Abstract
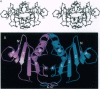
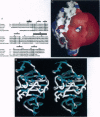
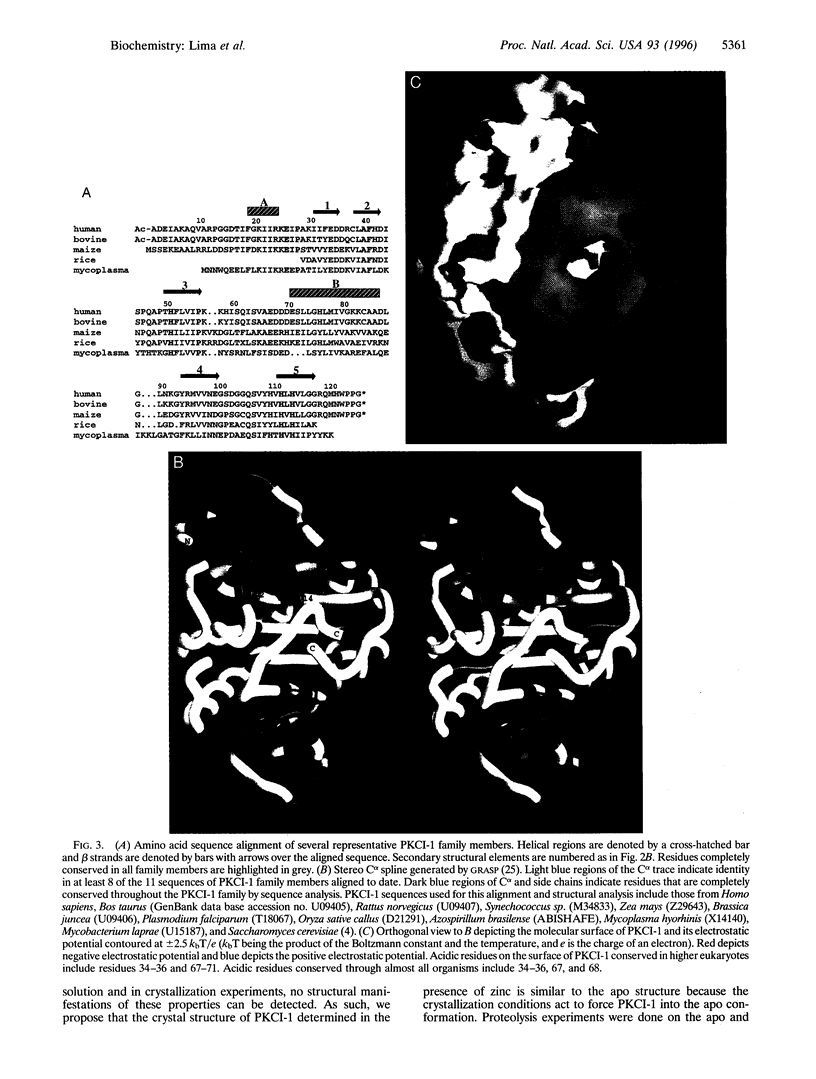
The three-dimensional structure of protein kinase C interacting protein 1 (PKCI-1) has been solved to high resolution by x-ray crystallography using single isomorphous replacement with anomalous scattering. The gene encoding human PKCI-1 was cloned from a cDNA library by using a partial sequence obtained from interactions identified in the yeast two-hybrid system between PKCI-1 and the regulatory domain of protein kinase C-beta. The PKCI-1 protein was expressed in Pichia pastoris as a dimer of two 13.7-kDa polypeptides. PKCI-1 is a member of the HIT family of proteins, shown by sequence identity to be conserved in a broad range of organisms including mycoplasma, plants, and humans. Despite the ubiquity of this protein sequence in nature, no distinct function has been shown for the protein product in vitro or in vivo. The PKCI-1 protomer has an alpha+beta meander fold containing a five-stranded antiparallel sheet and two helices. Two protomers come together to form a 10-stranded antiparallel sheet with extensive contacts between a helix and carboxy terminal amino acids of a protomer with the corresponding amino acids in the other protomer. PKCI-1 has been shown to interact specifically with zinc. The three-dimensional structure has been solved in the presence and absence of zinc and in two crystal forms. The structure of human PKCI-1 provides a model of this family of proteins which suggests a stable fold conserved throughout nature.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brzoska P. M., Chen H., Zhu Y., Levin N. A., Disatnik M. H., Mochly-Rosen D., Murnane J. P., Christman M. F. The product of the ataxia-telangiectasia group D complementing gene, ATDC, interacts with a protein kinase C substrate and inhibitor. Proc Natl Acad Sci U S A. 1995 Aug 15;92(17):7824–7828. doi: 10.1073/pnas.92.17.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Bustos S. A., Schaefer M. R., Golden S. S. Different and rapid responses of four cyanobacterial psbA transcripts to changes in light intensity. J Bacteriol. 1990 Apr;172(4):1998–2004. doi: 10.1128/jb.172.4.1998-2004.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S. V. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J Mol Graph. 1993 Jun;11(2):134-8, 127-8. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Fraser E. D., Walsh M. P. The major endogenous bovine brain protein kinase C inhibitor is a heat-labile protein. FEBS Lett. 1991 Dec 9;294(3):285–289. doi: 10.1016/0014-5793(91)81450-m. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kahn S. M., O'Driscoll K. R., Jiang W., Borner C., Xu D. B., Blackwood M. A., Zhang Y. J., Nomoto K., Weinstein I. B. Suppression of mitogenic activity by stable expression of the regulatory domain of PKC beta. Carcinogenesis. 1994 Dec;15(12):2919–2925. doi: 10.1093/carcin/15.12.2919. [DOI] [PubMed] [Google Scholar]
- Kalpana G. V., Marmon S., Wang W., Crabtree G. R., Goff S. P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994 Dec 23;266(5193):2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- McDonald J. R., Gröschel-Stewart U., Walsh M. P. Properties and distribution of the protein inhibitor (Mr 17,000) of protein kinase C. Biochem J. 1987 Mar 15;242(3):695–705. doi: 10.1042/bj2420695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. R., Walsh M. P. Ca2+-binding proteins from bovine brain including a potent inhibitor of protein kinase C. Biochem J. 1985 Dec 1;232(2):559–567. doi: 10.1042/bj2320559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozier N. M., Walsh M. P., Pearson J. D. Characterization of a novel zinc binding site of protein kinase C inhibitor-1. FEBS Lett. 1991 Feb 11;279(1):14–18. doi: 10.1016/0014-5793(91)80238-x. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 1984 Apr 19;308(5961):693–698. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]
- Ohta M., Inoue H., Cotticelli M. G., Kastury K., Baffa R., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996 Feb 23;84(4):587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- Orengo C. A., Thornton J. M. Alpha plus beta folds revisited: some favoured motifs. Structure. 1993 Oct 15;1(2):105–120. doi: 10.1016/0969-2126(93)90026-d. [DOI] [PubMed] [Google Scholar]
- Papov V. V., Gravina S. A., Mieyal J. J., Biemann K. The primary structure and properties of thioltransferase (glutaredoxin) from human red blood cells. Protein Sci. 1994 Mar;3(3):428–434. doi: 10.1002/pro.5560030307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. D., DeWald D. B., Mathews W. R., Mozier N. M., Zürcher-Neely H. A., Heinrikson R. L., Morris M. A., McCubbin W. D., McDonald J. R., Fraser E. D. Amino acid sequence and characterization of a protein inhibitor of protein kinase C. J Biol Chem. 1990 Mar 15;265(8):4583–4591. [PubMed] [Google Scholar]
- Robinson A. K., de la Peña C. E., Barnes L. D. Isolation and characterization of diadenosine tetraphosphate (Ap4A) hydrolase from Schizosaccharomyces pombe. Biochim Biophys Acta. 1993 Feb 13;1161(2-3):139–148. doi: 10.1016/0167-4838(93)90207-8. [DOI] [PubMed] [Google Scholar]
- Robinson K., Aitken A. Identification of a new protein family which includes bovine protein kinase C inhibitor-1. Biochem J. 1994 Dec 1;304(Pt 2):662–664. doi: 10.1042/bj3040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K., Jones D., Howell S., Soneji Y., Martin S., Aitken A. Expression and characterization of maize ZBP14, a member of a new family of zinc-binding proteins. Biochem J. 1995 Apr 1;307(Pt 1):267–272. doi: 10.1042/bj3070267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter D. B., Sommer S. S. The generation of radiolabeled DNA and RNA probes with polymerase chain reaction. Anal Biochem. 1989 Feb 15;177(1):90–94. doi: 10.1016/0003-2697(89)90019-5. [DOI] [PubMed] [Google Scholar]
- Séraphin B. The HIT protein family: a new family of proteins present in prokaryotes, yeast and mammals. DNA Seq. 1992;3(3):177–179. doi: 10.3109/10425179209034013. [DOI] [PubMed] [Google Scholar]