufficient to explain the observed changes in the number of annular lipids occurring during activation. Moreover, the fact that the number of lipids exchanged during activation is lower for PMCA2 than for PMCA4 suggest that, during the reaction cycle, PMCA2 suffer a more limited rearrangement among the extreme conformations E1A, E2 and E1I, a fact that may be related to its higher basal activity.
During the preparation of the present paper, Sønntag et al. [35], using low-resolution X-ray crystallography, found evidence that the lipid bilayer and membrane protein suffer a mutual adaptation during conformational changes associated with the transport cycle of SERCA. They conclude that the membrane adjusts to its phospholipidic environment by side-chain rotations and small helix tilt changes. These results are in agreement with the dynamic lipid–protein stoichiometry described in the present paper for PMCA.
Acknowledgments
We are greatly indebted to Dr J. Brunner for his gift of TTD-PC/16 (tin precursor) and to Dr Rolando Rossi for helpful comments.
FUNDING
The present work was supported by the National Institutes of Health Fogarty International Center [grant number R03TW006837] and by Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Secretaria de Ciencia y Técnica de la Universidad de Buenos Aires (UBACyT) from Argentina.
Abbreviations used
- C12E10
deca(ethylene glycol) dodecyl ether
- CaM
calmodulin
- DMPC
dimyristoyl phosphatidylcholine
- PMCA
plasma membrane Ca2+ -ATPase
- ePMCA
erythrocyte PMCA
- PS
L-α-phosphadidyl-L-serine
- SERCA
sarcoplasmic/endoplasmic reticulum Ca2+ -ATPase
- [125I]TID-PC/16
l-O-hexadecanoyl-2-O-[9-[[[2-[125I]iodo-4-(trifluoromemyl-3H-diazirin-3-yl)benzyl]oxy]carbonyl]nonanoyl]-sn-glycero-3-phosphocholine
- TTD-PC/16
l-O-hexadecanoyl-2-O-[9-[[[2-(tributylstannyl)-4-(trifluoromethyl-3H-diazirin-3-yl)benzyl]oxy]carbonyl]nonanoyl]-sn-glycero-3-phosphocholine
- YNB
yeast nitrogen base
Footnotes
AUTHOR CONTRIBUTION
Irene Mangialavori obtained the [125I]TID-PC16 probe and carried out the experiments in collaboration with Gerardo Corradi. She participated in the design and discussion of the experiments. Gerardo Corradi carried out the expression of the PMCA isoforms in yeast and collaborated with Irene Mangialavori. He also contributed to the discussion of the experiments. Débora Rinaldi collaborated in the purification of PMCAs. María Candelaria de la Fuente collaborated with the purification of the recombinant PMCAs. Hugo Adamo collaborated with Juan Pablo Rossi with the design, supervision and writing of the paper.
References
- 1.Strehler EE, Filoteo AG, Penniston JT, Caride AJ. Plasma-membrane Ca2+ pumps: structural diversity as the basis for functional versatility. Biochem Soc Trans. 2007;35:919–922. doi: 10.1042/BST0350919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 3.Enyedi A, Verma AK, Filoteo AG, Penniston JT. A highly active 120-kDa truncated mutant of the plasma membrane Ca2+ pump. J Biol Chem. 1993;268:10621–10626. [PubMed] [Google Scholar]
- 4.de Tezanos Pinto F, Adamo HP. Deletions in the acidic lipid-binding region of the plasma membrane Ca2+ pump: a mutant with high affinity for Ca2+ resembling the acidic lipid-activated enzyme. J Biol Chem. 2002;277:12784–12789. doi: 10.1074/jbc.M111055200. [DOI] [PubMed] [Google Scholar]
- 5.Strehler EE, Zacharias DA. Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol Rev. 2001;81:21–50. doi: 10.1152/physrev.2001.81.1.21. [DOI] [PubMed] [Google Scholar]
- 6.Hilfiker H, Guerini D, Carafoli E. Cloning and expression of isoform 2 of the human plasma membrane Ca2+ -ATPase: functional properties of the enzyme and its splicing products. J Biol Chem. 1994;269:26178–26183. [PubMed] [Google Scholar]
- 7.Elwess NL, Filoteo AG, Enyedi A, Penniston JT. Plasma membrane Ca2+ pump isoforms 2a and 2b are unusually responsive to calmodulin and Ca2+ J Biol Chem. 1997;272:17981–17986. doi: 10.1074/jbc.272.29.17981. [DOI] [PubMed] [Google Scholar]
- 8.Caride AJ, Penheiter AR, Filoteo AG, Bajzer Z, Enyedi A, Penniston JT. The plasma membrane calcium pump displays memory of past calcium spikes: differences between isoforms 2b and 4b. J Biol Chem. 2001;276:39977–39804. doi: 10.1074/jbc.M104380200. [DOI] [PubMed] [Google Scholar]
- 9.Palsdottir H, Hunte C. Lipids in membrane protein structure. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Lee AG. Lipid–protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi M, Kondou Y, Toyoshima C. Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors. Proc Natl Acad Sci USA. 2007;104:5800–5805. doi: 10.1073/pnas.0700979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 13.Toyoshima C, Mizutani T. Crystal structure of the calcium pump with a bound ATP analogue. Nature. 2004;430:529–535. doi: 10.1038/nature02680. [DOI] [PubMed] [Google Scholar]
- 14.Olesen C, Picard M, Winther AML, Gyrup C, Morth JP, Oxvig C, Moller JV, Nissen P. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–1042. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 15.Villamil Giraldo AM, Castello PR, González Flecha FL, Delfino JM, Rossi JPFC. Phospholipid distribution around the plasma membrane calcium pump: a hydrophobic photolabeling study. Cell Biochem Biophys. 2006;44:431–437. doi: 10.1385/cbb:44:3:431. [DOI] [PubMed] [Google Scholar]
- 16.Villamil Giraldo AM, Castello PR, González Flecha FL, Moeller JV, Delfino JM, Rossi JPFC. Stoichiometry of lipid–protein interaction assessed by hydrophobic photolabeling. FEBS Lett. 2006;580:607–612. doi: 10.1016/j.febslet.2005.12.078. [DOI] [PubMed] [Google Scholar]
- 17.Mangialavori I, Villamil Giraldo AM, Marino Buslje C, Ferreira Gomes M, Caride AJ, Rossi JPFC. A new conformation in sarcoplasmic reticulum calcium pump and plasma membrane Ca2+ pumps revealed by a photoactivatable phospholipidic probe. J Biol Chem. 2009;284:4823–4828. doi: 10.1074/jbc.M806912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mangialavori I, Ferreira-Gomes M, Pignataro MF, Strehler EE, Rossi JPFC. Determination of the dissociation constants for Ca2+ and calmodulin from the plasma membrane Ca2+ pump by a lipid probe that senses membrane domain changes. J Biol Chem. 2010;285:123–130. doi: 10.1074/jbc.M109.076679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunner J, Semenza G. Selective labeling of the hydrophobic core of membranes with 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine, a carbene-generating reagent. Biochemistry. 1981;20:7174–7182. doi: 10.1021/bi00528a019. [DOI] [PubMed] [Google Scholar]
- 20.Brunner J. New photolabeling and crosslinking methods. Annu Rev Biochem. 1993;62:483–514. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- 21.Weber T, Brunner J. 2-(Tributylstannyl)-4-[3-(trifluoro-methyl)-3H-diazirin-3-yl]benzyl alcohol: a building block for photo-labeling and cross-linking reagents of very high specific radioactivity. J Am Chem Soc. 1995;117:3084–3095. [Google Scholar]
- 22.Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. H+ -induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 23.Mangialavori I, Montes MR, Rossi RC, Fedosova NU, Rossi JP. Dynamic lipid–protein stoichiometry in E1 and E2 conformations of the Na+/K+ -ATPase. FEBS Lett. 2011;585:1153–1157. doi: 10.1016/j.febslet.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 24.Bredeston LM, Adamo HP. Loss of autoinhibition of the plasma membrane Ca2+ pump by substitution of aspartic 170 by asparagine: activation of plasma membrane calcium ATPase 4 without disruption of the interaction between the catalytic core and the C-terminal regulatory domain. J Biol Chem. 2004;279:41619–41625. doi: 10.1074/jbc.M403116200. [DOI] [PubMed] [Google Scholar]
- 25.Corradi GR, Adamo HP. Intramolecular fluorescence resonance energy transfer between fused autofluorescent proteins reveals rearrangements of the N-and C-terminal segments of the plasma membrane Ca2+ pump involved in the activation. J Biol Chem. 2007;282:35440–35448. doi: 10.1074/jbc.M703377200. [DOI] [PubMed] [Google Scholar]
- 26.Cura CI, Corradi GR, Rinaldi DE, Adamo HP. High sensibility to reactivation by acidic lipids of the recombinant human plasma membrane Ca2+ -ATPase isoform 4xb purified from Saccharomyces cerevisiae. Biochim Biophys Acta. 2008;1778:2757–2764. doi: 10.1016/j.bbamem.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Niggli V, Penniston JT, Carafoli E. Purification of the (Ca2+ -Mg2+ )-ATPase from human erythrocyte membranes using a calmodulin affinity column. J Biol Chem. 1979;254:9955–9958. [PubMed] [Google Scholar]
- 28.Filomatori CV, Rega AF. On the mechanism of activation of the plasma membrane Ca2+ -ATPase by ATP and acidic phospholipids. J Biol Chem. 2003;278:22265–22271. doi: 10.1074/jbc.M302657200. [DOI] [PubMed] [Google Scholar]
- 29.Fiske CH, Subbarow Y. The colorimetric determination of phosphorous. J Biol Chem. 1925;179:66–71. [Google Scholar]
- 30.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 31.Ball EH. Quantitation of proteins by elution of Coomassie Brilliant Blue R from stained bands after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal Biochem. 1986;155:26–27. doi: 10.1016/0003-2697(86)90218-6. [DOI] [PubMed] [Google Scholar]
- 32.Caride AJ, Elwess NL, Verma AK, Filoteo AG, Enyedi A, Bajzer Z, Penniston JT. The rate of activation by calmodulin of isoform 4 of the plasma membrane Ca2+ pump is slow and is changed by alternative splicing. J Biol Chem. 1999;274:35227–35232. doi: 10.1074/jbc.274.49.35227. [DOI] [PubMed] [Google Scholar]
- 33.Esmann M, Marsh D. Spin-label studies on the origin of the specificity of lipid–protein interactions in Na+, K+ -ATPase membranes from Squalus acanthias. Biochemistry. 1985;24:3572–3578. doi: 10.1021/bi00335a027. [DOI] [PubMed] [Google Scholar]
- 34.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 35.Sønntag Y, Musgaasd M, Olesen C, Sciøtt B, Møller JV, Nissen P, Thøgersen L. Mutual adaptation of a membrane protein and its lipid bilayer during conformational changes. Nat Commun. 2011;2:304. doi: 10.1038/ncomms1307. [DOI] [PubMed] [Google Scholar]