Abstract
Lysosomes require the presence of many specialized proteins to facilitate their roles in cellular maintenance. One such protein that has proven to be an important player in the lysosomal field is lysosomal integral membrane protein-2 (LIMP-2), encoded by the gene SCARB2. LIMP-2 is required for the normal biogenesis and maintenance of lysosomes and endosomes and has been identified as the specific receptor for glucocerebrosidase, the enzyme deficient in Gaucher disease. Research into LIMP-2 and the SCARB2 gene indicate that it may be a factor contributing to the clinical heterogeneity seen among patients with Gaucher disease. Mutations in SCARB2 have also been identified as the cause of action myoclonus renal failure (AMRF), and in some cases progressive myoclonic epilepsy. A total of 14 disease-causing SCARB2 mutations have been identified to date. The role of LIMP-2 in human pathology has expanded with its identification as a component of the intercalated disc in cardiac muscle and as a receptor for specific enteroviruses, two unanticipated findings that reaffirm the myriad roles of lysosomal proteins. Studies into the full impact of LIMP-2 deficiency and the LIMP2/glucocerebrosidase molecular pathway will lead to a better understanding of disease pathogenesis in Gaucher disease and AMRF, and to new insights into lysosomal processing, trafficking and function.
Keywords: Lysosomal Integral Membrane Protein-2, Lysosomal Membrane Protein, Gaucher disease, Action Myoclonus Renal Failure, Glucocerebrosidase, Transporter
1. Introduction
1.1. Biological functions of LIMP-2 protein and related disorders
Lysosomal integral membrane protein-2 (LIMP-2) is a type III glycoprotein (MIM: 602257, Locus: NM_005506) located in the limiting membranes of lysosomes and endosomes that contains two transmembrane domains. Among its known functions is its role in transporting the lysosomal protein glucocerebrosidase (GCase) from the endoplasmic reticulum (ER) through the Golgi system to lysosomes, the site of cellular degradation. Lysosomes are ubiquitous organelles crucial for the normal functioning of eukaryotic cells. They are responsible for the degradation and recycling of both extracellular and intracellular materials, relying on the presence of acid hydrolases and lysosomal membrane proteins (LMPs) to carry out this role. At least 50 lysosomal acid hydrolases have been identified, each of which is active at acidic pH and specifically targets its substrate for degradation [1]. These hydrolases include nucleases, lipases, glycosidases, phosphatases, sulfatases, and proteases, which together allow for the diverse catabolic capacity of lysosomes [2].
Although acid hydrolases are immediately responsible for the breakdown of complex molecules, the lysosomal membrane, along with the LMPs embedded within it, are necessary precursors for a functioning lysosome. Most notably, the membrane must maintain an acidic lumen, selectively transport materials, and mediate lysosomal fusion with other membrane-bound organelles [3,4]. To carry out these functions, the lysosomal membrane contains as many as 100 LMPs, of which about 25 have been actively studied, and all of which are likely crucial to lysosomal biogenesis and function [1,5,6].
Deficiencies of lysosomal hydrolases or LMPs contribute to the accumulation of substrate that cannot be properly transported into or degraded within the lysosome. Depending on the nature of the substrate involved, this accumulation can have broad physiological effects, although the path from storage to pathogenesis remains largely unclear [7]. Together, these metabolic disorders are known as lysosomal storage disorders (LSDs). The first LSD described was Tay-Sachs disease in 1881, preceding the discovery of the lysosome [8]. It was not until 1963 that the first disease (Pompe) was classified as an LSD, but the lysosomal storage disorders now encompass a group of more than 50 inherited disorders [9,reviewed in 10]. Although most LSDs are autosomal recessive, some such as Fabry, Hunter, and Danon diseases are inherited in an X-linked manner [8,11]. As a whole, the incidence of LSDs in the general population is estimated to be around 1 in 5000, and specific disorders are far more common within certain ethnic groups [12].
Although LSDs are relatively rare in occurrence, there is an increasing awareness of their importance in the understanding of molecular and cellular pathways as well as clinical medicine. Disorders affecting lysosomal function represent a paradigm in biological studies in which the elucidation of a highly specific pathway can lead to unanticipated connections between more prevalent illnesses. This proved true for the study of the most common LSD, Gaucher disease (GD), when mutations in the glucocerebrosidase gene (GBA1) were found to be a risk factor for parkinsonism and other Lewy body disorders [13,14]. Mutations in GBA1 are now accepted as the most common genetic risk factor for developing Parkinson disease (PD), a disorder affecting one percent of Americans over age 60 [15]. This unexpected connection between GD and PD has elevated current interest in lysosomal disorders and in GD in particular [16]. Other LSDs may similarly provide insights into aspects of aging and Alzheimer disease, where lysosomal pathways may play an important role [17].
GD is traditionally considered a single-gene disorder, and as such, GD research has generally focused on the GBA1 gene and the translated lysosomal hydrolase, GCase. This has led to much progress in the study of GD, culminating in the first successful treatment of an LSD with enzyme replacement therapy (ERT) for GD in the early 1990s [8]. GCase, however, does not act in isolation, a point emphasized by the broad phenotypic diversity observed in patients with GD and the same GBA1 genotype [18]. Even monozygotic twins have been reported with different clinical manifestations [19]. With such clinical heterogeneity associated with a given genotype, GD is clearly more than a simple, single-gene disorder and other modifying factors must play a role. When considering genetic modifiers, a logical focus is on the entire network involved in lysosomal processing.
Progress in this direction was made in 2007, when Reczek et al. discovered that LIMP-2 is a receptor for GCase and is involved in targeting the enzyme to late endosomes and lysosomes [20]. Identification of LIMP-2 as a specific receptor of GCase offers a new direction for GD research that could help to explain the phenotypic diversity associated with different GBA1 mutations, and could have applicability to the study of other LSDs. This review aims to summarize the current knowledge of the structure and function of LIMP-2, highlighting its disease associations and pleiotropic functions that lend insight into the study of LSDs.
1.2. A brief history of LIMP-2 research
LIMP-2 was discovered in 1985 by Lewis et al. in an immunological study of rat liver lysosomal membranes [3]. The authors discussed in detail their discovery of three lysosomal glycoproteins (LGPs) of molecular weights 120, 100, and 80 kDa. A fourth, 85 kDa protein was precipitated from the aqueous phase and recognized by their polyclonal antibody [3]. Lewis et al. suggested that this 85 kDa protein could be a soluble fragment of the previously mentioned 100 kDa LGP, but it was more likely a distinct LIMP, namely the 85 kDa LIMP-2 protein [3]. The following year, Barriocanal et al. published a paper elucidating the transport of LIMP-2 (in addition to LIMP-1 and LIMP-3) in normal rat kidney cells. Using time-course studies, Barriocanal et al. determined the glycosylation and transport pathway of LIMP-2 from its point of synthesis in the ER to the trans-Golgi network (TGN) [21]. It was not until the early 1990s that LIMP-2 was structurally defined, following the cloning and sequencing of the rat and human cDNAs [22]. Since then, the study of LIMP-2 has focused on its structural characterization and role in overall lysosomal maintenance.
A decade after its isolation, the first LIMP-2 over-expression studies in COS1 and other cell lines were undertaken, revealing that LIMP-2 over-expression results in swollen vacuoles and impaired trafficking out of the enlarged compartments, providing further evidence implicating a role for LIMP-2 in the biogenesis of the lysosome [23]. The following year, the same group published the first LIMP-2 knockout mouse model, demonstrating the phenotypic consequences of LIMP-2 deficiency in vivo [24]. LIMP-2 deficient mice were shown to develop obstruction of the ureteric pelvic junction, deafness, and a peripheral demyelinating neuropathy [24].
The spectrum of manifestations associated with LIMP-2 expanded further in 2007, when LIMP-2 was identified as a regulator of the cardiac intercalated disc in humans [25]. That same year, LIMP-2 was identified as a receptor for GCase [20]. LIMP-2 was most recently found to be associated with action myoclonus renal failure (AMRF), with its deficiency causing myoclonic epilepsy and glomerulosclerosis [26]. Each of these disease associations will be discussed in greater detail after a comprehensive description of the LIMP-2 protein.
1.3. LIMP-2 among the LMPs
In order for lysosomes to function as an acidic compartment capable of intracellular degradation of endogenous and exogenous macromolecules, they require a specific lysosomal membrane structure, which serves both to separate their catabolic activity from other cellular functions, and to maintain the necessary acidic lumenal pH. The passage of material into and out of the lysosome is typically controlled by a single phospholipid bilayer membrane containing diverse lysosomal proteins [6,27]. LMPs have proven to be instrumental in different aspects of lysosomal function, facilitating their biogenesis and its interaction with other organelles. Furthermore, LMPs provide a means for the selective trafficking of materials, and for protecting the lysosomal membrane from degradation [4]. In all, at least 12 diseases have been directly linked to deficiencies of LMPs, with a broad range of associated phenotypes that vary in both the age of onset and severity of symptoms [6,9].
The most abundant LMPs are the well-characterized lysosomal associated membrane proteins (LAMP)-1 and LAMP-2, which account for more than fifty percent of all lysosomal membrane proteins [6,9,28]. Many more LMPs were identified using proteomic approaches, with one study revealing a total of 124 proteins that are enriched in the lysosomal membranes of placentas [5]. It is difficult to determine how many of the proteins identified are actual LMPs without extensive validation, but it is clear that there are a wide range of proteins involved in the structural and trafficking support of lysosomal membranes, with our knowledge of their complexity just beginning.
In general, LMPs contain a highly glycosylated luminal region important for withstanding the acidity of the lysosomal compartment [21,29]. The glycosylated regions are thought to form a continuous glycoprotein layer within the lysosome, serving as a barrier to acid hydrolases and lending to lysosomal stability [27]. The luminal region structurally contains the largest portion of LMPs, and is attached to the transmembrane domain, a region that spans the membrane one or more times [29]. All LMPs are thought to reach the lysosomes by one of two ways: via the direct route where LMPs are shuttled to the lysosomes directly from the TGN; or alternatively, the transport may be indirect, with the LMP first secreted to the plasma membrane, and then internalized by endocytosis [6]. Also contributing to the targeting of LMPs to lysosomes are the inclusion of sorting signals in the short cytoplasmic tail, typically consisting of either a tyrosine or di-leucine motif that interacts with adaptor protein complexes [27]. LIMP-2 happens to follow the direct route to the lysosome, with a di-leucine residue crucial for its targeting to lysosomes [21,27,30].
The categorization of LMPs are based on structural homology and the location of the NH2 and COOH terminal domains with regard to the lysosomal membrane [28]. The abundant LAMP-1 and LAMP-2 proteins share 37% amino acid sequence homology and each contain one transmembrane domain and a conserved COOH cytoplasmic tail [28]. The LIMPs are not homologous to the LAMPs, but the luminal region of LIMP-2 does share 36% homology with the extracellular domain of CD36, accounting for its inclusion in the CD36 superfamily, an evolutionarily conserved group of proteins that play major roles in cell adhesion and signal transduction. Like CD36, LIMP-2 has been shown to bind the adhesive glycoprotein thrombospondin-1, an interaction that may affect platelet adhesion, angiogenesis, and the uptake of apoptotic cells, though this interaction has not been studied in detail [31]. Although less abundant than the LAMP proteins, LIMP-2 accounts for 4% of all lysosomal membrane proteins, and as such, represents a major component of lysosomes that is crucial for normal development [32].
2. LIMP-2 Gene, Protein Structure and Function
2.1. The genomic and protein structure
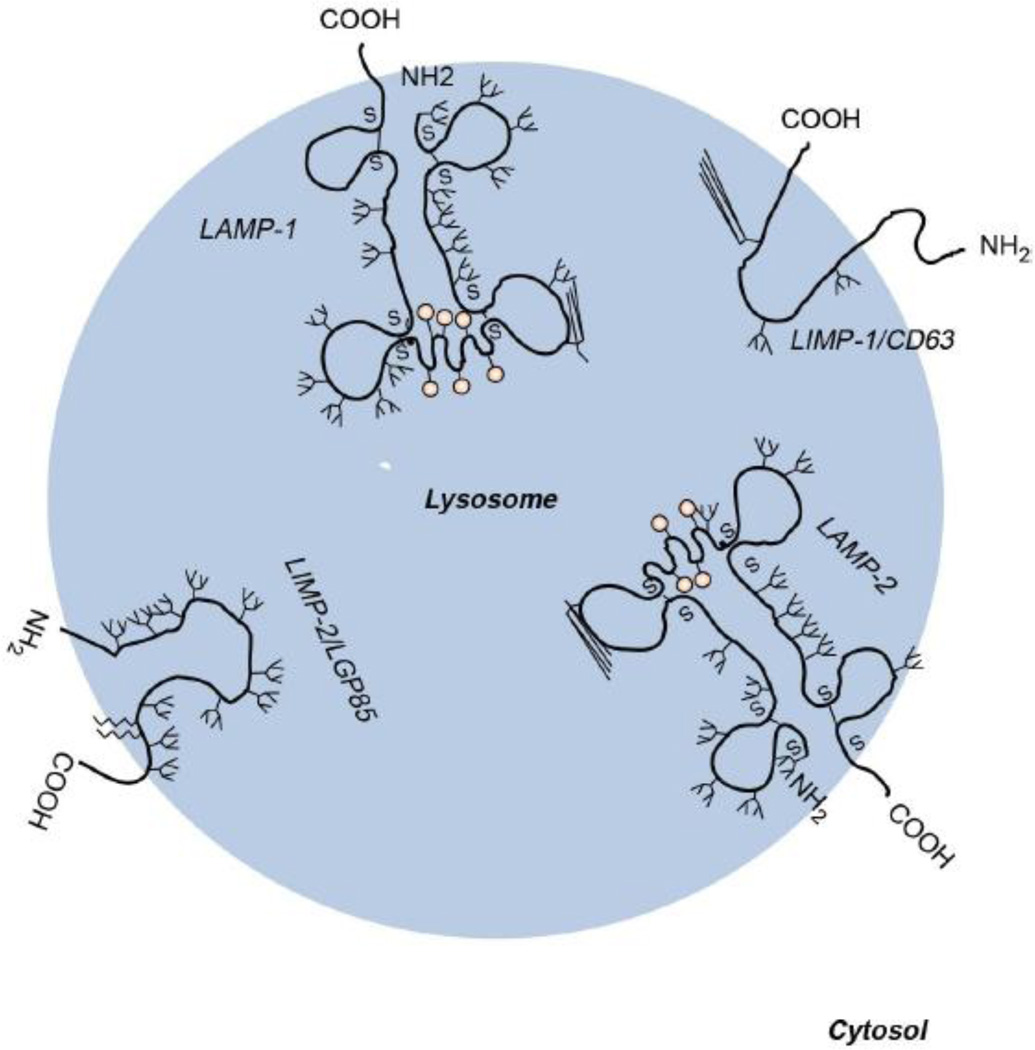
LIMP-2 has been referred to in the literature by a number of names including lysosomal glycoprotein 85 (LGP85), scavenger receptor class B membrane protein-2 (SR-BII), and cluster of differentiation 36 like-2 (CD36L2) (http://www.ncbi.nlm.nih.gov). The protein is encoded by the human SCARB2 gene, which encompasses twelve exons and is located on chromosome 4q21.1 (http://www.ncbi.nlm.nih.gov). LIMP-2 cDNA (http://genome.ucsc.edu), initially isolated from rat liver, contains 2,065 nucleotides resulting in a 478 amino acid protein that contains 11 potential N-glycosylation sites [22]. Due to its many glycosylation sites, the molecular weight of LIMP-2 ranges from 60–85 kDa. Sequencing of human LIMP-2 cDNA revealed 86% amino acid similarity and 79% nucleic acid homology to rat LIMP-2, with only 10 N-glycosylation sites [33]. The protein has two hydrophobic regions corresponding to its two transmembrane domains, with both the C- and N- termini located in the cytoplasmic region [22,34]. This results in a protein with two cytoplasmic tails and a large luminal region (Figure 1) [22,35]. Fujita et al. characterized the transmembrane, cytoplasmic, and luminal domains of LIMP-2 through hydropathy mapping, which closely matched a similar structural characterization shown by Vega et al, also performed on rat liver [34]. Amino acid residues 1–3 make up the NH2-terminal cytoplasmic tail [22]. The NH2 terminal transmembrane region spans amino acids 4–26 and contains the uncleavable signal peptide of LIMP-2 [22]. The large luminal domain is composed of amino acids 27–432 and contains 11 possible N-glycosylation sites [22]. This luminal domain also contains a conserved coiled-coil motif between amino acids 150–167 that is important for substrate binding [20]. Finally, the COOH-terminal cytoplasmic tail spans amino acid residues 458–478 [22]. This C-terminus cytoplasmic tail contains the leucine-isoleucine motif, which is critical for the sorting of LIMP-2 to lysosomes [27].
Figure 1. Schematic illustration of the four major lysosomal integral membrane proteins, LAMP-1, LAMP-2, LIMP-1 (CD63) and LIMP-2 (LGP85/CD36).
Possible N- and O-glycosylation sites are indicated with antennas and pink circles, respectively. Both the C- and the N-terminal ends of the LIMPs are located in the cytoplasm, whereas with the LAMPs, only the C-terminal ends are cytoplasmic tails. (Adapted from Eskelinen, E.-L., et al: At the acidic edge: emerging functions for lysosomal membrane proteins. Trends in Cell Biology 13, 137–145 (2003).
Recently, the crystal structure of LIMP-2 was determined, uncovering new potential functions for LIMP-2 and the homologous proteins SR-BI and CD36 through homology mapping [36]. LIMP-2 was found to have a distinct helical bundle at the site where GCase binds, and interestingly, to contain a cavity that spans the entire protein [36]. This cavity forms a channel through which the authors suggest cholesterol is transported, given the proposed role of SR-BI in the trafficking of cholesterol [36]. Although LIMP-2 has not been shown to be involved in lipid transport or hydrolysis, the conserved cavity discovered from its crystal structure may stimulate further analyses in this area. Until then, the emphasis will remain on elucidating the molecular pathway of LIMP-2 and its known roles in disease pathology.
2.2. Molecular trafficking of LIMP-2
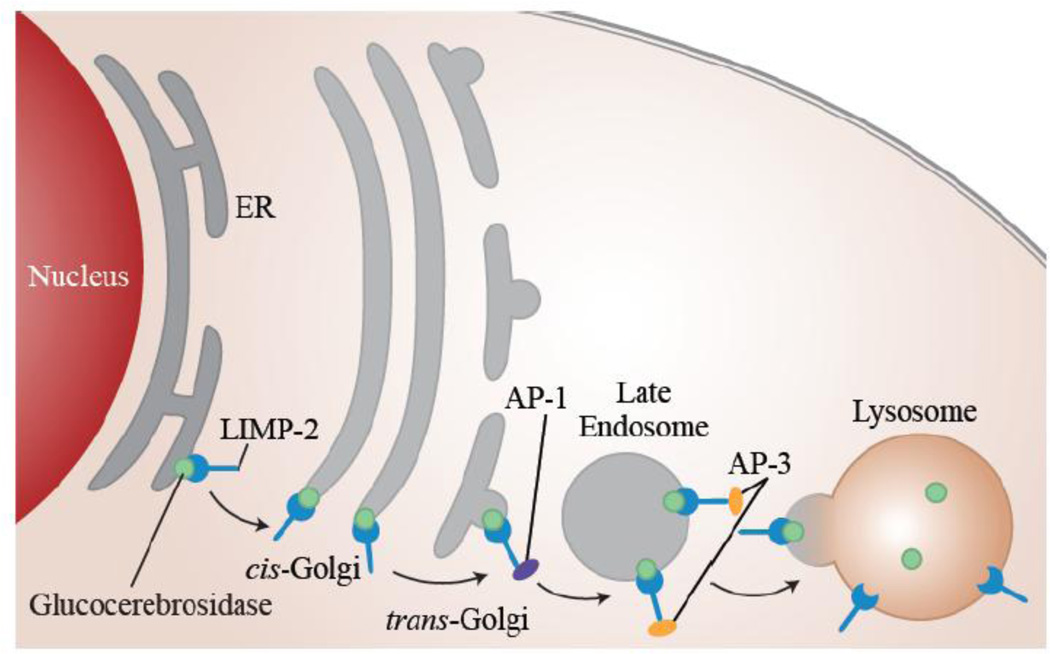
Time-course studies by Barriocanal et al. demonstrated that after its synthesis in the ER, LIMP-2 is transported through the entire TGN in a cis to trans manner (Figure 2) [21]. Upon reaching the trans-Golgi, LIMP-2 acquires endonuclease-H resistance, with the conversion of its N-linked oligosaccharide chain to sialylated carbohydrates [21]. LIMP-2 is then sorted and directly transported to late endosomes and lysosomes, without passing through the plasma membrane [34]. The targeting of LIMP-2 to lysosomes occurs independently of its acquisition of N-linked carbohydrates, and does not involve the prevalent mannose-6-phosphate (M6P) mechanism observed with other lysosomal proteins [34]. Instead, LIMP-2 targeting relies on the di-leucine motif (Leu-475 and Ile-476) in its carboxyl cytoplasmic tail [27,34,37]. The cytoplasmic tail of LIMP-2 is conserved across rat, human, and mouse species, further highlighting its significance [22,33,35,38].
Figure 2. LIMP-2 assists in trafficking GCase from the ER to the lysosome.
GCase (green) binds to the luminal domain of LIMP-2 (blue) in the ER, and moves through the Golgi in a cis-to-trans manner. In the trans-Golgi the adaptor protein (AP1) binds to the GCase/LIMP-2 complex and assists in its post-Golgi trafficking to the late endosome. AP3 is thought to assist in moving the complex from the late endosome to the lysosome. LIMP-2 and GCase dissociate upon reaching the lysosome due to the acidic pH.
The proper targeting of LIMP-2 to lysosomes relies on the interaction between its carboxyl cytoplasmic tail and the ubiquitously expressed adaptor protein (AP)-3 [39]. This interaction requires the presence of two acidic amino acid residues, Asp-470 and Glu-471, located just downstream of the di-leucine motif [39–41]. The presence of two casein kinase II phosphorylation sites in the LIMP-2 cytoplasmic tail may also play a role in regulating the di-leucine signal [42]. Honing et al. showed that interaction with AP-3 accounts for nearly all of the specific binding activity to the LIMP-2 cytoplasmic tail, and observed no measurable interaction between either AP-1 or AP-2 with LIMP-2 [39]. However, Fujita et al. showed that LIMP-2 interacts with both AP-1 and AP-3, and that the presence of both complexes is necessary for lysosomal targeting [43]. These conflicting findings have led to two differing characterizations of the association of LIMP-2 and APs. Honing et al. proposes that the LIMP-2/AP-3 interaction is initiated in the TGN due to the localization of AP-3 to this region as well as endosomal membranes [39]. Fujita et al., however, suggests the more probable sequential interaction of LIMP-2 with AP-1 in the TGN and then AP-3 in late endosomes (Figure 2) [43]. In Fujita’s model, it is postulated that LIMP-2 is packed into clathrin coated vesicles (CCVs) in the TGN and then transported to late endosomes, since the AP-1 complex is restricted to CCVs from the TGN [21,43,44].
In addition to the carboxyl cytoplasmic tail, the NH2-terminal transmembrane and luminal domains were shown to be necessary for the targeting of LIMP-2 to late endosomes and lysosomes [45]. Although the domains crucial for lysosomal targeting and protein association have been identified, the intracellular route of LIMP-2 remains to be fully characterized. There is still much to be learned about cytoplasmic signaling and associations with adaptor proteins.
3. The functions of LIMP-2
It is becoming clear that LIMP-2 has several discrete functions. It is now appreciated that the protein is involved in general lysosomal maintenance, serves as a receptor for GCase, and is the critical receptor for all EV71 strains and Coxsackieviruses A7,A14, and A16. [20,23,46,55].
3.1. General lysosomal maintenance
The initial over-expression studies of LIMP-2, performed in COS1 cells by Kuronita et al., revealed a role for LIMP-2 in the biogenesis and maintenance of endosomes and lysosomes [23]. Over-expression of LIMP-2 was shown to result in the enlargement of early/late endosomes and lysosomes, whereas overexpression of LAMP-1 and LAMP-2 had no such effect [23]. It was also observed that overexpression of LIMP-2 led to impaired trafficking out of the affected compartments, which likely correlates with the cholesterol accumulation observed, although the nature of this interaction is not completely understood [23].
3.2. Receptor for GCase
The targeting of most lysosomal enzymes involves the well-studied M6P pathway [47]. Via this mechanism, M6P residues added to the translated proteins in the TGN are specifically recognized by M6P receptors that target the proteins to late endosomes and lysosomes [47]. M6P targeting is defective in patients with I-cell disease, due to mutations in the phosphotransferase gene responsible for adding M6P residues [20,48]. In such patients, however, normal levels of some lysosomal hydrolases are still observed, suggesting the likelihood of M6P-independent targeting mechanisms [47]. One of the hydrolases that is unaffected in patients with I-cell disease is the membrane-associated hydrolase GCase [49,50]. Although it has been known since 1988 that GCase was targeted to lysosomes via a distinct mechanism, it wasn't until recently that the targeting mechanism was identified [20].
In 2007, Reczek et al. discovered that LIMP-2 is a receptor for GCase, stimulating research into LIMP-2 as a new lysosomal targeting pathway [20,51]. The connection between GCase and LIMP-2 was identified through an affinity chromatography study on mouse liver using GCase affinity resin [20]. This revealed the presence of a 75 kDa protein, absent in the control resin, which was identified as LIMP-2 based on its amino acid sequence [20]. Further studies, both in vivo and in vitro, showed that GCase and LIMP-2 co-localize in lysosomal compartments, and that GCase activity levels directly correlate with the presence of LIMP-2 [20]. In the LIMP-2 knockout mouse, different tissues were found to have a dramatic decrease both in GCase activity and protein levels, whereas increased GCase activity and protein were detected in the sera of these LIMP-2 knockouts [20]. These data are consistent with LIMP-2 playing a role in targeting GCase to the lysosome.
The interaction between LIMP-2 and GCase was shown to involve the luminal domain of LIMP-2, and to respond in a pH dependent manner [20]. The pH dependence was first characterized by affinity studies at varying pH, which showed that the association between LIMP-2 and GCase is favored at neutral pH, and is terminated at lysosomal pH [20]. Further studies showed that a conserved coiled-coil motif within the luminal domain of LIMP-2 (amino acids 152–167) is crucial for GCase binding, with one study highlighting the importance of amino acid 166 in this association [20]. In addition, a region between amino acids 178–288 is a coiled-coil conserved domain which may contribute to GCase binding [52]. The most recent analysis suggests an association with an alpha helix between amino acids 180–190 [36]. In another evaluation the histidine residue (H171) of LIMP-2, located in close proximity to the coiled-coil domain, was shown to act as a sensor for the pH dependent binding of GCase [53].
The association between LIMP-2 and GCase and the coupled trafficking pathway were further elucidated by analyzing the effect of GBA1 mutations on binding and GCase expression in lysosomes [20,52]. The GBA1 L444P mutation, which is known to be retained in the ER, was shown to localize in lysosomes after over-expression of LIMP-2, supporting an association between LIMP-2 and GCase in the ER [20]. Reczek et al. suggested that this LIMP-2/GCase interaction occurs independently of glycosylation, a process that occurs after ER transit [20]. In all, these data indicate a highly specific, pH dependent interaction between LIMP-2 and GCase involving the coiled-coil domain of LIMP-2, starting in the ER and terminating upon proper targeting of GCase to late endosomes and lysosomes [20,54].
3.3. Receptor for Enterovirus 71 and Coxsackieviruses A16, A14, and A7
LIMP-2 was also found to be a receptor for several enteroviruses that are associated with hand foot and mouth (HFMD) and neurological diseases [46,55,56]. These viruses include enterovirus 71 (EV71), coxsackievirus A16 (CVA16), CVA14, and CVA7. EV71 and CVA16 are two of the major causes of HFMD, and infection by EV71 is a common cause of sporadic and epidemic HFMD outbreaks [55]. EV71 and CVA7 are also associated with neurological symptoms including encephalitis, meningitis, and acute flacid paralysis [46,55]. The LIMP-2 binding domain for EV71 was isolated to amino acids 144–204, which overlaps with the coiled-coil motif involved in GCase binding [56]. It has therefore been postulated that EV71 can negatively influence the LIMP-2/GCase association, although no further research into such an interaction is available [6]. It is intriguing to speculate that past exposure to enteroviruses might contribute to the clinical phenotype in GD. Even without a role in GCase binding, the identification of LIMP-2 as a receptor of enteroviruses has important implications for the study of virus receptor interactions and will inform research into the spread of HFMD [46,56].
4. Disease Manifestations Associated with LIMP-2 Deficiency
4.1. LIMP-2 deficiency in mice
The generation of LIMP-2 knockout mice revealed a range of phenotypic alterations suggesting a pleiotropic function of LIMP-2 [24]. Gamp et al. studied mice homozygous and heterozygous for LIMP-2 deficiency, but did not notice a difference in growth, weight development, or fertility between the two groups [24]. The primary disease manifestations in the mice included ureteric pelvic junction obstruction, deafness, and peripheral demyelinating neuropathy, which together led to increased postnatal mortality [24]. Pathological assessments of the mice revealed that most displayed hydronephrosis (unilateral or bilateral) as a result of the pelvic junction obstruction [24]. A more detailed study of hearing loss in LIMP-2 knockout mice demonstrated that impairment of the potassium channel KCNQ1/KCNE1 in the stria vascularis was the primary cause of deafness in the mice [57]. The progressive neuropathy in LIMP-2 deficient mice was restricted to the peripheral nervous system resulting from an apparent down-regulation of peripheral myelin proteins [24]. The presence of neurological and renal manifestations in LIMP-2 deficient mice has important implications for LIMP-2 involvement in several well-studied disorders.
4.2. LIMP-2 and Gaucher disease
The identification of LIMP-2 as a receptor of GCase has clear implications for its role in the pathogenesis of GD. Although mutations in LIMP-2 do not appear to be directly responsible for GD, it is likely one of the many factors that contribute to the phenotypic heterogeneity observed in patients with the same GBA1 genotype [58]. GD is traditionally classified into three distinct types depending on the presence and progression of neurological symptoms [59]. Type 1 GD, the most common form, is defined by the lack of neuronopathic manifestations, although type 1 GD is still associated with an increased incidence of parkinsonism [18,60]. Even among patients with type 1 GD there is a wide range of phenotypic variation; some patients are asymptomatic whereas others may present with significantly enlarged organs, bone manifestations, and anemia, among other symptoms [59]. Type 2 GD, the rare acute neuronopathic form, has its onset in infancy and rapidly progresses with neurologic deterioration, usually resulting in death by two years of age [18,59]. Chronic neuronopathic GD is classified as type 3 and is associated with diverse manifestations including myoclonus, ataxia, seizures, dementia, and abnormal saccadic eye movements [59]. While over 300 distinct mutations in GBA1 have been identified to date, the vast phenotypic heterogeneity observed in GD patients is still not solely explained by genotype, as differing manifestations and response to therapy are observed in patients sharing the same genotype [59,61]. Additional modifiers must therefore influence GD phenotype, and at the present time, LIMP-2 and other proteins involved in LIMP-2 pathways are promising candidates to be evaluated.
Moreover, the study of LIMP-2 may offer deeper insights into GD pathogenesis and the molecular basis of GCase deficiency. In their characterization of the LIMP-2/GCase interaction, Reczek et al. studied the ER-retained GBA1 mutations N370S, G202R, and L444P, noting that none of these mutations had an effect on LIMP-2 affinity, but that over-expression of LIMP-2 reversed the ER retention observed with the L444P mutation [20]. Even with over-expression of LIMP-2, the other two mutants remained localized in the ER, suggesting that the targeting pathway is not the only cause of the ER retention of these mutations [20]. The study of another ER-retained GBA1 mutation, P415R, revealed that this mutation, unlike the others studied, interfered with the GCase/LIMP-2 interaction [20].
A more direct association between LIMP-2 deficiency and GD was made through the study of two discordant siblings with the same GBA1 mutations, yet drastically different phenotypes [58]. One sibling (P1) was diagnosed with type 3 GD after presenting with progressive myoclonic epilepsy (PME) and dementia, while the other (P2), was found to have GD, yet was asymptomatic [58]. Sequencing of SCARB2 revealed that P1 was heterozygous for the mutation E471G, shown to cause extracellular excretion of GCase, while P2 lacked this mutation [58]. The mother was heterozygous for the same SCARB2 mutation, so it is evident that while LIMP-2 deficiency alone does not cause the observed phenotype, it is likely an important modifier of GD, potentially turning a patient with type 1 GD into a type 3 [58]. As far as can be determined, this work by Velayati et al. is the only study to explicitly examine GD heterogeneity in the context of SCARB2 mutations.
4.3. LIMP-2 and progressive myoclonic epilepsy
AMRF is a form of PME with associated renal failure that typically presents with neurological symptoms including tremor, seizures, ataxia, and action myoclonus [26]. These symptoms frequently overlap with those observed in type 3 GD, which might suggest a common genetic and molecular basis for these two disorders. Through gene-expression studies of three unrelated patients with AMRF, Berkovic et al. in 2008 identified mutations in SCARB2 as the major cause of AMRF [26]. It is interesting to note the lack of correlation between symptoms observed in LIMP-2 deficient mice and in patients with AMRF. Patients with AMRF show no clinical evidence of deafness or peripheral neuropathy, and LIMP-2 knockout mice show no evidence of action myoclonus or seizures [24,26]. Furthermore, brain pathology of patients with AMRF showed extraneuronal storage, whereas in mice the storage was primarily intraneuronal [26].
Patients with AMRF do share the ataxia and renal involvement observed in the mice, to differing degrees [26]. Glomerular lesions were found in both humans and mice, leading to collapsing glomerulopathy in humans and more mild proteinuria in mice [26]. However, while there are discrepancies between the phenotypes associated with LIMP-2 deficiency in mice and patients with AMRF, the study of LIMP-2 deficiency in vitro has provided deeper insight into the pathogenesis of AMRF.
To date, a total of 14 disease-causing mutations in SCARB2 have been identified, most of which are associated with either AMRF or PME (Table 1). The independent expression of the LIMP-2 mutants Q288X, W146SfsX16, and W178X in COS7 cells indicated that these mutated proteins were all retained in the ER and showed no lysosomal localization, in contrast to wild type LIMP-2 [52]. Of these mutants, only Q288X was able to bind GCase as effectively as wild type, while W146SfsX16 and W178X were unable to bind GCase [52]. A surprising increased effect on GCase binding was observed in COS7 cells expressing the H363N mutation [52]. This mutation likely evolved a new glycosylation site that contributed to its ER retention and prolonged half-life, leading to increased GCase binding [52]. The observation that LIMP-2 mutations differentially affect LIMP-2 localization and GCase binding may have an impact on future drug design, emphasizing the importance of the targeting pathway of GCase in disease pathology.
Table 1.
Disease-causing mutations in SCARB2
| Disease Association(s) | Mutation | Type | Reference |
|---|---|---|---|
| AMRF; PME | Q288X c.862C>T |
Nonsense |
Blanz et al. 2010; Dibbens et al. 2011; Berkovic et al. 2008 |
| AMRF; PME | W178X c.533G>A |
Nonsense |
Blanz et al. 2010; Balreira et al. 2008 |
| PME; AMRF | H363N c.1087C>A |
Missense |
Dardis et al. 2009; Dibbens et al. 2009; Blanz et al. 2010 |
| PME and GD type 3 | Q471G c.1412A>G |
Missense | Velayati et al. 2011 |
| PME | c. 1116-2A>C | Splice-site mutation | Dibbens et al. 2009 |
| PME | c. 704+1G>C | Splice-site mutation | Dibbens et al. 2009 |
| PME | E420RfsX5 c. 1258delG |
Frameshift | Dibbens et al. 2009 |
| PME | Y222X c.666delCCTTA |
Nonsense |
Dibbens et al. 2009 Hopfner et al. 2011 |
| PME | c. 424-2A>C | Splice-site mutation | Dibbens et al. 2009 |
| AMRF | c.1239+1G>T | Splice-site mutation; premature truncation |
Berkovic et al. 2008 |
| AMRF | W146SfsX16 c.435_436insAG |
Frameshift |
Berkovic et al. 2008; Blanz et al. 2010 |
| AMRF | N99IfsX34 c. 296 delA |
Frameshift | Berkovic et al. 2008 |
| PME | c.1187 + 3insT | Splice-site mutation | Dibbens et al. 2011 |
| AMRF | c.111delC | Deletion; frameshift | Hopfner et al. 2011 |
There have been numerous case studies reporting the phenotypes of patients with PME and the associated SCARB2 mutation identified [62]. Two siblings who were homozygous for the SCARB2 mutation W178X presented with PME and nephrotic syndrome, and also developed horizontal saccadic eye movement, myoclonus, and thrombocytopenia, findings characteristic of type 3 GD [63]. The siblings developed normally until the ages of 15 and 17, when symptoms rapidly progressed, resulting in death at the ages of 23 and 26, respectively [63]. GD was ruled out when no Gaucher cells could be identified, and no GBA1 mutations were detected [63]. When GCase activity was measured, it was found that although patient fibroblasts only had 10% of control GCase activity, leukocytes and plasma showed normal activity levels, suggesting that there may be a secondary targeting mechanism by which GCase is transported to lysosomes [63]. A similar patient with PME that was heterozygous for the LIMP-2 mutation H363N had onset of symptoms at 26 years consisting of myoclonus, tremors, and ataxia [51]. In contrast to the siblings, this patient had normal GCase activity in fibroblasts [51]. The relatively late onset of symptoms observed in these patients, suggests that cells may become more sensitive to LIMP-2 deficiency later in life [26,51]. A broader screen of subjects with unexplained PME identified five new SCARB2 mutations that were believed to contribute to pathogenesis, although detailed molecular studies have not been conducted [64]. These mutations are included in Table 1. Of note, only one SCARB2 mutation, W178X, has been associated with both AMRF and PME, and this patient also had renal involvement. Thus ultimately it might be possible to draw some genotype/phenotype correlations based on the mutations identified.
The evaluation of additional patients with PME shows that certain SCARB2 mutations may also result in a demyelinating peripheral neuropathy. One such patient was a young man diagnosed with PME without renal failure who was found to be compound heterozygous for the SCARB2 mutations Q288X and c.11187+3insT [64]. Like the other patients described, he initially developed normally, with the onset of his first symptoms at 16 years of age, followed by rapid disease progression with myoclonus, seizures, dysphagia, and wheelchair-dependence within 4 years [64]. Reevaluation of this patient in light of his defined SCARB2 mutations showed that symptoms were consistent with a peripheral demyelinating neuropathy, similar to that observed in LIMP-2 deficient mice. Another study by Hopfner et al. examined a German family with AMRF, showing that three affected members who were homozygous for the SCARB2 mutation c.111delC also had a demyelinating peripheral neuropathy [65].
4.4. LIMP-2 and the heart
An interesting and unexpected role of LIMP-2 is its involvement in the intercalated disc [25]. In cardiac muscle, intercalated discs connect individual cardiomyocytes, enabling them to function as a unified organ. LIMP-2 was found to influence the binding affinity of N-cadherin and β-catenin, an interaction that is crucial for the hypertrophic response to cardiac loading [25]. Two of the affected members of the German AMRF family described by Hopfner et al. developed cardiomyopathy, which may be significant given this important finding, but further analyses are necessary to elucidate this connection [25,65].
4.5 LIMP-2 and Parkinson disease
The known link between GD and PD has led to the consideration of LIMP-2 as a protein of potential interest in PD research. Parkinson disease, as well as other synucleinopathies, is characterized by an accumulation of alpha-synuclein (α-syn) [66]. It has been shown that α-syn and GCase have a reciprocal relationship, where GCase deficiency appears to result in the accumulation of glucosylceramide, which leads to increased α-syn aggregation and further depletion of GCase [67]. Given the relationship between these two proteins, Gegg et al. examined whether LIMP-2 might fit into the pathway of α-syn aggregation via a direct interaction with α-syn [68]. They found that there is no direct interaction between α-syn and LIMP-2, and no alteration in LIMP-2 levels in PD brain samples [68].
The gene SCARB2 has also been of considerable interest to PD research. A candidate-gene study, conducted in Greece, screened for SCARB2 mutations in patients with PD and identified a potential association between the SCARB2 polymorphism rs6825004 and PD [69]. A second polymorphism just upstream of the SCARB2 gene, rs6812193, was identified as a PD susceptibility factor by a genome-wide association study looking at 3,426 cases with PD and 29,624 controls [70]. The connection between PD and polymorphism rs6812193 was studied further by Chen et al. in an association study that showed no significant difference in the genotype distribution of rs6812193, and later by Hopfner et al. in a study that supported the association between rs6812193 and PD [71,72]. A fourth association study on polymorphism rs6812193 concluded that there was a statistically significant difference in genotype frequency of polymorphism rs6812193 among male PD patients only [73]. These conflicting findings emphasize the need for gene validation. One such validation study focused on polymorphisms rs6812193 and rs6825004 using quantitative PCR and Western blots in control subjects [74]. No significant differences in SCARB2 expression or LIMP-2 levels were found that correlated with the genotypes of either polymorphism [74]. Based on the inconsistent association studies and lack of correlation between identified polymorphisms and SCARB2/LIMP-2 expression, there is not conclusive evidence that SCARB2 is a susceptibility factor in PD, however, additional research into the connection between SCARB2 and PD is warranted.
5. Conclusion
As a receptor for both GCase and different enteroviruses, the causative factor implicated in AMRF, and with its newly appreciated role in the intercalated disc, LIMP-2 is proving to be far more than simply a component of lysosomal membranes. Its unexpected identification as a receptor for GCase has generated attention among researchers investigating the pathogenesis of both Gaucher disease and Parkinson disease. It is exciting to consider that LIMP-2 and/or other proteins involved in the LIMP-2 pathway may serve as modifiers of GD, helping to account for the clinical diversity seen in patients with GD sharing the same GBA1 genotype. However, the contribution of these proteins to Gaucher phenotypes remains to be fully elucidated. Furthermore, patients with SCARB2 mutations that disrupt GCase binding do not necessarily develop GD, suggesting the possibility of additional or alternate pathways involved in targeting GCase to the lysosome. It will be interesting to look more closely at other members of the CD36 superfamily as potential receptors for GCase. A wider screen of LIMP-2 protein levels among patients with GD with the same genotype might also shed light on the relationship between LIMP-2 and GCase levels. The continued study of lysosomal proteins such as LIMP-2 represent an important area of research that can provide insights into disease pathogenesis and assist in the elucidation of the entire lysosomal processing network.
Highlights.
LIMP-2 is required for the biogenesis and maintenance of lysosomes and endosomes.
As the glucocerebrosidase transporter, LIMP-2 may play a role in Gaucher disease
Lysosomal proteins can serve multiple diverse roles.
LIMP-2 deficiency provides insights into lysosomal processing, trafficking and function.
Acknowledgements
This work was supported by the Intramural Research Programs of the National Human Genome Research Institute, and the National Institutes of Health. The authors thank Dr. Elma Aflaki and Julia Fekecs for their assistance with the figures.
Abbreviations Used
- LIMP-2
lysosomal integral membrane protein-2
- ER
endoplasmic reticulum
- LAMP
lysosomal associated membrane protein
- LMP
lysosomal membrane protein
- PME
progressive myoclonic epilepsy
- GCase
glucocerebrosidase
- M6P
mannose-6-phosphate
- AMRF
action myoclonus renal failure
- LSD
lysosomal storage disorder
- GD
Gaucher disease
- GBA1
glucocerebrosidase gene
- LGP85
lysosomal glycoprotein 85
- SR-BII
scavenger receptor class B membrane protein-2
- CD36L2
CD36-like2 protein (LIMP-2)
- SCARB2
scavenger receptor class B2 gene (encodes LIMP-2)
- TGN
trans-Golgi network
- AP
adaptor protein
- CCV
clathrin coated vesicles
- EV71
enterovirus 71
- COS7
Cercopithecus aethiops cell line (SV40 transformed African green monkey kidney fibroblast-like cell line)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 2.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis V, Green SA, Marsh M, Vihko P, Helenius A, Mellman I. Glycoproteins of the lysosomal membrane. Journal Cell Biol. 1985;100:1839–1847. doi: 10.1083/jcb.100.6.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saftig P, Schröder B, Blanz J. Lysosomal membrane proteins: life between acid and neutral conditions. Biochem. Soc. Trans. 2010;38:1420–1423. doi: 10.1042/BST0381420. [DOI] [PubMed] [Google Scholar]
- 5.Schröder B, Wrocklage C, Pan C, Jäger R, Kösters B, Schäfer H, et al. Integral and associated lysosomal membrane proteins. Traffic. 2007;8:1676–1686. doi: 10.1111/j.1600-0854.2007.00643.x. [DOI] [PubMed] [Google Scholar]
- 6.Schwake M, Schröder B, Saftig P. Lysosomal membrane proteins and their central role in physiology. Traffic. 2013;14:739–748. doi: 10.1111/tra.12056. [DOI] [PubMed] [Google Scholar]
- 7.Ballabio A, Gieselmann V. Lysosomal disorders: from storage to cellular damage. Bba. 2009;1793:684–696. doi: 10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Vellodi A. Lysosomal storage disorders. Br. J. Haematol. 2005;128:413–431. doi: 10.1111/j.1365-2141.2004.05293.x. [DOI] [PubMed] [Google Scholar]
- 9.Schröder BA, Wrocklage C, Hasilik A, Saftig P. The proteome of lysosomes. Proteomics. 2010;10:4053–4076. doi: 10.1002/pmic.201000196. [DOI] [PubMed] [Google Scholar]
- 10.Ruivo R, Anne C, Sagné C, Gasnier B. Molecular and cellular basis of lysosomal transmembrane protein dysfunction. Bba. 2009;1793:636–649. doi: 10.1016/j.bbamcr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Fuller M, Meikle P, Hopwood J. Fabry disease: perspectives from 5 years of FOS. In: A M, M B, G S-P, editors. Epidemiology of Lysosomal Storage Diseases: an Overview. Oxford PharmaGenesis; 2002. [PubMed] [Google Scholar]
- 12.Fletcher JM. Screening for lysosomal storage disorders--a clinical perspective. J Inherit Metab Dis. 2006;29:405–408. doi: 10.1007/s10545-006-0246-7. [DOI] [PubMed] [Google Scholar]
- 13.Swan M, Saunders-Pullman R. The association between β-glucocerebrosidase mutations and parkinsonism. Curr Neurol Neurosci Rep. 2013;13:368. doi: 10.1007/s11910-013-0368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lwin A, Orvisky E, Goker-Alpan O, LaMarca ME, Sidransky E. Glucocerebrosidase mutations in subjects with parkinsonism. Mol Genet Metab. 2004;81:70–73. doi: 10.1016/j.ymgme.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N. Engl. J. Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shachar T, Lo Bianco C, Recchia A, Wiessner C, Raas-Rothschild A, Futerman AH. Lysosomal storage disorders and Parkinson's disease: Gaucher disease and beyond. Mov. Disord. 2011;26:1593–1604. doi: 10.1002/mds.23774. [DOI] [PubMed] [Google Scholar]
- 17.Nixon RA, Yang D-S, Lee J-H. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4:590–599. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- 18.Rosenbloom BE, Weinreb NJ. Gaucher disease: a comprehensive review. Crit Rev Oncog. 2013;18:163–175. doi: 10.1615/critrevoncog.2013006060. [DOI] [PubMed] [Google Scholar]
- 19.Biegstraaten M, van Schaik IN, Aerts JMFG, Langeveld M, Mannens MMAM, Bour LJ, et al. A monozygotic twin pair with highly discordant Gaucher phenotypes. Blood Cells Mol Dis. 2011;46:39–41. doi: 10.1016/j.bcmd.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reczek D, Schwake M, Schröder J, Hughes H, Blanz J, Jin X, et al. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell. 2007;131:770–783. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Barriocanal JG, Bonifacino JS, Yuan L, Sandoval I. Biosynthesis, glycosylation, movement through the Golgi system, and transport to lysosomes by an N-linked carbohydrate-independent mechanism of three lysosomal integral membrane proteins. J Biol Chem. 1986;261:16755–16763. [PubMed] [Google Scholar]
- 22.Fujita H, Ezaki J, Noguchi Y, Kono A, Himeno M, Kato K. Isolation and sequencing of a cDNA clone encoding 85kDa sialoglycoprotein in rat liver lysosomal membranes. Biochem Biophys Res Commun. 1991;178:444–452. doi: 10.1016/0006-291x(91)90127-s. [DOI] [PubMed] [Google Scholar]
- 23.Kuronita T, Eskelinen E-L, Fujita H, Saftig P, Himeno M, Tanaka Y. A role for the lysosomal membrane protein LGP85 in the biogenesis and maintenance of endosomal and lysosomal morphology. J. Cell. Sci. 2002;115:4117–4131. doi: 10.1242/jcs.00075. [DOI] [PubMed] [Google Scholar]
- 24.Gamp A-C, Tanaka Y, Lüllmann-Rauch R, Wittke D, D'Hooge R, De Deyn PP, et al. LIMP-2/LGP85 deficiency causes ureteric pelvic junction obstruction deafness and peripheral neuropathy in mice. Hum Mol Genet. 2003;12:631–646. [PubMed] [Google Scholar]
- 25.Schroen B, Leenders JJ, van Erk A, Bertrand AT, van Loon M, van Leeuwen RE, et al. Lysosomal integral membrane protein 2 is a novel component of the cardiac intercalated disc and vital for load-induced cardiac myocyte hypertrophy. J. Exp. Med. 2007;204:1227–1235. doi: 10.1084/jem.20070145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkovic SF, Dibbens LM, Oshlack A, Silver JD, Katerelos M, Vears DF, et al. Array- based gene discovery with three unrelated subjects shows SCARB2/LIMP-2 deficiency causes myoclonus epilepsy and glomerulosclerosis. Am J Hum Genet. 2008;82:673–684. doi: 10.1016/j.ajhg.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogata S, Fukuda M. Lysosomal targeting of Limp II membrane glycoprotein requires a novel Leu-Ile motif at a particular position in its cytoplasmic tail. J. Biochem. 1994;269:5210–5217. [PubMed] [Google Scholar]
- 28.Eskelinen E-L, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda M. Lysosomal membrane Structure glycoproteins biosynthesis and intracellular trafficking. J. Biochem. 1991;266:21327–21330. [PubMed] [Google Scholar]
- 30.Niwa K, Tanaka R, Murase H, Ishikawa T, Fujita H, Himeno M, et al. Two lysosomal membrane proteins, LGP85 and LGP107, are delivered to late endosomes/lysosomes through different intracellular routes after exiting from the trans-Golgi network. Biochem Biophys Res Commun. 2003;301:833–840. doi: 10.1016/s0006-291x(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 31.Crombie R, Silverstein R. Lysosomal integral membrane protein II binds thrombospondin-1. Structure-function homology with the cell adhesion molecule CD36 defines a conserved recognition motif. J. Biochem. 1998;273:4855–4863. doi: 10.1074/jbc.273.9.4855. [DOI] [PubMed] [Google Scholar]
- 32.Akasaki K, Kinoshita H, Fukuzawa M, Maeda M, Yamaguchi Y, Furuno K, et al. Isolation and characterization of a novel membrane glycoprotein of 85,000 molecular weight from rat liver lysosomes. Chem. Pharm. Bull. 1992;40:170–173. doi: 10.1248/cpb.40.170. [DOI] [PubMed] [Google Scholar]
- 33.Fujita H, Takata Y, Kono A, Tanaka Y, Takahashi T, Himeno M, et al. Isolation and sequencing of a cDNA clone encoding the 85 kDa human lysosomal sialoglycoprotein (hLGP85) in human metastatic pancreas islet tumor cells. Biochem Biophys Res Commun. 1992;184:604–611. doi: 10.1016/0006-291x(92)90632-u. [DOI] [PubMed] [Google Scholar]
- 34.Vega MA, Rodriguez F, Seguí B, Calés C, Alcalde J, Sandoval IV. Targeting of lysosomal integral membrane protein LIMP II. The tyrosine-lacking carboxyl cytoplasmic tail of LIMP II is sufficient for direct targeting to lysosomes. J. Biochem. 1991;266:16269–16272. [PubMed] [Google Scholar]
- 35.Vega MA, Segui-Real B, Garcia JA, Cales C, Rodriguez F, Vanderkerckhove J, et al. Cloning sequencing expression of a cDNA encoding rat LIMP II a novel 74-kDa lysosomal membrane protein related to the surface adhesion protein CD36. J Biol Chem. 1991;266:16818–16824. [PubMed] [Google Scholar]
- 36.Neculai D, Schwake M, Ravichandran M, Zunke F, Collins RF, Peters J, et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nat. 2013 doi: 10.1038/nature12684. [DOI] [PubMed] [Google Scholar]
- 37.Sandoval IV, Arredondo JJ, Alcalde J, Gonzalez Noriega A, Vandekerckhove J, Jimenez MA, et al. The residues Leu(Ile)475-Ile(Leu Val Ala) 476 contained in the extended carboxyl cytoplasmic tail are critical for targeting of the resident lysosomal membrane protein LIMP II to lysosomes. J. Biochem. 1994;269:6622–6631. [PubMed] [Google Scholar]
- 38.Tabuchi N, Akasaki K, Sasaki T, Kanda N, Tsuji H. Identification and characterization of a major lysosomal membrane glycoprotein, LGP85/LIMP II in mouse liver. J. Biochem. 1997;122:756–763. doi: 10.1093/oxfordjournals.jbchem.a021820. [DOI] [PubMed] [Google Scholar]
- 39.Honing S, Sandoval IV, von Figura K. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. Embo J. 1998;17:1304–1314. doi: 10.1093/emboj/17.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandoval IV, Martinez-Arca S, Valdueza J, Palacios S, Holman GD. Distinct reading of different structural determinants modulates the dileucine-mediated transport steps of the lysosomal membrane protein LIMPII and the insulin-sensitive glucose transporter GLUT4. J. Biochem. 2000;275:39874–39885. doi: 10.1074/jbc.M006261200. [DOI] [PubMed] [Google Scholar]
- 41.Tabuchi N, Akasaki K, Tsuji H. Two acidic amino acid residues, Asp(470), and Glu(471), contained in the carboxyl cytoplasmic tail of a major lysosomal membrane protein, LGP85/LIMP II, are important for its accumulation in secondary lysosomes. Biochem Biophys Res Commun. 2000;270:557–563. doi: 10.1006/bbrc.2000.2448. [DOI] [PubMed] [Google Scholar]
- 42.Sandoval IV, Bakke O. Targeting of membrane proteins to endosomes and lysosomes. Trends Cell Biol. 1994;4:292–297. doi: 10.1016/0962-8924(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 43.Fujita H, Saeki M, Yasunaga K, Ueda T, Imoto T, Himeno M. In vitro binding study of adaptor protein complex (AP-1) to lysosomal targeting motif (LI-motif) Biochem Biophys Res Commun. 1999;255:54–58. doi: 10.1006/bbrc.1998.0140. [DOI] [PubMed] [Google Scholar]
- 44.Hirst J, Robinson MS. Clathrin and adaptors. Bba. 1998;1404:173–193. doi: 10.1016/s0167-4889(98)00056-1. [DOI] [PubMed] [Google Scholar]
- 45.Kuronita T, Hatano T, Furuyama A, Hirota Y, Masuyama N, Saftig P, et al. The NH(2)- terminal transmembrane and lumenal domains of LGP85 are needed for the formation of enlarged endosomes/lysosomes. Traffic. 2005;6:895–906. doi: 10.1111/j.1600-0854.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- 46.Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, et al. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 2009;15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- 47.Coutinho MF, Prata MJ, Alves S. A shortcut to the lysosome: the mannose-6-phosphate- independent pathway. Mol Genet Metab. 2012;107:257–266. doi: 10.1016/j.ymgme.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Kollmann K, Pohl S, Marschner K, Encarnação M, Sakwa I, Tiede S, et al. Mannose phosphorylation in health and disease. Eur. J. Cell Biol. 2010;89:117–123. doi: 10.1016/j.ejcb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Aerts JMFG, Schram AW, Strijland A, van Weely S, Jonsson LMV, Tager JM, et al. Glucocerebrosidase, a lysosomal enzyme that does not undergo oligosaccharide phosphorylation. Bba. 1988;964:303–308. doi: 10.1016/0304-4165(88)90030-x. [DOI] [PubMed] [Google Scholar]
- 50.Rijnboutt S, Aerts HM, Geuze HJ, Tager JM, Strous GJ. Mannose 6-phosphate- independent membrane association of cathepsin D, glucocerebrosidase, and sphingolipid-activating protein in HepG2 cells. J. Biochem. 1991;266:4862–4868. [PubMed] [Google Scholar]
- 51.Dardis A, Filocamo M, Grossi S, Ciana G, Franceschetti S, Dominissini S, et al. Biochemical and molecular findings in a patient with myoclonic epilepsy due to a mistarget of the beta-glucosidase enzyme. Mol Genet Metab. 2009;97:309–311. doi: 10.1016/j.ymgme.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 52.Blanz J, Groth J, Zachos C, Wehling C, Saftig P, Schwake M. Disease-causing mutations within the lysosomal integral membrane protein type 2 (LIMP-2) reveal the nature of binding to its ligand beta-glucocerebrosidase. Hum Mol Genet. 2010;19:563–572. doi: 10.1093/hmg/ddp523. [DOI] [PubMed] [Google Scholar]
- 53.Zachos C, Blanz J, Saftig P, Schwake M. A critical histidine residue within LIMP-2 mediates pH sensitive binding to its ligand β-glucocerebrosidase. Traffic. 2012;13:1113–1123. doi: 10.1111/j.1600-0854.2012.01372.x. [DOI] [PubMed] [Google Scholar]
- 54.Griffiths GM. Gaucher disease: forging a new path to the lysosome. Cell. 2007;131:647–649. doi: 10.1016/j.cell.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 55.Yamayoshi S, Iizuka S, Yamashita T, Minagawa H, Mizuta K, Okamoto M, et al. Human SCARB2-dependent infection by coxsackievirus A7, A14, and A16 and enterovirus 71. J. Virol. 2012;86:5686–5696. doi: 10.1128/JVI.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamayoshi S, Koike S. Identification of a human SCARB2 region that is important for enterovirus 71 binding and infection. J. Virol. 2011;85:4937–4946. doi: 10.1128/JVI.02358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knipper M, Claussen C, Rüttiger L, Zimmermann U, Lüllmann-Rauch R, Eskelinen E-L, et al. Deafness in LIMP2-deficient mice due to early loss of the potassium channel KCNQ1/KCNE1 in marginal cells of the stria vascularis. J. Physiol. (Lond.) 2006;576:73–86. doi: 10.1113/jphysiol.2006.116889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velayati A, DePaolo J, Gupta N, Choi JH, Moaven N, Westbroek W, et al. A mutation in SCARB2 is a modifier in Gaucher disease. Hum Mutat. 2011;32:1232–1238. doi: 10.1002/humu.21566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sidransky E. Gaucher disease: complexity in a “simple” disorder. Mol Genet Metab. 2004;83:6–15. doi: 10.1016/j.ymgme.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 60.Rosenbloom B, Balwani M, Bronstein JM, Kolodny E, Sathe S, Gwosdow AR, et al. The incidence of Parkinsonism in patients with type 1 Gaucher disease: data from the ICGG Gaucher Registry. Blood Cells Mol Dis. 2011;46:95–102. doi: 10.1016/j.bcmd.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29:567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 62.Dibbens LM, Michelucci R, Gambardella A, Andermann F, Rubboli G, Bayly MA, et al. SCARB2 mutations in progressive myoclonus epilepsy (PME) without renal failure. Ann Neurol. 2009;66:532–536. doi: 10.1002/ana.21765. [DOI] [PubMed] [Google Scholar]
- 63.Balreira A, Gaspar P, Caiola D, Chaves J, Beirão I, Lima JL, et al. A nonsense mutation in the LIMP-2 gene associated with progressive myoclonic epilepsy and nephrotic syndrome. Hum Mol Genet. 2008;17:2238–2243. doi: 10.1093/hmg/ddn124. [DOI] [PubMed] [Google Scholar]
- 64.Dibbens LM, Karakis I, Bayly MA, Costello DJ, Cole AJ, Berkovic SF. Mutation of SCARB2 in a patient with progressive myoclonus epilepsy and demyelinating peripheral neuropathy. Arch. Neurol. 2011;68:812–813. doi: 10.1001/archneurol.2011.120. [DOI] [PubMed] [Google Scholar]
- 65.Hopfner F, Schormair B, Knauf F, Berthele A, Tölle TR, Baron R, et al. Novel SCARB2 mutation in action myoclonus-renal failure syndrome and evaluation of SCARB2 mutations in isolated AMRF features. BMC Neurol. 2011;11:134. doi: 10.1186/1471-2377-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Recchia A, Debetto P, Negro A, Guidolin D, Skaper SD, Giusti P. Alpha-synuclein and Parkinson's disease. Faseb J. 2004;18:617–626. doi: 10.1096/fj.03-0338rev. [DOI] [PubMed] [Google Scholar]
- 67.Mazzulli JR, Xu Y-H, Sun Y, Knight AL, McLean PJ, Caldwell GA, et al. Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gegg ME, Burke D, Heales SJR, Cooper JM, Hardy J, Wood NW, et al. Glucocerebrosidase deficiency in substantia nigra of parkinson disease brains. Ann Neurol. 2012;72:455–463. doi: 10.1002/ana.23614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michelakakis H, Xiromerisiou G, Dardiotis E, Bozi M, Vassilatis D, Kountra P-M, et al. Evidence of an association between the scavenger receptor class B member 2 gene and Parkinson's disease. Mov. Disord. 2012;27:400–405. doi: 10.1002/mds.24886. [DOI] [PubMed] [Google Scholar]
- 70.Do CB, Tung JY, Dorfman E, Kiefer AK, Drabant EM, Francke U, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet. 2011;7:e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen S, Zhang Y, Chen W, Wang Y, Liu J, Rong T-Y, et al. Association study of SCARB2 rs6812193 polymorphism with Parkinson's disease in Han Chinese. Neurosci Lett. 2012;516:21–23. doi: 10.1016/j.neulet.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 72.Hopfner F, Schulte EC, Mollenhauer B, Bereznai B, Knauf F, Lichtner P, et al. The role of SCARB2 as susceptibility factor in Parkinson's disease. Mov. Disord. 2013;28:538–540. doi: 10.1002/mds.25349. [DOI] [PubMed] [Google Scholar]
- 73.Li K, Tang B-S, Yu R-L, Lv Z-Y, Sun Q-Y, Li Q, et al. Association study between two novel single nucleotide polymorphisms and sporadic Parkinson's disease in Chinese Han population. Neurosci Lett. 2012;517:56–59. doi: 10.1016/j.neulet.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 74.Maniwang E, Tayebi N, Sidransky E. Is Parkinson disease associated with lysosomal integral membrane protein type-2?: challenges in interpreting association data. Mol Genet Metab. 2013;108:269–271. doi: 10.1016/j.ymgme.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]