Key Points
BCL6 and BACH2 cooperatively regulate GC B-cell development.
The cooperative action of BCL6 and BACH2 is through both transcriptional and biochemical mechanisms.
Abstract
The transcriptional repressors BCL6 and BACH2 are crucial regulators of germinal center (GC) B-cell fate, and are known to interact and repress transcription of PRDM1, a key driver of plasma cell differentiation. How these factors cooperate is not fully understood. Herein, we show that GC formation is only minimally impaired in Bcl6+/− or Bach2+/− mice, although double heterozygous Bcl6+/−Bach2+/− mice exhibit profound reduction in GC formation. Splenic B cells from Bcl6+/− Bach2+/− mice display accelerated plasmacytic differentiation and high expression of key plasma cell genes such as Prdm1, Xbp1, and CD138. Chromatin immunoprecipitation sequencing revealed that in B cells, BACH2 is mostly bound to genes together with its heterodimer partner MAFK. The BACH2-MAFK complex binds to sets of genes known to be involved in the GC response, 60% of which are also targets of BCL6. Approximately 30% of BACH2 peaks overlap with BCL6, including cis-regulatory sequences of the PRDM1 gene. BCL6 also modulates BACH2 protein stability and their protein levels are positively correlated in GC B cells. Therefore, BCL6 and BACH2 cooperate to orchestrate gene expression patterning in GC B cells through both transcriptional and biochemical mechanisms, which collectively determine the proper initiation and timing of terminal differentiation.
Introduction
Germinal centers (GCs) are transient microstructures that form within B-cell follicles of secondary lymphoid tissues to enable immunoglobulin affinity maturation in response to T-cell–dependent antigens.1-3 Within GCs, B cells undergo clonal expansion, somatic hypermutation, and class-switch recombination. Once this process is complete, B cells expressing high affinity immunoglobulin are selected for terminal differentiation into plasma or memory cells.1,2 The timing of the transition from GC B cells to plasma cells is considered to play a crucial role in determining the magnitude of the GC response.4,5 The molecular mechanism underlying this cell-fate decision is highly complex and tightly regulated, and is controlled, at least in part, through various lineage-restricted transcription regulators including BCL6, BACH2, and PRDM1.5,6
BCL6 is a BTB-zinc finger family transcription repressor and a master regulator of the GC response.7-10 BCL6 protein is highly upregulated in GC B cells,11,12 where it regulates a broad network of direct target genes involved in various cellular processes.9,13-16 A critical biological function of BCL6 in GC B cells is to facilitate rapid replication and tolerance of genomic damage occurring during clonal expansion and somatic hypermutation by directly repressing DNA damage sensing and checkpoint genes such as ATR17 and TP53.18 Another important function of BCL6 is to suppress expression of the key plasma cell transcription factor PRDM1 to maintain the GC phenotype during affinity maturation and prevent premature differentiation.14,19-21 Bcl6-deficient (Bcl6−/−) B cells fail to form GC B cells in vivo8,10 and Bcl6−/− splenic B cells are prone to differentiate into plasma cells.19
The transcription factor BACH2 is widely expressed within the B-lymphoid lineage, except in plasma cells.22,23 As with BCL6, BACH2 contains an N-terminal BTB domain involved in its transcriptional function, but instead of having zinc fingers at the C-terminus, it binds to DNA through a basic leucine zipper motif.22 BACH2 binds DNA consensus sequences, termed MARE motifs (5′TGAG/CTCA3′), by forming a heterodimer with small leucine zipper MAF proteins such as MAFK, MAFG, and MAFF.22,23 The importance of Bach2 for mature B cells was revealed in Bach2-deficent mice (Bach2−/−), which is similar to Bcl6 −/− mice, display a loss of GCs, and are unable to produce high-affinity antibodies after T-cell–dependent antigen challenge.24 Also, as with Bcl6, Bach2 prevents mature B-cell differentiation into plasma cells in vitro, at least in part, by inhibiting Prdm1.25,26
Functional collaboration between transcription factors plays a key role in cell fate decisions. In the B-cell lineage, EBF1 and E2A cooperatively regulate early B-cell differentiation by coregulating a complex transcriptional network.27,28 Combinatorial actions of transcriptional activators have also been suggested to occur during GC B-cell differentiation5,6 and may help explain how GC B cells maintain their phenotypes, as well as potentially contribute to the pathogenesis of B-cell lymphomas, most of which originate from GC B cells. Several lines of evidence suggest that BCL6 and BACH2 cooperate in GC B cells: (1) Both are required for GC formation; (2) they physically interact in the 18-81 pre-B-cell line29; and (3) both repress PRDM1 transcription and plasma differentiation.14,19,25 These factors offer the opportunity to understand the cooperation between transcriptional repressors in GC B-cell differentiation. Here, we combined a genetic model with transcriptional functional assays to explore the cooperation of BACH2 and BCL6 in orchestrating the GC B-cell fate.
Methods
Mice and immunization
Bcl6−/− mice and Bach2−/− were kindly provided by H. Ye (Albert Einstein Medical College) and K. Igarishi (Tohoku University), respectively. To generate the µMT mixed chimera (Figure 2E), a mixture of 4 × 106 bone marrow cells from µMT (The Jackson Laboratory, stock number 002288) and wild-type (WT) or Bcl6+/−Bcl6+/− mice at a 4:1 ratio were intravenously transferred into sublethally irradiated Rag1−/− mice (The Jackson Laboratory, stock number 002612). For analysis of GC formation, mice were immunized intraperitoneally with sheep red blood cells (SRBC) (108 cells/mouse) and spleens were collected for analysis at 10 days after immunization. Mice were housed in the specific pathogen free animal facility at Weill Cornell Medical College and the animal experiments were performed using protocols approved by Institutional Animal Care and Use Committee.
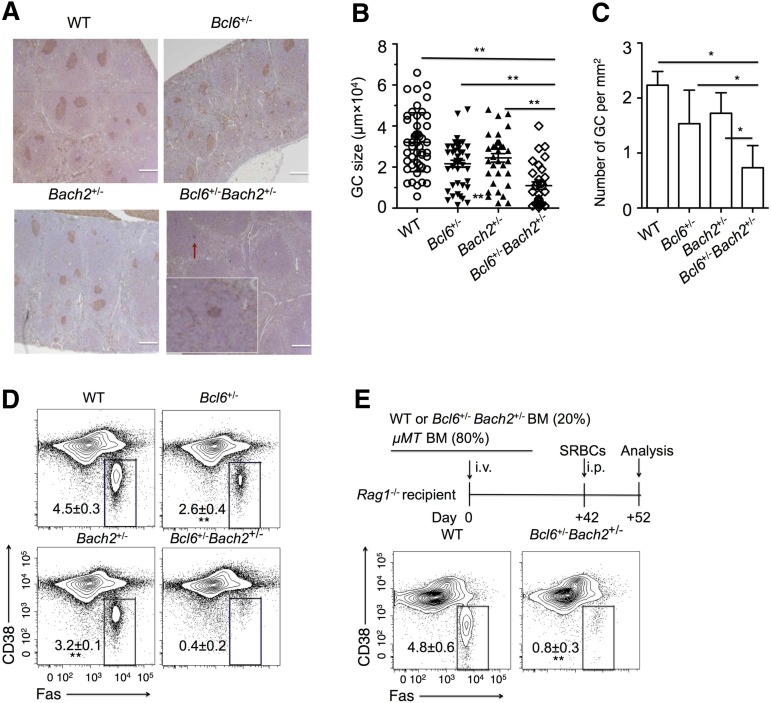
Figure 2.
Bcl6+/−Bach2+/− mice display a marked reduction of GCs. WT, Bach2+/−, Bcl6+/−, and Bach2+/−Bcl6+/− mice (n = 4/group) were immunized intraperitoneally with SRBC and euthanized after 10 days to evaluate GC formation. (A) Representative peanut agglutinin staining of splenic sections from immunized mice. A small GC in the Bcl6+/−Bach2+/− splenic section is indicated by the red arrow and shown as inset (original magnification ×20). Bars represent 200 μm (B-C). The size (B) and number (C) of GCs in spleen sections of immunized mice with the indicated genotypes. Individual dots represent each GC. (D) Representative flow cytometric plots of GC B cells (FAS+ CD38lo-neg, boxed) gated on live splenic B220+ lymphocytes from immunized mice. (E) μMT chimeras were generated by transferring either 20% WT or Bcl6+/−Bach2+/− with 80% μMT bone marrow cells. Seven weeks later, these chimeras were immunized with SRBCs and were euthanized 10 days later for analysis. Flow cytometry plots are shown depicting the percent of FAS+ CD38low/− GC B cells among viable splenic B220+ cells from the indicated μMT chimeras. Data are shown as mean ± standard error of the mean from 2 independent experiments. *P < .05 and **P < .01; 2-tailed Student t test.
Primary cell isolation, culture, and stimulation
Splenic B cells were isolated using a murine B cell negative selection kit (Miltenyi Biotech) according to the manufacturer’s protocol. B-cell purity was determined by flow cytometry and populations >95% were used for further experiments. B cells were grown in the medium containing 90% RPMI 1640 and 10% fetal calf serum supplemented with antibodies, l-glutamine, nonessential amino acids, N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, and 2-mercaptoethanol. B cells were treated with 10 µg/mL lipopolysaccharide (LPS) to induce differentiation before collection for analysis of gene expression and the generation of plasma cells. Normal human GC B cells and naïve B cells were isolated from fresh tonsils as previously described.15
Statistical analysis
A Student t test was performed for statistical analysis. The software GraphPad Prism 5 was used for this analysis. P <.05 is considered significant.
Additional experimental procedures are provided in the supplemental Methods, available on the Blood Web site.
Results
BACH2 expression is positively correlated with BCL6 in human GC B cells
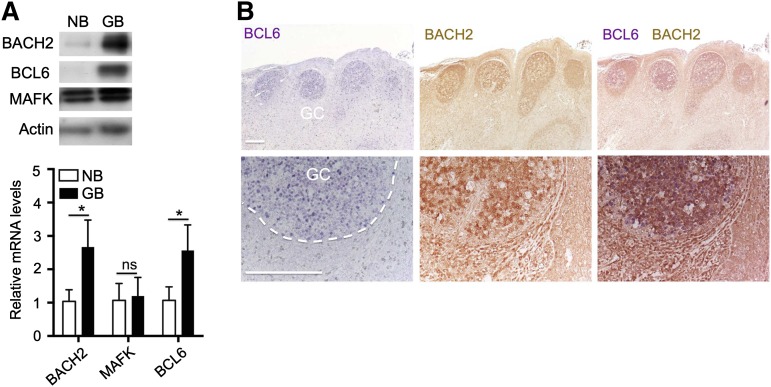
During transition from naïve B cells to GC B cells in both humans and mice, BCL6 messenger RNA (mRNA) is moderately increased, whereas BCL6 protein levels are more dramatically upregulated.11,12 BACH2 mRNA and protein were reported to be expressed in splenic IgM+ cells in unimmunized mice,23,24 however, it remains unknown whether their protein levels change significantly in GC B cells. To this aim, we first raised rabbit polyclonal antibodies against the BACH2 protein. Immunoblot analysis demonstrated that these antibodies specifically recognized exogenous expressed and endogenous BACH2 protein (data not shown). We then isolated naïve B cells and GC B cells from human tonsils and examined expressions of BACH2, MAFK, and BCL6 using qRT-PCR and western blot analysis. BACH2 mRNA abundance was only modestly increased, whereas its protein levels were markedly upregulated in GC B cells as compared with naïve B cells, although BACH2 protein was still detected at a low level in naïve B cells (Figure 1A). Immunohistochemistry in human tonsil sections showed strongly positive staining for BCL6 and BACH2 in GC B cells (Figure 1B). In contrast, MAFK expression remained unchanged at either mRNA or the protein level. Hence, BCL6 is coexpressed and its protein levels markedly induced together with BACH2 in normal GC B cells from human tonsils.
Figure 1.
Positive correlation of BCL6 and BACH2 expression in human GC B cell. (A) Immunoblot and quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis of BCL6, BACH2, and MAFK expression in naïve B cells (NB) and GC B cells (GB) purified from human tonsils. For qRT-PCR analysis, data are shown as mean ± standard error of the mean from 3 different sets of NBs and GBs. *P < .01; ns, not significant; 2-tailed Student t test. (B) Immunohistochemistry analysis of BCL6 (blue) and BACH2 (brown) in human tonsil sections. Bars represent 200 μm.
Bcl6+/−Bach2+/− mice exhibit a marked reduction of GCs
We wished to determine whether BCL6 and BACH2 cooperate biologically in mediating the GC phenotype. Given that Bcl6+/− or Bach2+/− (heterozygous null allele) mice exhibit minimal defects in GC response and/or plasma differentiation,8,24,26 we postulated that if these transcription factors cooperate in B cells, mice heterozygous for deletion of both factors might display a more severe phenotype. Bcl6+/− and Bach2+/− mice were crossed to generate litters containing WT, Bcl6, and Bach2 single or double heterozygous pups. All of these mice were viable, born at Mendelian frequencies and did not display developmental retardation or evidence of sickness. Similar to the single heterozygous Bach2+/− or Bcl6+/− mice, Bach2+/−Bcl6+/− mice formed normal primary splenic lymphoid follicles (supplemental Figure 1A). Analysis of early B-cell maturation in Bach2+/−Bcl6+/− mice indicated that B-cell development was normal in these animals (supplemental Figure 1B). Bach2−/− and Bcl6−/− mice display abnormal T-cell development.7,8,30 However, the proportions of T cells in splenic lymphocytes of Bach2+/−Bcl6+/− mice did not differ from those in WT mice (supplemental Figure 1C). Next, we analyzed formation of GCs in WT, Bcl6+/−, Bach2+/−, and Bcl6+/−Bach2+/− mice, 10 days after immunization with SRBC, a T-cell dependent antigen. Examination of spleens by immunohistochemistry showed that both Bach2+/− and Bcl6+/− mice displayed a mild reduction in GC size and number compared with WT mice at day 10 after SRBC immunization. However, Bach2+/−Bcl6+/− mice developed a more severe phenotype with fewer and smaller GCs (Figure 2A). Both GC size and number in spleens of Bach2+/−Bcl6+/− mice were significantly reduced as compared with those in Bach2+/−, Bcl6+/−, or WT mice after immunization with SRBCs (Figure 2B-C). We also used flow cytometry to compare the frequency of GC B cells (CD38lo-negFas+B220+) among splenic B220+ cells isolated from the spleens of WT, Bcl6+/−, Bach2+/−, and Bcl6+/−Bach2+/− mice 10 days after SRBC immunization. This analysis confirmed the mild reduction of GC B cells in Bcl6+/− (2.6 ± 0.4%) and Bach2+/− (3.2 ± 0.1%) mice compared with WT mice (4.5 ± 0.3%) (Figure 2D). In contrast, Bcl6+/−Bach2+/− mice displayed almost complete loss of GC B cells (0.4 ± 0.2%) 10 days postimmunization (Figure 2D). Finally, to determine whether deficiency in GC formation in Bcl6+/−Bach2+/− mice was intrinsic to B cells, we generated mixed chimeras by transplanting a mixture composed of 20% of either WT or Bcl6+/−Bach2+/− bone marrow cells with 80% μMT bone marrow cells into sublethally irradiated Rag1−/− mice. All B cells in these chimeras are derived from either the WT or Bcl6+/−Bach2+/− bone marrow cells, respectively. Bcl6+/−Bach2+/− chimeras exhibited profound reduction of GC B cells (0.8 ± 0.3%) compared with WT chimeras (4.8 ± 0.6%) after SRBC immunization (P < .01) (Figure 2E). Collectively, the data suggest that BACH2 and BCL6 cooperation may play a critical, cell-intrinsic role in the development of GC B cells in vivo.
Splenic Bcl6+/−Bach2+/− B cells are more prone to differentiate into plasma cells
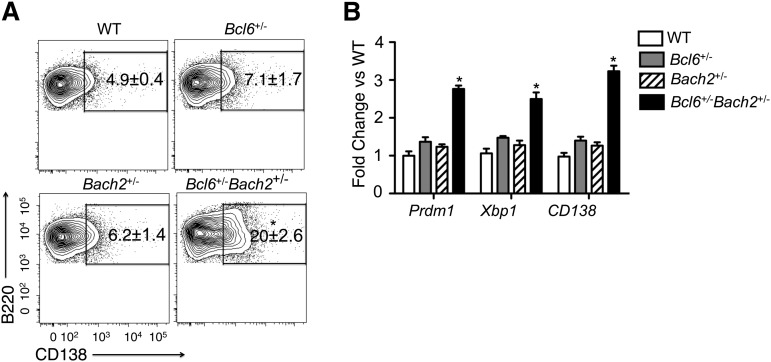
Previous studies showed that Bcl6−/− and Bach2−/− mice display accelerated plasma cell differentiation due to upregulation of Prdm1, which may explain, in part, the failure of these mice to form GCs.14,19,25,26 We explored whether reduction of the GC compartment in double heterozygous Bcl6+/−Bach2+/− mice is also associated with this phenotype. Plasma cell frequency was assessed among splenic B cells sorted from WT, Bcl6+/−, Bach2+/−, and Bcl6+/−Bach2+/− mice after 2 days of LPS stimulation in vitro to induce plasma cell differentiation31,32 by activating expression of Prdm1, Xbp1, and CD138.33 After LPS stimulation, the frequency of plasma cells in WT, Bcl6+/−, and Bach2+/− was 4.9 ± 0.4%, 7.1 ± 1.7%, and 6.2 ± 1.4%, respectively, but were markedly increased to 20 ± 2.6% among Bach2+/− Bcl6+/− B cells (P < .05; 2-tailed Student t test) (Figure 3A). Furthermore, qRT-PCR revealed significant increase in the abundance of Prdm1, Xbp1, and CD138 transcripts in Bach2+/−Bcl6+/− B cells compared with single mutant cells (P < .05; 2-tailed Student t test) (Figure 3B). Taken together, these experiments suggest cooperative functional interaction between BACH2 and BCL6 in suppressing the terminal differentiation of mature B cells.
Figure 3.
Bcl6+/−Bach2+/− splenic B cells undergo rapid differentiation ex vivo. (A) Flow cytometry was performed on mouse splenic B220+ cells treated with LPS for 48 hours. Numbers in outlined areas indicate percent CD138+ (plasma) cells. (B) qRT-PCR analysis of Prdm1, Xbp1, and CD138 mRNA levels in LPS-treated splenic B cells. Data are presented as mean ± standard error of the mean from 2 independent experiments (A-B). *P < .01 when compared with single heterozygous or WT mice; 2-tailed Student t test.
The BACH2 cistrome overlaps with MAFK and is linked to GC-related gene sets
The complement of direct target genes of BACH2 in B cells is unknown and must be elucidated to understand its immediate effect on transcriptional programming. To identify direct BACH2 target genes, we first evaluated our anti-BACH2 antibodies in chromatin immunoprecipitation (ChIP) experiments. BACH2 was reported to bind the PRDM1 promoter through a putative MARE element.25 ChIP quantitative polymerase chain reaction (QChIP) analysis showed robust and specific enrichment of the PRDM1 promoter region in BACH2-positive OCI-LY1 diffuse large B-cell lymphoma (DLBCL) cells, but not BACH2-negative Karpas422 DLBCL cells (supplemental Figure 2A-B), which confirms the suitability and specificity of these antibodies for ChIP experiments.
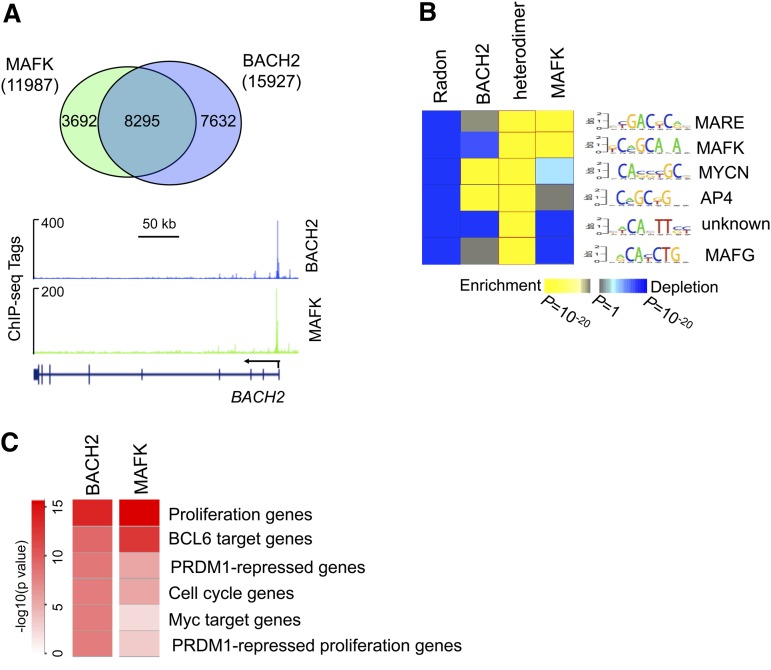
To identify the cistrome of BACH2 and its cofactor MAFK, we next performed ChIP sequencing (ChIP-seq) for these proteins in the GCB-type DLBCL cell line OCI-LY7, which expresses abundant BACH2, MAFK as well as BCL6 proteins (supplemental Figure 2A). A total 15 927 BACH2 and 11 987 MAFK DNA binding sites were identified. These binding sites corresponded to 5107 genes bound by BACH2 and 4039 genes bound by MAFK in OCI-LY7 cells (supplemental Figure 3A-B). Although 34% BACH2 and 36% MAFK binding sites were located within promoters, most of the remaining binding sites were associated with either introns or intergenic regions (supplemental Figure 3A-B). Notably, approximately 60% of the BACH2-bound sites (8295 out of 15 927 sites) were also occupied by MAFK (Figure 4A), consistent with the notion that BACH2 forms heterodimers with MAFK to regulate most of its target genes. Of note, BACH2-MAFK complexes bound to the proximal promoter region of the BACH2 gene (Figure 4A), suggesting potential autoregulation of BACH2.
Figure 4.
Genome-wide occupation of BACH2 and MAFK in B cells. (A) Union analysis of BACH2 and MAFK peaks. Numbers indicate unique and overlapping peaks. ChIP-seq tracks for BACH2 and MAFK ChIP-seq are shown for the BACH2 gene. (B) The heat map represents the relative enrichment of transcription factor motifs overpresented in BACH2, MAFK, and BACH2/MAFK heterodimer peaks relative to random genomic regions. (C) A heat map is shown representing the relative enrichment of gene signatures among BACH2-MAFK target genes. The statistical significance is shown in the color key. P values were calculated by Fisher’s exact test with Benjamini-Hochberg correction.
Next, we used an unbiased DNA motif discovery algorithm to identify DNA sequences enriched at BACH2-, MAFK- and heterodimer-bound sites. Notably, the most highly enriched sequence (5′ T/CGACT/CCA3′; P < 10−20) bound by both BACH2 and MAFK, is highly similar to the canonical MARE motif (5′TGAG/CTCA3′) (Figure 4B). This motif may represent a more physiological BACH2/MAFK MARE in B cells, as opposed to sequences identified by in vitro screening.22 Other DNA elements that were highly overrepresented at sites with BACH2-MAFK complexes included MAFK, MYCN, AP4, and MAFG (Figure 4B). We next explored the association of BACH2 and MAFK target genes with lymphoid-specific gene signatures.34 The BACH2-MAFK target genes are highly enriched in cell proliferation genes and BCL6 target genes identified by ChIP-on-ChIP (P < 10−10 and P < 10−9, respectively) (Figure 4C). A more complete description of these signatures is provided in supplemental Table 4.
Widespread coordination of BACH2 and BCL6 binding to the genome
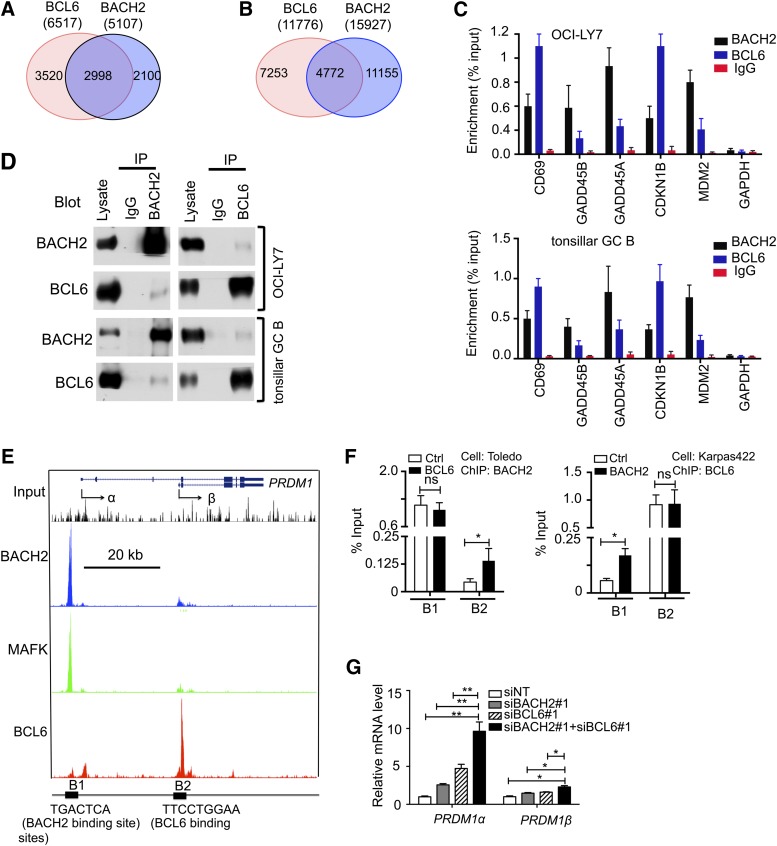
Cooperation between transcription factors has been associated with binding to overlapping target gene sets, and coordinated, at least in part, by recruitment to common or closely linked DNA regulatory elements.28 Therefore, we examined whether BACH2 and BCL6 might broadly colocalize throughout the genome. We performed ChIP-seq for BCL6 in OCI-Ly7 cells and compared BCL6 and BACH2 target gene sets. There were 2998 of 5107 BACH2 target genes (approximately 60%) also present within the 6517 BCL6 targets (Figure 5A). Next, we aligned the BACH2 and BCL6 peaks. There were 4772 of 15 927 BACH2 peaks (30%) overlapped with BCL6 peaks (Figure 5B). Examples of genes containing overlapping peaks included PRDM1 and CD69, as well as checkpoint genes such as GADD45A, GADD45B, CDKN1B, and MDM2. The presence of BCL6 and BACH2 at these loci was confirmed by QChIP in OCI-LY7 cells and primary GC B cells isolated from human tonsils (Figure 5C).
Figure 5.
BACH2 co binds with BCL6 to sets of genes involved in GC B-cell development including PRDM1. (A-B) Venn diagram representations illustrate the number of overlapping and unique target genes (A) and binding sites (B) of BACH2 and BCL6 identified by ChIP-seq in OCI-Ly7 cells. (C) QChIP was performed to determine enrichment of BCL6, BACH2, and MAFK at the indicated gene loci in OCI-LY7 (top panel) and primary human GC B cells (bottom panel). (D) Reciprocal coimmunoprecipitations were performed to detect the interaction of endogenous BCL6 and BACH2 in OCI-LY7 and primary human GC B cells. (E) The illustration depicts ChIP-seq tracks of BACH2, MAFK, and BCL6 at the PRDM1 locus. The transcription start sites of PRDM1α and PRDM1β are indicated by the arrows. Consensus DNA binding elements located at the major binding sites (B1 and B2) are shown at the bottom. (F) QChIP was performed to detect enrichment of BACH2 at B1 and B2 sites in Toledo DLBCL cell infected with a control virus (Ctrl) or virus expressing BCL6 (left panel), and reciprocally to detect BCL6 binding in Karpas422 DLBCL cells infected with a control (Ctrl) or virus expressing BACH2 (right panel). (G) qRT-PCR analysis was performed to assess the mRNA abundance of PRDM1α and PRDM1β in OCI-LY1 cells transfected with either scrambled non-specific short interfering RNA (siNT), BACH2 small interfering RNA (siRNA) (siBACH2#1), BCL6 siRNA (siBCL6#1), or siBACH2, together with siBCL6 after 72 hours. Data are presented as fold change of relative PRDM1 mRNA levels normalized to HPRT. Data are representative of 3 independent experiments (C,F,G). *P < .05; ns, not significant; 2-tailed Student t test.
The broad colocalization of BACH2 and BCL6 may be caused by at least 2 possible mechanisms. Specifically, BACH2 and BCL6 may bind 2 different, but proximal, DNA elements; whereas, at other loci these factors might physically interact and recruit each other to their respective DNA binding sites. The PRDM1 gene is of particular interest because it is a key target of BCL6 and BACH2. BCL6 has been reported to interact with BACH2 in HEK293 cells exogenously overexpressing these proteins and in 18-81 pre-B cells.29 Indeed in coimmunoprecipitation assays, we observed an endogenous protein complex formed by BACH2 and BCL6 in primary human tonsillar GC B cells, as well as in several GC-derived DLBCL cell lines including OCI-LY7 (Figure 5D and supplemental Figure 5). This finding prompted us to explore whether the corecruitment model could explain binding of these factors to the PRMD1 locus. ChIP-seq data revealed that BCL6 and BACH2 were bound and colocalized to 2 major sites across the PRDM1 locus in OCI-Ly7 cells (Figure 5E). Analysis of the DNA sequence of these regions revealed the presence of a MARE motif-containing region (B1) at the promoter of long PRDM1 isoform (PRDM1α), and a BCL6 consensus binding element-containing region (B2) within intron 3, which is also the promoter for the short PRDM1 isoform (PRDM1β). These locations are consistent with previous reports showing binding of BACH2 and BCL6 to the PRDM1 locus.25,35 We also confirmed this binding pattern in primary human tonsillar GC B cells (supplemental Figure 4A).
To determine whether there was any interdependency of BCL6 or BACH2 binding, we next compared the enrichment of these 2 factors by QChIP in 3 DLBCL cell lines with differential expression of these 2 transcription factors. We observed that BACH2 enrichment was markedly lower at the PRDM1 intron 3 locus in Toledo cells, which express almost no BCL6, compared with the highly BCL6-postive OCI-LY7 cell line, although its binding at the PRDM1 promoter was similar in the 2 cell lines (supplemental Figure 4B). In contrast, the DNA binding activity of BCL6 at the promoter site (B1) was significantly reduced in the BACH2-negative Karpas422 cell line compared with OCI-LY1 cells, although BCL6 showed similar enrichment at the intron 3 (B2) locus in 2 cell types (supplemental Figure 4B). Recruitment of BCL6 to the PRMD1 promoter site could be rescued in Karpas422 cells by ectopic expression of BACH2, whereas BACH2 occupancy of intron 3 was rescued in Toledo cells after transfection with a BCL6 expression vector (Figure 5F). Thus, the binding of BCL6 to the PRDM1 promoter is indirectly mediated through BACH2, whereas the binding of BACH2 to intron 3 is dependent on BCL6. Finally, qRT-PCR revealed significant increase of the mRNA abundance of PRDM1α and PRDM1β, more strikingly for PRDM1α, in the OCI-LY1 cell with simultaneous knockdown of BACH2 and BCL6 compared with those with knockdown of each gene alone (Figure 5G). Taken together, these results demonstrate that these factors cooperatively repress PRDM1 expression.
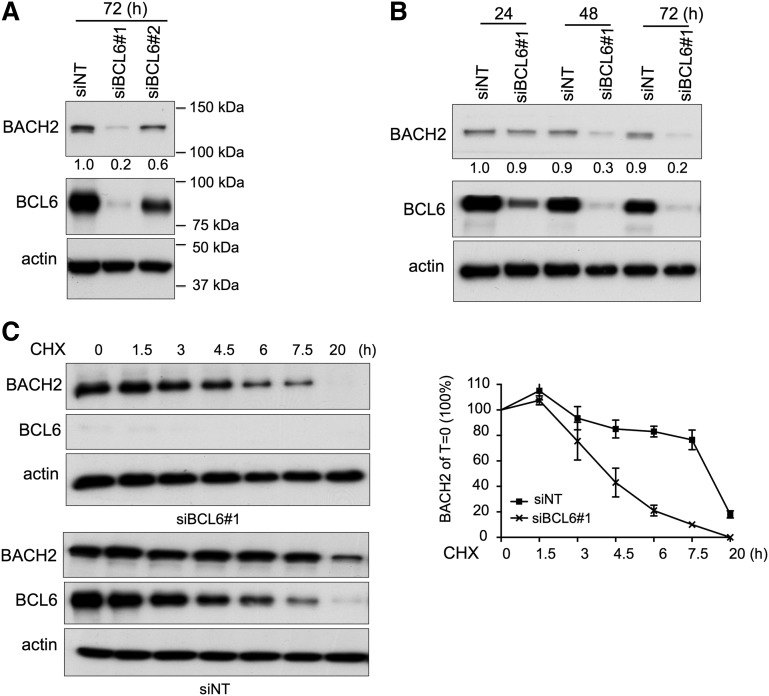
BCL6 is required for BACH2 protein stability
To better understand how BACH2 and BCL6 cooperate functionally, we investigated additional aspects of the significance of their interactions at the protein level. First, we depleted BCL6 protein in OCI-LY1 cells using siRNA and then we examined the BACH2 expression. BCL6 knockdown did not affect BACH2 mRNA expression (supplemental Figure 6A), but resulted in marked reduction of BACH2 protein at 48 and 72 hours after siRNA treatment (Figure 6A-B). This effect was reproducible in additional DLBCL cell lines (supplemental Figure 6B). However, BACH2 knockdown did not affect BCL6 protein levels (supplemental Figure 6C). To further investigate the temporal association between BCL6 and BACH2 protein levels, we examined BACH2 protein levels in OCI-Ly1 cells that were exposed to cycloheximide (a protein synthesis inhibitor) starting at 24 hours after BCL6 siRNA, a time point at which BCL6 protein is decreased, but BACH2 protein is not changed. Relative BACH2 protein abundance decreased from 100% to 10% in BCL6 siRNA transfected cells compared with 80% in control siRNA treated cells at 7.5 hours after cycloheximide treatment (Figure 6C). Hence, BCL6 maintains BACH2 protein stability in B cells.
Figure 6.
BCL6 stabilizes BACH2 protein. (A) Immunoblot analysis of BACH2 and BCL6 protein levels in OCI-LY1 cells treated with siNT or siBCL6 (#1 and #2) for 72 hours. (B) Immunoblot analysis of BACH2 and BCL6 protein levels in OCI-LY1 cells treated with siNT or siBCL6#1 at 3 different time points. (A-B) The relative amount of BACH2 to actin was quantified by densitometry (indicated by numbers below the BACH2 immunoblot). (C) OCI-LY1 cells were treated with siNT or siBACL6#1 for 24 hours, followed by incubation with 5 μM cycloheximide (CHX) for indicated time periods. Immunoblot analysis was performed to determine BCL6 and BACH2 protein levels. The relative amount of BACH2 to actin was quantified by densitometry (Y-axis) and plotted with respect to time (X-axis). The BACH2 to actin level in CHX-untreated cells is calculated as 100%.
Discussion
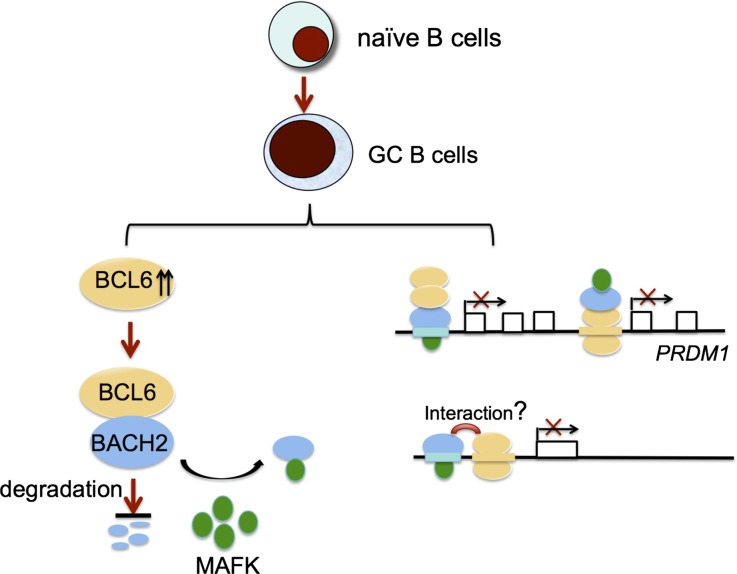
The BCL6 transcriptional repressor is a master regulator of GC B-cell development. Its canonical mechanism of action involves binding to its cognate DNA binding sequence, recruiting co-repressors, and directly repressing gene expression. However, BCL6 may also interact with other transcription factors, which may contribute to specific aspects of BCL6-driven cellular functions. For example, BCL6 was shown to interact with AP-1 in Raji lymphoma cells,20 PPARδ in macrophages,36 T-bet in T cells,37 and with BACH2 in B cells.29 The latter finding is of particular interest given that Bach2 homozygous deletion induces a GC defect that is reminiscent of that observed in Bcl6 null mice. These studies prompted us to explore whether BCL6 and BACH2 truly and functionally cooperate in GC formation. Such results would suggest a model, whereby the striking effects of BCL6 in B cells might represent a more complex process than its independent binding and repression to target genes. Indeed, our data collectively support the notion that BCL6 actions in GC B cells are, in part, driven through extensive cooperation with BACH2. Consistent with this notion, single Bcl6+/− or Bach2+/− heterozygous mice display only minimal perturbation in GC formation, whereas, in contrast double heterozygous Bcl6+/−Bach2+/− mice displayed a marked impairment of GC formation and reduction of GC B cells in response to T-cell dependent antigen immunization. Moreover, Bcl6+/−Bach2+/− B cells were impaired in their ability to maintain the GC phenotype, as demonstrated in plasma cell differentiation assays and their significantly greater expression of plasma cell genes. The functional link between these 2 proteins is further supported by the fact that they are coordinately upregulated in GC B cells. Cooperative functions of transcription factors are known to occur in early B-cell development. For example, EBF1 collaborates with E2A to orchestrate the pro-B–cell fate.27 Our study extends this paradigm to transcriptional repressors that control GC phenotype, and shows that although BCL6 is individually required for GC B-cell development in vivo, Bcl6 cooperates with Bach2 to control and maintain the GC phenotype.
BCL6 blocks GC B cells from undergoing plasma cell differentiation, at least in part, through direct repression of the PRDM1 locus,38-41 but the mechanism of BCL6-mediated transcriptional repression of PRDM1, in fact, is much more complex. BCL6 is most abundantly associated with intron 3 of PRDM1 via a canonical BCL6 binding site.35 However, BCL6 has been suggested to indirectly bind to the PRDM1 promoter by association with AP-1 in Raji lymphoma cells.20 Here, we found that BACH2 is also responsible for BCL6 indirect binding at the PRDM1 promoter. The PRDM1 promoter contains a classic MARE motif and BACH2 has been suggested to bind this region in the BA17 mature B cell line.25 Moreover, coimmunoprecipitation experiments revealed that BACH2 interacts with BCL6 in GC–derived B cells, consistent with a previous report showing BCL6/BACH2 interactions in a pre-B–cell line.29 Mostly importantly, BCL6 binding to the PRDM1 promoter is dependent on BACH2. Thus, we propose that BCL6 is indirectly recruited to the BACH2 binding site at the PRDM1 promoter region through physical interaction with BACH2. Reciprocally, BACH2 is indirectly targeted to the BCL6 binding site at the PRDM1 intron 3 via BACH2/BCL6 interaction (Figure 7). We did not observe BACH2 and BCL6 binding at intron 5 of PRDM1, which was reported in a murine pre-B–cell line,29 suggesting that BACH2/BCL6 binding may be species-context dependent. Collectively, this dually reciprocal binding pattern of BCL6 and BACH2 likely explains their cooperative repression of PRDM1 transcription.
Figure 7.
The molecular mechanisms of functional cooperation between BCL6 and BACH2. BCL6 maintains BACH2 protein stability (left). BCL6 and BACH2 recruit each other to their own DNA binding sites (PRDM1) through their physical interactions, or they bind to 2 different, but proximal DNA elements, and their binding is potentially promoted by their interactions (right).
The significant overlap between the BCL6 and BACH2 cistromes in GC derived B cells suggests a much broader functional collaboration. It is well established that transcriptional factors can be linked together by networks of cis-regulatory elements, allowing them to cooperatively regulate gene expression throughout the genome. In additional to PRDM1, among the loci that show overlapping BCL6 and BACH2 binding sites were a large set of loci known to be involved in cell proliferation, survival, DNA damage, and differentiation, all of which are features of the GC B-cell phenotype. It is plausible that BCL6 and BACH2 cooperatively affect the phenotype of GC B cells in additional ways, not limited to differentiation. The overlapping cistrome of BCL6 and BACH2 suggests at least 2 potential mechanisms through which BCL6 and BACH2 could cooperate in regulating the GC transcriptional program: (1) by forming a complex and recruiting each other to their respective DNA binding sites, as in the case of the dual reciprocal mechanism shown for the PRDM1 gene; and (2) by binding to 2 different but proximal DNA elements at specific gene loci (Figure 7). In the latter case, the BCL6-BACH2 direct interactions may still facilitate their binding to their own DNA binding sites, although our study was not designed to address this point. Either way, the interdependency between BCL6 and BACH2 is further underlined by our finding that BCL6 is required for BACH2 protein stability by regulating its turnover (Figure 7). The coordinated expression of BCL6 and BACH2 protein may be needed for the establishment of their functional cooperation in GCs.
Of note, although ChIP-seq demonstrated that BACH2- and MAFK-binding sites largely overlap (Figure 3A), MAFK was not essential to GC B cells. MAFK knockdown did not up regulate PRDM1 mRNA levels in OCI-LY1 cells, and MAFK knockout mice have normal GC (data not shown). BACH2 is able to form a heterodimer with other small leucine zipper MAF proteins including MAFG and MAFF.22,23 Thus, MAFK may function in a redundant manner with other MAF proteins to heterodimerize with BACH2 to bind to DNA and regulate GC formation.
Overall, this study provides evidence for widespread and functionally significant cooperation between BCL6 and BACH2 in determining aspects of the GC B-cell phenotype. Deregulation of GC B-cell differentiation has been linked to human and mouse B-cell lymphomas.5,40,41 Further studies will be needed to reveal whether functional collaboration of BCL6 and BACH2 may also be relevant to the pathogenesis of B-cell lymphomas.
Supplementary Material
Acknowledgments
We thank H. Ye from the Albert Einstein College of Medicine for sharing Bcl6−/− mice and K. Igarish from Tohoku University for Bach2−/− mice.
This work was supported by the National Institutes of Health, National Cancer Institute (NCI-R01 104348) (A.M.), and this work was facilitated by the Sackler Center for Biomedical and Physical Sciences at Weill Cornell Medical College.
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: C.H. designed the experiments, interpreted data, and wrote the first version of the manuscript; H.G. analyzed ChIP-seq data; C.H., I.B., and L.W. did the experiments; and A.M. conceived the project and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ari Melnick, Division of Hematology and Oncology, Department of Medicine, Weill Cornell Medical College, 1300 York Ave, New York, NY 10065; e-mail: amm2014@med.cornell.edu.
References
- 1.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 2.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27(2):190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 4.Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11(8):681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- 5.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8(1):22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5(3):230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 7.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276(5312):589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 8.Ye BH, Cattoretti G, Shen Q, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16(2):161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 9.Huang C, Hatzi K, Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat Immunol. 2013;14(4):380–388. doi: 10.1038/ni.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuda T, Yoshida T, Okada S, et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186(3):439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cattoretti G, Chang CC, Cechova K, et al. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86(1):45–53. [PubMed] [Google Scholar]
- 12.Allman D, Jain A, Dent A, et al. BCL-6 expression during B-cell activation. Blood. 1996;87(12):5257–5268. [PubMed] [Google Scholar]
- 13.Hatzi K, Jiang Y, Huang C, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013;4(3):578-588. [DOI] [PMC free article] [PubMed]
- 14.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13(2):199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 15.Ci W, Polo JM, Cerchietti L, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113(22):5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basso K, Saito M, Sumazin P, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115(5):975–984. doi: 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranuncolo SM, Polo JM, Dierov J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8(7):705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 18.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432(7017):635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 19.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173(2):1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 20.Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169(4):1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- 21.Shaffer AL, Lin KI, Kuo TC, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 22.Oyake T, Itoh K, Motohashi H, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16(11):6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muto A, Hoshino H, Madisen L, et al. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3′ enhancer. EMBO J. 1998;17(19):5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muto A, Tashiro S, Nakajima O, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429(6991):566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 25.Ochiai K, Katoh Y, Ikura T, et al. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem. 2006;281(50):38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- 26.Muto A, Ochiai K, Kimura Y, et al. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010;29(23):4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11(1):21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 28.Lin YC, Jhunjhunwala S, Benner C, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11(7):635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochiai K, Muto A, Tanaka H, Takahashi S, Igarashi K. Regulation of the plasma cell transcription factor Blimp-1 gene by Bach2 and Bcl6. Int Immunol. 2008;20(3):453–460. doi: 10.1093/intimm/dxn005. [DOI] [PubMed] [Google Scholar]
- 30.Tsukumo S, Unno M, Muto A, et al. Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proc Natl Acad Sci USA. 2013;110(26):10735–10740. doi: 10.1073/pnas.1306691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafrenz D, Koretz S, Stratte PT, Ward RB, Strober S. LPS-induced differentiation of a murine B cell leukemia (BCL1): changes in surface and secreted IgM. J Immunol. 1982;129(3):1329–1335. [PubMed] [Google Scholar]
- 32.Schliephake DE, Schimpl A. Blimp-1 overcomes the block in IgM secretion in lipopolysaccharide/anti-mu F(ab’)2-co-stimulated B lymphocytes. Eur J Immunol. 1996;26(1):268–271. doi: 10.1002/eji.1830260142. [DOI] [PubMed] [Google Scholar]
- 33.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21(1):81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Shaffer AL, Wright G, Yang L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 35.Parekh S, Polo JM, Shaknovich R, et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110(6):2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takata Y, Liu J, Yin F, et al. PPARdelta-mediated antiinflammatory mechanisms inhibit angiotensin II-accelerated atherosclerosis. Proc Natl Acad Sci USA. 2008;105(11):4277–4282. doi: 10.1073/pnas.0708647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208(5):1001–1013. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 39.Falini B, Fizzotti M, Pucciarini A, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 2000;95(6):2084–2092. [PubMed] [Google Scholar]
- 40.Mandelbaum J, Bhagat G, Tang H, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18(6):568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calado DP, Zhang B, Srinivasan L, et al. Constitutive canonical NF-κB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell. 2010;18(6):580–589. doi: 10.1016/j.ccr.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.