Abstract
Purpose
Dabrafenib is a selective inhibitor of V600-mutant BRAF kinase, which recently demonstrated improved progression free survival (PFS) as compared with dacarbazine, in metastatic melanoma patients. The current study examined potential genetic markers associated with response and PFS in the phase I study of dabrafenib.
Experimental Design
Baseline (pre-treatment or archival) melanoma samples were evaluated in 41 patients using a custom genotyping melanoma-specific assay, sequencing of PTEN, and copy number analysis using multiplex ligation amplification and array based comparative genomic hybridization. Nine patients had on-treatment and/or progression samples available.
Results
All baseline patient samples had BRAFV600E/K confirmed. Baseline PTEN loss/mutation was not associated with best overall response (BOR) to dabrafenib, but it showed a trend for shorter median progression free survival (PFS) (18.3 [95% confidence interval (CI) 9.1–24.3] vs. 32.1 weeks [95% CI 24.1–33], p=0.059). Higher copy number of CCND1 (p=0.009) and lower copy number of CDKN2A (p=0.012) at baseline were significantly associated with decreased PFS. Although no melanomas had high level amplification of BRAF, the two patients with progressive disease as their best response had BRAF copy gain in their tumors.
Conclusions
Copy number changes in CDKN2A, CCND1, and mutation/copy number changes in PTEN correlated with the duration of PFS in patients treated with dabrafenib. The results suggest that these markers should be considered in the design and interpretation of future trials with selective BRAF inhibitors in advanced melanoma patients.
Keywords: Melanoma, BRAF inhibitors, correlative studies, BRAF mutation, PTEN
Introduction
Melanoma is the most lethal form of skin cancer. In the United States, it is estimated that there will be 76,250 new cases and 9,180 deaths related to melanoma in 2012, and the incidence in increasing (1). Melanomas are characterized by a high rate of single base mutations, as compared to other solid cancer types, associated with a UV light induced signature, as well as a characteristic profile of genomic amplifications and deletions (2, 3). Mutations in BRAF are found in ~45% of melanomas (4, 5), the majority of which are at codon 600 and result in constitutive kinase activity of BRAF and subsequent downstream signaling through the MAP kinase pathway (6). Seventy to ninety percent of BRAF mutations are due to the substitution of glutamic acid for valine (V600E mutation; c.1799T>A), and 10–30% are due to the substitution with lysine (V600K) (4, 5, 7). Common genomic changes in melanoma include deletion of PTEN, and CDKN2A and amplifications of KIT, MITF, TERT, CCND1, among others (3). PTEN deletions are most commonly observed in conjunction with BRAF mutations, in approximately 30%, whereas CDKN2A mutations are seen across all mutational (BRAF/NRAS/WT) sub-types of melanomas (8, 9). The frequency of these genomic changes allows their study in conjunction with outcome upon treatment of melanoma.
Until recently, most systemic therapy options available for patients with advanced stage melanoma were ineffective, and the five year survival rate was less than 15%. In addition to immunotherapeutic agents, therapies directed at the MAPK signaling pathway have been developed, in particular targeted inhibition of the mutant V600E BRAF protein. Dabrafenib is a reversible ATP-competitive inhibitor that selectively inhibits mutant BRAFV600E, with an IC50 (concentration required for 50% inhibition) fivefold lower than for wildtype BRAF or CRAF (10). Treatment of BRAF-mutant metastatic melanoma with the RAF inhibitors dabrafenib or vemurafenib, results in response rates of approximately 50% and significantly improved progression free survival (PFS) and overall survival (OS) as compared with dacarbazine chemotherapy (11–14). The emergence of acquired resistance in the majority of patients, for which multiple mechanisms have been described, involving both reactivation of the MAPK signaling pathway and by-pass mechanisms, remains the greatest barrier to better clinical outcomes (15–22).
Pre-clinical studies have also identified several mechanisms of de novo or intrinsic resistance to BRAF inhibitors, including PTEN loss (alone or in conjunction with Rb1 loss), MET and SRC activation (associated with amplification of MET, CTNNB1 and CCND1), activation of P70S6K and S6, and HGF mediated MET expression (21–26). In order to improve our understanding of the clinical significance of these DNA-based aberrations implicated in intrinsic resistance, and other candidate genes previously identified as aberrant in melanoma, we report here the molecular analysis of a large collection of human melanoma tumor samples from patients treated on the phase I clinical trial of the BRAF inhibitor dabrafenib (11).
Patients and Methods
Patients and clinical outcome
Melanoma tumor samples were collected from patients enrolled on the first-time-in-human BRF112680 phase I trial of dabrafenib (GSK2118436; clinical trial number NCT00880321) between May 27, 2009 and March 20, 2012, at eight study centers in Australia and the United States (11). Samples were collected either at baseline (‘pre-treatment’ and/or archival samples), ‘on-treatment’, and when possible, at time of progression. Informed consent was obtained from all patients before the start of treatment and collection of tumor samples; the study complied with all local guidelines. Patients commenced on variable doses of dabrafenib (35mg – 300mg) (Table 1); all were escalated to a total daily dose of ≥300mg/day. The clinical outcome measures used in this study included the objective response and progression-free survival using computed tomography (CT) and RECIST 1.0 as previously reported (11).
Table 1.
Patient description
| Patient characteristic | Total (n = 41)* | |
|---|---|---|
| Dabrafenib starting dose | < 150 mg BID | 17 (41%) |
| 150 mg BID | 15 (37%) | |
| > 150 mg BID | 9 (22%) | |
|
| ||
| BRAF V600 mutation | V600E | 34 (83%) |
| V600K | 7 (17%) | |
|
| ||
| Distant metastasis | M1a/M1b | 7 (17%) |
| M1c | 34 (83%) | |
|
| ||
| ECOG performance | 0 | 28 (68%) |
| 1 | 12 (29%) | |
| 2 | 1 (2%) | |
|
| ||
| LDH level | < 1xULN | 19 (46%) |
| 1–2xULN | 13 (32%) | |
| > 2xULN | 7 (17%) | |
| Missing | 2 (5%) | |
|
| ||
| Number of prior systemic therapies | 0 | 17 (41%) |
| 1 | 12 (29%) | |
| ≥ 2 | 12 (29%) | |
Only patients with baseline (pre-treatment or archival) samples are included
Tumor QC, DNA extraction
All samples were formalin-fixed and paraffin embedded (FFPE) and processed for hemotoxylin and eosin (H&E) staining and pathologist review of tumor content at the ACC Histology Core Facility by QCY. Samples with over 70% tumor content were processed for DNA extraction directly, while those with less than 70% tumor underwent H&E-guided macrodissection prior to DNA extraction. DNA was extracted using standard methods.
iPlex genotyping and PTEN sequencing
Genotyping performed using a custom iPlex (Sequenom, Inc.) single nucleotide extension panel Genotyping was done at the Perelman School of Medicine Molecular Profiling Facility. Analysis of data was done using the iSeq™ software; we assessed the ratio of wildtype:mutant (T:A) nucleotide in BRAFV600E using the peak height chromatograms. Sanger sequencing of PTEN exons 1 through 9 was performed using standard methods and published primers (27). Detailed methods are included in Supplementary Methods.
Copy number analysis
Multiplex ligation dependent probe amplification (MLPA) was used to detect copy number aberrations of genes located on chromosome 10q23 (MLPA kit P225-B2 PTEN, MRC-Holland). MLPA was performed according to the manufacturer’s instructions and analyzed with their software - MRC-Coffalyser Stand Alone Alpha Version 1.0.0.43 software. The fragments were analyzed on ABI 3130xl capillary sequencer using Genemapper software (Applied Biosystems, Inc.). Variation in peak height was evaluated by comparing each test sample to three normal controls present in the same experiment. Normalization was done intra-sample by dividing the peak area of each probe’s amplification product by the total area of only the reference probes in this probe mix. Single regression for control and tumor data slope correction was performed. Normal ratio limits were set at −0.70 and 1.2. This program identifies a peak as deleted when showing a ratio < | 0.7 | and amplified when showing a ratio ≥ 1.2. The copy number of PTEN was measured in 17 samples using both MLPA and aCGH for cross-validation; all samples had the same copy number profiles.
Array based comparative genomic hybridization was done using the Agilent SurePrint G3 Human CGH 1x1M microarrays following manufacturer’s instructions. Arrays were scanned using Agilent’s High-Resolution C Scanner. Extracted data was analyzed using BioDiscovery’s Nexus 6 copy-number software (Nexus Genomics Inc., Mountain View, CA, USA). Copy number variation was assessed using the CBS-like Rank Segmentation algorithm provided with Nexus 6; genes mapping was done to hg19, Feb 2009 build. Copy number gain was defined as log2 scale value ≥ 0.3 and loss as log2 scale value ≤ −0.3, with at least three contiguous SNPs needed. Segments were particularly examined for the presence of high copy gains (log2 scale value ≥ 1.14) and homozygous loss (log2 scale value ≤ −1.1). Additionally, only segments derived from > 16 probes were included in subsequent analysis.
Chromosomal instability analysis
Characterizing cytogenetic instability was used as an alternative to analyzing specific, recurring copy number changes across the tumor set. To this end, copy number alterations were first identified in the segmented data for each tumor and mapped to a specific chromosomal arm. Considering gains and losses separately, the total accumulation of copy number altered regions was calculated for each arm (i.e. total bases altered) and the fraction of the arm altered (total bases gained or lost/total size of arm).
Statistical Analysis
Progress free survival (PFS) was compared in patients whose tumors had wild-type or non-deleted/mutant PTEN status versus those with deleted or mutant PTEN using the log-rank test and Kaplan-Meier analysis. Correlation between copy number values among 36 genes was assessed using Spearman’s correlation analysis. Association between copy number values and PFS was evaluated using proportional hazards regression. In this analysis, p<0.05 was considered statistically significant. Cytogenetic instability was analyzed between patients with pre or early dose tissue. Comparisons were made using the median PFS of 24 weeks, with 12 patients having a PFS over 24 weeks and 11 with a PFS less than 24 weeks, using a t-test.
Results
Description of patient population
A total of 91 samples were available from 77 patients. Eleven patients had multiple samples available. Twenty-seven samples were determined to have no tumor or in an amount too small to allow for adequate DNA extraction. We obtained BRAF mutational data on 57 melanoma tumor samples from 45 patients. Of these samples, two were on-treatment and eleven were progression samples, with the remaining 44 baseline (pre-treatment or archival) samples. Patient characteristics for the 41 patients with baseline samples available are in Table 1.
Mutational information
In the 41 patients with baseline samples available, six had BRAFV600K, 35 BRAFV600E mutations (Table 2). Two melanoma samples carrying the BRAFV600E mutation exhibited a T(wildtype):A(mutant) ratio favoring the mutant allele, one suggestive of amplification of that allele (patient 32, 1:4). Of the seven BRAFV600K mutations observed, two samples showed T:A ratio favoring the mutant allele, suggestive of amplification of the mutant allele (patients 11, 19). The highest ratio favoring the mutant allele was seen in the archival sample from a patient also with a pre-treatment sample, which had a ratio favoring the wild-type allele. Two patients with V600E mutations had additional concurrent mutations, one with CTNNB1 (beta-catenin) p.S45del and another with MAP2K2 (MEK2) p.Q60P. The biopsy with the mutation in MAP2K2 was taken at time of progression on trametinib (MEK inhibitor), prior to treatment with dabrafenib. Melanoma tumor samples, all taken from the same lesion, at multiple time points - archival, prior to treatment, and on-treatment with trametinib - were available for this patient and did not show the MAP2K2 p.Q60P mutation, suggesting it arose after treatment with trametinib (Infante, Nathanson, unpublished data). This patient had progressive disease as their best response to treatment with dabrafenib.
Table 2.
Mutational status using targeted assays and copy number analysis of PTEN
| Patient # | Sample type | BRAF | WT/mutant (T:A) Ratios | PTEN Mutation Status and Copy # | Additional mutations |
|---|---|---|---|---|---|
| 1 | Pre-treatment | V600E | 1:1 | WT/no CN* | |
| 1 | Progression | V600E | 1:1 | WT | |
| 2 | Progression | K601E | ND | Homo-Del† | |
| 3 | Pre-treatment | V600E | 1.5:1 | WT | |
| 4 | Archival | V600K | 1.5:1 | Hemi-Del‡ | |
| 4 | Progression | V600K | 2:1 | WT | |
| 5 | Archival | V600E | 2:1 | AF | |
| 6 | Archival | V600E | 1:1 | Dup Ex 1α | |
| 7 | Archival | V600E | 3:1 | WT | |
| 7 | On-treatment | V600E | 1:1 | Hemi-Del | |
| 7 | Progression | V600E | 3:1 | WT | |
| 8 | Archival | V600E | 1:1 | Hemi-Del | |
| 9 | Archival | V600E | 1:1 | WT | CTNNB1 S45del |
| 10 | Pre-treatment | V600E | 1.5:1 | Homo-Del | |
| 10 | Progression | V600E | 2:1 | Homo-Del | |
| 10 | Progression | V600E | 3:1 | Hemi-Del | |
| 11 | Archival | V600K | 1:3 | Amplified | |
| 11 | Pre-treatment | V600K | 1.5:1 | WT | |
| 12 | Archival | V600E | 1.5:1 | WT | |
| 13 | Archival | V600E | 3:1 | Amp/Delβ | |
| 14 | Archival | V600E | 1:1 | WT/no CN | |
| 14 | Pre-treatment | V600E | 1.5:1 | WT | |
| 14 | Progression | V600E | 1:1 | WT | |
| 15 | Archival | V600E | 1.5:1 | WT | |
| 16 | Archival | V600E | 1:1 | WT | |
| 16 | Pre-treatment | V600E | 2:1 | Mutant | |
| 17 | Archival | V600E | 1.5:1 | WT/no CN | |
| 18 | Archival | V600K | 1.5:1 | WT | |
| 19 | Pre-treatment | V600K | 1:1.5 | WT | |
| 19 | On-treatment | V600K | 2:1 | WT | |
| 19 | Progression | V600K | 1:1.5 | WT | |
| 20 | Archival | V600E | 1:1 | Hemi-Del | |
| 21 | Pre-treatment | V600E | 2:1 | WT | |
| 22 | Archival | V600E | 2:1 | WT | |
| 23 | Archival | V600E | 1:1 | WT | |
| 24 | Archival | V600E | 1:1 | Hemi-Del | |
| 25 | Archival | V600E | 1.5:1 | WT | |
| 26 | Pre-treatment | V600K | 2:1 | Homo-Del | |
| 27 | Archival | V600K | 1:1 | Dup Ex 1 | |
| 28 | Archival | V600E | 2:1 | Hemi-Del | |
| 29 | Archival | V600E | 2:1 | Hemi-Del | |
| 30 | Archival | V600E | 1.5:1 | Amp/Del | |
| 31 | Archival | V600E | 3:1 | WT/no CN | |
| 32 | Archival | V600E | 1:4 | AFγ | |
| 33 | Archival | V600E | 1:2 | Hemi-Del | |
| 34 | Progression | V600E | 1:1 | Homo-Del | |
| 35 | Progression | V600E | 3:1 | Homo-Del | |
| 36 | Archival | V600E | 2:1 | Hemi-Del | |
| 37 | Progression | WT | ND | WT | NRAS Q61K |
| 38 | Pre-treatment | V600E | 1:1 | WT | MEK2 Q60P |
| 39 | Archival | V600E | 1:1 | WT | |
| 40 | Archival | V600E | 1:1 | WT | |
| 41 | Archival | V600E | 2.5:1 | WT | |
| 42 | Archival | V600E | 5:1 | Hemi-Del | |
| 43 | Pre-treatment | V600E | 2:1 | AF | |
| 44 | Archival | V600E | 2:1 | WT | |
| 45 | Archival | V600E | 3:1 | AF |
WT/No CN – Sequence wild-type, copy number could not be obtained;
Homo-Del – homozygous deletion;
Hemi-Del – hemizygous deletion;
Dup Ex 1 – duplication of exon 1;
Amp/Del – duplication of exon 1, deletion of other exons;
AF - assay failed
Data were available from two on-treatment and 11 progression samples from ten patients (Table 2). Both of the on-treatment samples were from patients who also had progression samples. The ten patients had melanomas carrying the following mutations; BRAFV600E (6), BRAFV600K (2), BRAFK601E (1), and NRASQ61K (1). The progression sample with the NRAS mutation did not have a concurrent BRAF mutation, so it is possible that the patient had more than one primary melanoma or that the BRAF V600E mutation was present but undetectable based on the sensitivity of the assay. Six patients had matched pre-treatment/archival and progression samples. In general, the ratio of mutant to wild-type allele was the same in both samples.
Association of baseline PTEN status with PFS
As we were particularly interested in the relationship of PTEN status to PFS upon treatment with dabrafenib, we evaluated it genetically with copy number analysis, using MLPA and/or aCGH, and sequencing of all exons. For this analysis, we focused on pre-treatment and archival samples; data from the pre-treatment sample was used preferentially for all analyses. Three patients had both archival and pre-treatment samples. One patient had an archival sample with amplified PTEN; the pre-treatment sample showed diploid PTEN. In one patient, a mutation in PTEN was observed in the pre-treatment, but not archival sample. This mutation was the only one observed in PTEN, p.P95L, and has been previously reported in association with cancer (27–29). For four samples, we obtained only mutational, but not copy number data; these samples were removed from further analysis. Thus, 34 patients had melanoma samples with PTEN data available for analysis (Table 2); patient characteristics are reported in Supplementary Table 1.
PTEN status was available for 11 progression and two on-treatment samples. Five progression samples had matched archival or pre-treatment samples; in all but one case the copy number status was the same in both samples. Of note, homozygous deletion of PTEN was more frequently observed in progression tumor samples (4/10) than in archival or pre-treatment samples (2/34), p=0.017.
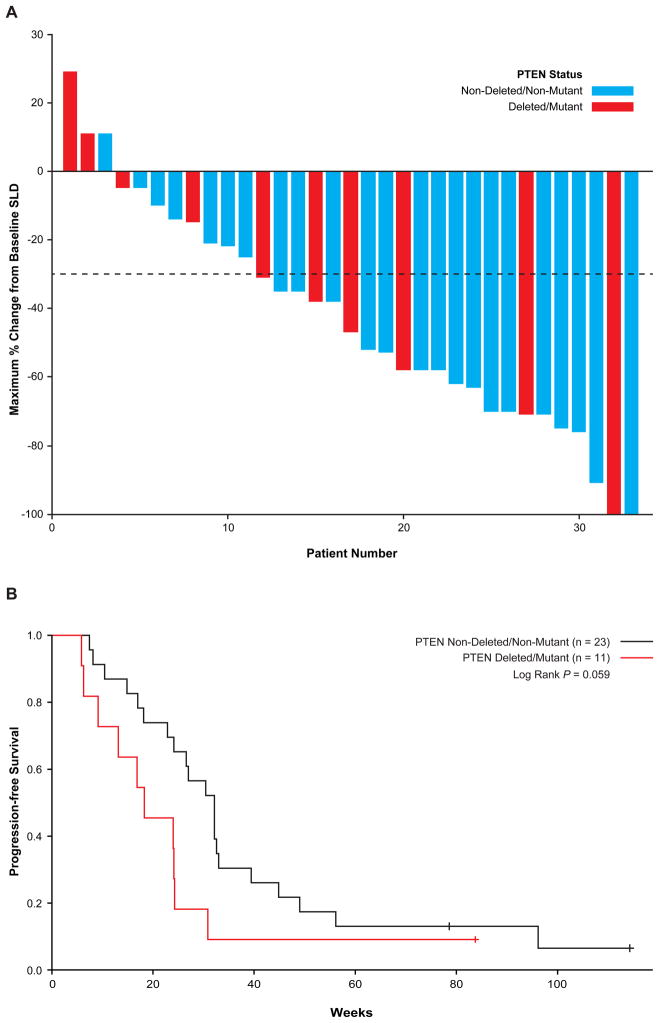
The relationship between best overall response (BOR) and PFS were evaluated. No association was seen between BOR and PTEN status with patients with melanomas having wild-type or non-deleted/non-mutant PTEN status having a response rate (CR+PR) of 43% (10/23; 95% confidence interval [CI] 21–65.9%) versus those with deleted or mutant PTEN of 36% (4/11; 95% CI 3.4–69.3%), p=0.059 (Figure 1A). However, the patients with wild-type or non-deleted/non-mutant PTEN status had a longer PFS (32.1 weeks, 95% CI 24.1–33) and those with deleted/mutant PTEN (18.3 weeks, 95% CI 9.1–24.3, p=0.059), p=0.059 (Figure 1B). As the PTEN status for four samples with exonic amplification could not be precisely determined to be either wild-type or deleted, as potentially they could indicate rearrangements leading to PTEN loss, as in prostate cancer (30), they were removed for a secondary analysis. The PFS in the 18 patients with wild-type PTEN status was 32.1 weeks (95% CI 24.1–39.4), longer than those with mutant/deleted PTEN, but this difference did not reach statistical significance (p=0.066).
Figure 1. Association with PTEN genetic status with response and progression free survival upon treatment with dabrafenib.
A. Waterfall plot. One patient with stable disease was not included because tumor percent change was unknown. B. Median PFS in patients with tumors with mutations or deletions of PTEN (red line) vs. all others (black line).
PTEN immunohistochemistry was performed on a subset (n=17) of the samples with complete genetic data (Supplementary Table 2). The correlation between IHC data and genetic status was 75% (95% CI: 50.5–89.8); presence/absence of staining data was not predictive of PFS in this small subset of patients (data not shown).
aCGH data
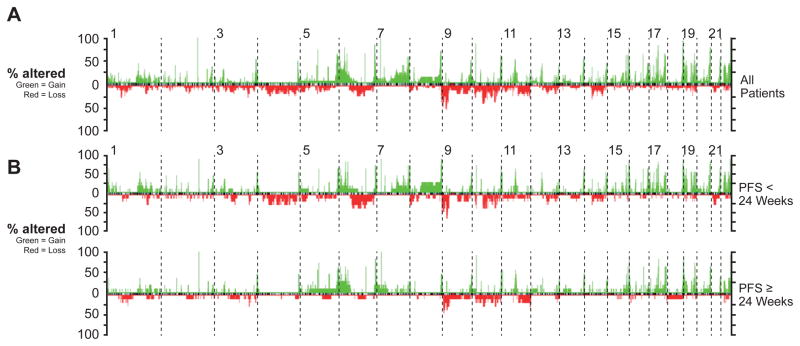
aCGH data was generated on 34 samples from 26 patients. Twenty-four archival and pre-treatment samples were analyzed from 23 patients (one patient had both archival and pre-treatment samples). Eight patients had progression samples analyzed (two for which we also had on-treatment samples). Three patients had both archival or pre-treatment samples and progression samples. Overall, the copy number analysis (Figure 2A) was similar to that previously published for BRAF mutant cutaneous melanoma, with frequent copy number gains of 6p, 7q and 22q, and frequent losses of 6q, 9p, and 10q (31–34). We also examined the overall genomic profile of pre-treatment melanomas in relationship to PFS (Figure 2B), BOR (Supplementary Figure 1), and BRAFV600E vs. BRAFV600K mutation status (Supplementary Figure 2). The percentage of individual chromosome arms and total fraction of the genome that was altered in patients with PFS > 24 weeks (n = 12) was compared to those with PFS ≤24 weeks (n = 11). While the overall fraction of the genome gained or lost did not significantly differ between groups (p = 0.19 and 0.11, respectively), there were some notable qualitative differences. Most notably, when considering large alterations defined as those encompassing > 25% of the size of a chromosomal arm (indicative of broad instability), gains of 6p appeared more frequent in the patients demonstrating > 24 vs. ≤24 week PFS (58% vs. 27% respectively). Similarly, large losses of 11q appeared more frequent in those patients with PFS > 24 vs. ≤ 24 weeks (33% vs. 9% of patients respectively). Conversely, losses of 9p (25% vs. 55%), and 4q (0 % vs. 27%) were qualitatively more frequent in those with shorter PFS. While none of these differences achieved statistical significance (p > 0.05 in all cases), the overall rarity of such large genomic changes could suggest relationships with response in a subset of patients.
Figure 2. array based Comparative Genomic Hybridization analysis of melanoma samples.
Specific genes from aCGH data
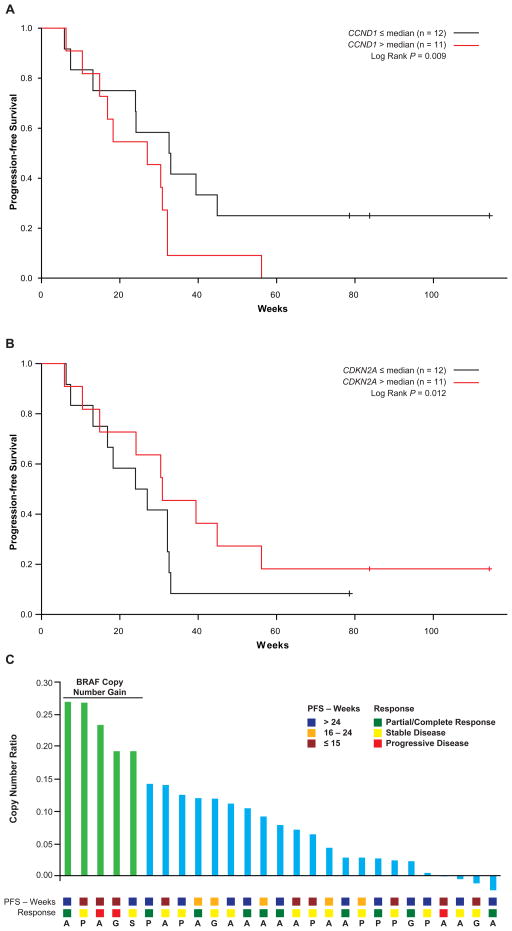
We selected 36 genes (Supplementary Table 3) implicated in melanoma pathogenesis for copy number variation (CNV) characterization. A correlation analysis between copy number gains and losses of various genes, both positive and negative (i.e. gain-gain, gain-loss) was done, and is shown in Supplementary Table 4. Correlation of CNV results to PFS demonstrated that copy gain of CCND1 (p=0.009) and loss of CDKN2A (p=0.012) were independently predictive of shorter PFS. For visualization purposes of the association between copy number and outcome, the samples were divided by median copy number and Kaplan-Meier curves for PFS were generated (Figure 3A, B). For CCND1, patients whose tumors had greater than the median copy number (n=11) had a median PFS of 27 weeks (95% CI, 15–32 weeks), which was shorter (Hazard ratio [HR]=2.22) than those with a lower copy number (n=12, median PFS 33 weeks, 95% CI, 24–45 weeks). Patients with less than median copy number of CDKN2A (n=11) had a median PFS of 25 weeks (95% CI, 17–32 weeks), whereas patients with a higher copy number had a median PFS of 31 weeks (95% CI, 15–56 weeks, HR=0.59). Of note, neither copy number of CDKN2A nor CCND1 was associated with BOR. High level amplification of the BRAF locus (> 5 copies) was not observed in these 25 patients, and BRAF copy number did not significantly correlate with BOR or PFS (Figure 3c).
Figure 3. Association between gene copy number and outcome.
For visualization of the association between copy number and outcome, the samples were divided by median copy number and Kaplan-Meier curves for PFS were generated A. CCND1 B. CDKN2A C. BRAF copy gain and correlation with PFS, response and time of sample collection (A – archival; P – pre-treatment; S – on-treatment; G – progression)
Discussion
To the authors’ knowledge, this study represents the largest genetic and genomics based examination of human melanoma tissue from patients treated with BRAF inhibitors, specifically patients treated with dabrafenib on the phase I/II trial (11), to identify correlates associated with response. We focused on the genes encoding the proteins of the MAPK (Ras/Raf/MEK/ERK) and PI3K/Akt pathways, as they are the principal signaling pathways shown to be crucial in melanoma initiation and progression (6, 35). Cell cycle regulatory proteins, such as p16, CKD4 and cyclin D1, also have been shown to play important roles in melanoma, and those genetic and genomic aberrations involving these also were specifically interrogated (36–38). We also took a more comprehensive un-biased approach, using aCGH, to identify additional genomic aberrations which might be associated with response.
All baseline (pre-treatment and archival) samples had confirmed V600 mutations in BRAF, a study eligibility requirement. High level copy (over five) amplification of BRAF was not observed in any of the samples. A few samples demonstrated relative increases in the ratio of the BRAF V600 mutant to wild-type allele, either in the pre-treatment samples, or at the time of progression; most samples demonstrated equivalent or increased wild-type allele. Of note, two of the three samples from patients with progressive disease as best response had copy number gain of BRAF, however one was from a progression sample without a matching pre-treatment specimen. Although our numbers are too small to draw any definitive conclusion from, they are consistent with prior findings suggesting that increased copy number of BRAF is associated with progressive disease in some patients who are treated with BRAF or MEK inhibitors (15, 39). Importantly, preclinical studies suggest that this mechanism of resistance may potentially be overcome with increased doses of selective BRAF inhibitors (15).
We performed detailed analysis of PTEN genetic status in association with response to BRAF inhibition, as several pre-clinical studies have suggested that PTEN loss contributes to intrinsic BRAF resistance (23, 24). Our data suggest that PTEN loss is associated with a shorter PFS in patients treated with BRAF inhibitors, although is not predictive of best overall clinical response. This finding is not unexpected, as degree of response is not necessarily correlated with duration with targeted agents. Interestingly, PFS correlated with overall survival in the phase 3 study of vemurafenib, the only study able to use OS as a primary endpoint (40). It has been suggested that PTEN loss, as measured through immunohistochemistry (IHC), is a negative prognostic factor for melanoma, independent of BRAF inhibition (41, 42). PTEN loss also is found more frequently in brain than lung or liver metastases, which are associated with worse prognosis (35). However, genetic analysis of PTEN status has not been previously assessed as a predictive or prognostic marker in melanoma, and has not been correlated with immunochemistry analyses. Although our data suggest that analyses using genetic methods may ultimately be better predictors of outcome than immunochemistry measurements, analyses of larger cohorts of patients with parallel genetics and proteomic analysis are needed to make definitive conclusions.
Copy number analysis of 36 genes previously observed to be altered in melanoma was performed on samples from 23 dabrafenib-treated patients. Among those genes, lower copy number of CDKN2A and higher copy number of CCND1 were significantly associated with shorter PFS. The regions containing these genes also were implicated in our overall (unbiased) analysis of aCGH data, as CCND1 is located on 11q and CDKNA on 9p. Both deletion/loss of CDKN2A (p16) and amplification/overexpression of CCND1 (cyclin D1) previously have been implicated as poor prognostic markers in melanoma (43–47). Thus, these genetic changes may function as either prognostic or predictive markers in response to BRAF inhibition. Amplification of CCND1 also has been associated with intrinsic resistance to BRAF inhibition in pre-clinical studies (25, 48). Other genes which have been implicated in intrinsic resistance to BRAF inhibition, including Rb1 loss, MET and SRC amplification, did not emerge as associated with PFS in our analysis (24, 25).
Although this study contains a large set of melanoma tissue samples from patients treated with dabrafenib, samples were not available from all patients enrolled in the clinical trial, potentially limiting the generalizability of our results. However, the characteristics of the patients included in the correlative studies, as reviewed in Table 1, did not differ from those on the trial as a whole (data not shown). We were not able to determine whether the genetic alterations identified as correlates of clinical outcome were predictive, prognostic or both, and prior clinical and pre-clinical data support both interpretations of the data. Some tissue samples collected at progression lacked a paired baseline specimen, limiting our interpretation of the genetic and genomic findings in the progression samples. Several studies have demonstrated intertumoral heterogeneity using massively parallel sequencing (49, 50). Based on our own studies using massively parallel sequencing in a similar sample set (data not shown), low level point mutations (<5–10% allele frequency), such as in NRAS, could have been missed. In addition, copy number changes (other than high level amplification) are more difficult to interpret without matched germline samples, particularly for capture based methods. Thus, it is less likely that massively parallel sequencing would have provided greatly improved sensitivity in this context. Finally, as we mainly focused on known genetic alterations and genomic aberrations associated with melanoma, it is possible that we did not identify novel predictors of clinical outcome in this data set.
In summary, we identified PTEN loss, CDKN2A deletion and CCND1 amplification as associated with decreased PFS upon treatment with dabrafenib. An exploratory analysis (data not shown) showed that carriage of two, or all three together, was associated with a trend towards worsening PFS. Although the interpretation of this analysis was limited by the small number of patients in each group, this approach should be considered for larger sample sets in the future. Ultimately, if validated in future patients, genetic analysis of PTEN, CDKN2A, and CCND1 may be used to identify patients who should be considered for frontline combinatorial approaches, particularly utilizing agents that are activated by aberrations in these genes. Thus, these data suggest where potential combination therapies in conjunction with dabrafenib might be most effectively targeted. These studies also emphasize the continued importance of trials with associated biopsies on a large number of patients, so that we may gain insights into predictors of clinical outcome.
Supplementary Material
Statement of translational relevance.
Multiple studies have evaluated acquired mutations in melanomas from patients who progress on BRAF inhibitor therapies. The current study focuses on pre-treatment predictors of outcome when treated with the BRAF inhibitor dabrafenib. Common genetic mutation and genomic aberrations in melanoma were examined, with those in CDKN2A, CCND1, and PTEN found to correlate with the duration of PFS. Interestingly, these genetic changes have been previously implicated as associated with outcome in natural history studies of melanoma, and pre-clinically, in response to treatment with BRAF inhibition, so we cannot discriminate if they are predictive or prognostic markers. Our data suggest genetic analysis of PTEN, CDKN2A, and CCND1 may be used to identify patients who should be considered for frontline combinatorial approaches, particularly utilizing agents targeting pathways activated by aberrations in these genes. This study also emphasizes the continued importance of trials with associated biopsies on a large number of patients, so that we may gain insights into predictors of clinical outcome.
Acknowledgments
The authors wish to thank Madhavi Vaddi at the Penn Molecular Profiling Facility and Dr. Quian-Chen Yu at the Histology Core Facility for excellent technical assistance at the University of Pennsylvania. Services provided in the Penn Molecular Profiling and Histology Core Facilities are supported by the Abramson Cancer Center core grant P30CA016520. They also would like to thank the staff of Sydney West Cancer Trials Centre, and the surgical, dermatology and Anatomical Pathology staff of Westmead Hospital and Melanoma Institute Australia. This work was performed in the U.T. MD Anderson Cancer Center Clinical and Translational Research Center (CTRC) and was supported by the Center for Clinical and Translational Sciences, which is funded by National Institutes of Health Clinical and Translational Science Award UL1 RR024148 and by the National Institutes of Health Cancer Center Support Grant (CCSG) award CA016672 to MD Anderson Cancer Center.
Grant Support
Supported by research grants from GlaxoSmithKline, Inc., Melanoma Research Alliance Team Science Award to KLN and MAD, ASCO Career Development Award to MAD, Cancer Institute New South Wales Fellowship program to GL.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Conflicts of interest: KLN, SO’D and H-TA have received research funding from GlaxoSmithKline; A-MM, JG, RG, BM and MC are employees of, and have stock ownership of, GlaxoSmithKline; BW, RL, KD’A and AP have no conflicts of interest to declare; JRI has acted as an uncompensated consultant or advisor for GlaxoSmithKline; GSF has received research funding and other remuneration from GlaxoSmithKline; MM has acted as a compensated consultant or advisor for GlaxoSmithKline; MPB has acted as a compensated consultant or advisor for, and has received research funding from, GlaxoSmithKline; MAD has served on advisory boards for GlaxoSmithKline, Genentech and Novartis, and has received research funding from GlaxoSmithKline, Genentech, AstraZeneca, Merck, Myriad and Oncothyreon; PFL was an employee of GlaxoSmithKline; RK has acted as a compensated consultant or advisor for GlaxoSmithKline; GVL has acted as a compensated consultant or advisor for GlaxoSmithKline, Roche, Novartis, Amgen and Bristol-Myers Squibb, and has received honoraria from Roche.
References
- 1.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Altekruse SF, et al. SEER Cancer Statistics Review, 1975–2009 (Vintage 2009 Populations) 2012. Bethesda, MD: National Cancer Institute; 2012. based on November 2011 SEER data submission, posted to the SEER web site. [Google Scholar]
- 2.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jakob JA, Bassett RL, Jr, Ng CS, Curry JL, Joseph RW, Alvarado GC, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2011;118:4014–23. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29:1239–46. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 6.Fecher LA, Amaravadi RK, Flaherty KT. The MAPK pathway in melanoma. Curr Opin Oncol. 2008;20:183–9. doi: 10.1097/CCO.0b013e3282f5271c. [DOI] [PubMed] [Google Scholar]
- 7.Lovly CM, Dahlman KB, Fohn LE, Su Z, Dias-Santagata D, Hicks DJ, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS One. 2012;7:e35309. doi: 10.1371/journal.pone.0035309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gast A, Scherer D, Chen B, Bloethner S, Melchert S, Sucker A, et al. Somatic alterations in the melanoma genome: a high-resolution array-based comparative genomic hybridization study. Genes Chromosomes Cancer. 2010;49:733–45. doi: 10.1002/gcc.20785. [DOI] [PubMed] [Google Scholar]
- 9.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GlaxoSmithKline. BRF113683 Clinical Study Report. 2012. [Google Scholar]
- 11.Falchook GS, Long GV, Kurzrock R, Kim KB, Arkenau TH, Brown MP, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379:1893–901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–65. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 13.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–60. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing F, Persaud Y, Pratilas CA, Taylor BS, Janakiraman M, She QB, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene. 2012;31:446–57. doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vergani E, Vallacchi V, Frigerio S, Deho P, Mondellini P, Perego P, et al. Identification of MET and SRC activation in melanoma cell lines showing primary resistance to PLX4032. Neoplasia (New York, NY) 2011;13:1132–42. doi: 10.1593/neo.111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng W, Gopal YN, Scott A, Chen G, Woodman SE, Davies MA. Role and therapeutic potential of PI3K-mTOR signaling in de novo resistance to BRAF inhibition. Pigment Cell Melanoma Res. 2012;25:248–58. doi: 10.1111/j.1755-148X.2011.00950.x. [DOI] [PubMed] [Google Scholar]
- 27.Levine RL, Cargile CB, Blazes MS, van Rees B, Kurman RJ, Ellenson LH. PTEN mutations and microsatellite instability in complex atypical hyperplasia, a precursor lesion to uterine endometrioid carcinoma. Cancer Res. 1998;58:3254–8. [PubMed] [Google Scholar]
- 28.Tashiro H, Blazes MS, Wu R, Cho KR, Bose S, Wang SI, et al. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res. 1997;57:3935–40. [PubMed] [Google Scholar]
- 29.Yaginuma Y, Yamashita T, Ishiya T, Morizaki A, Katoh Y, Takahashi T, et al. Abnormal structure and expression of PTEN/MMAC1 gene in human uterine cancers. Mol Carcinog. 2000;27:110–6. doi: 10.1002/(sici)1098-2744(200002)27:2<110::aid-mc6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 30.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greshock J, Nathanson K, Medina A, Ward MR, Herlyn M, Weber BL, et al. Distinct patterns of DNA copy number alterations associate with BRAF mutations in melanomas and melanoma-derived cell lines. Genes Chromosomes Cancer. 2009;48:419–28. doi: 10.1002/gcc.20651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonsson G, Dahl C, Staaf J, Sandberg T, Bendahl PO, Ringner M, et al. Genomic profiling of malignant melanoma using tiling-resolution arrayCGH. Oncogene. 2007;26:4738–48. doi: 10.1038/sj.onc.1210252. [DOI] [PubMed] [Google Scholar]
- 33.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–42. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 34.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 35.Davies MA, Stemke-Hale K, Lin E, Tellez C, Deng W, Gopal YN, et al. Integrated Molecular and Clinical Analysis of AKT Activation in Metastatic Melanoma. Clin Cancer Res. 2009;15:7538–46. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev. 2012;26:1131–55. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 38.Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet. 1996;12:97–9. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 39.Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal. 2010;3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer S, Fuchs TJ, Bosserhoff AK, Hofstadter F, Pauer A, Roth V, et al. A seven-marker signature and clinical outcome in malignant melanoma: a large-scale tissue-microarray study with two independent patient cohorts. PLoS One. 2012;7:e38222. doi: 10.1371/journal.pone.0038222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikhail M, Velazquez E, Shapiro R, Berman R, Pavlick A, Sorhaindo L, et al. PTEN expression in melanoma: relationship with patient survival, Bcl-2 expression, and proliferation. Clin Cancer Res. 2005;11:5153–7. doi: 10.1158/1078-0432.CCR-05-0397. [DOI] [PubMed] [Google Scholar]
- 43.Gammon B, Ali L, Guitart J, Gerami P. Homogeneous staining regions for cyclin D1, a marker of poor prognosis in malignant melanoma. Am J Dermatopathol. 2012;34:487–90. doi: 10.1097/DAD.0b013e31823894f8. [DOI] [PubMed] [Google Scholar]
- 44.Gerami P, Jewell SS, Pouryazdanparast P, Wayne JD, Haghighat Z, Busam KJ, et al. Copy number gains in 11q13 and 8q24 [corrected] are highly linked to prognosis in cutaneous malignant melanoma. J Mol Diagn. 2011;13:352–8. doi: 10.1016/j.jmoldx.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karim RZ, Li W, Sanki A, Colman MH, Yang YH, Thompson JF, et al. Reduced p16 and increased cyclin D1 and pRb expression are correlated with progression in cutaneous melanocytic tumors. Int J Surg Pathol. 2009;17:361–7. doi: 10.1177/1066896909336177. [DOI] [PubMed] [Google Scholar]
- 46.Conway C, Beswick S, Elliott F, Chang YM, Randerson-Moor J, Harland M, et al. Deletion at chromosome arm 9p in relation to BRAF/NRAS mutations and prognostic significance for primary melanoma. Genes Chromosomes Cancer. 2010;49:425–38. doi: 10.1002/gcc.20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallagher SJ, Thompson JF, Indsto J, Scurr LL, Lett M, Gao BF, et al. p16INK4a expression and absence of activated B-RAF are independent predictors of chemosensitivity in melanoma tumors. Neoplasia (New York, NY) 2008;10:1231–9. doi: 10.1593/neo.08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smalley KS, Lioni M, Dalla Palma M, Xiao M, Desai B, Egyhazi S, et al. Increased Cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF-V600E mutated melanomas. Mol Cancer Ther. 2008;7:2876–83. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.