Abstract
Hydrogels are hydrophilic polymer-based materials with high water content and physical characteristics that resemble the native extracellular matrix. Because of their remarkable properties, hydrogel systems are used for a wide range of biomedical applications, such as three-dimensional (3D) matrices for tissue engineering, drug-delivery vehicles, composite biomaterials, and as injectable fillers in minimally invasive surgeries. In addition, the rational design of hydrogels with controlled physical and biological properties can be used to modulate cellular functionality and tissue morphogenesis. Here, the development of advanced hydrogels with tunable physiochemical properties is highlighted, with particular emphasis on elastomeric, light-sensitive, composite, and shape-memory hydrogels. Emerging technologies developed over the past decade to control hydrogel architecture are also discussed and a number of potential applications and challenges in the utilization of hydrogels in regenerative medicine are reviewed. It is anticipated that the continued development of sophisticated hydrogels will result in clinical applications that will improve patient care and quality of life.
1. Introduction
Hydrogels are three-dimensional (3D) networks consisting of hydrophilic polymer chains, which are crosslinked to form matrices with high water content (up to thousand of times their dry weight).[1] Due to their remarkable characteristics, including tunable physical, chemical, and biological properties, high biocompatibility, versatility in fabrication, and similarity to native extracellular matrix (ECM), hydrogels have emerged as promising materials in the biomedical field.[1–3] Significant progress has been made in the synthesis and fabrication of hydrogels from both natural and synthetic sources for various applications; these include regenerative medicine, drug/gene delivery, stem cell and cancer research, and cell therapy.[4–6] Naturally-derived hydrogels, such as collagen, chitosan, hyaluronic acid (HA), alginate, gelatin, elastin, chondroitin sulfate, and heparin, are appealing for biological applications due to their cell signaling and cell-interactive properties, and biodegradability.[7] However, their limitations include low mechanical properties, inability to control their degradation and structure, and potential immunogenicity. On the other hand, synthetic hydrogels, such as poly(ethylene glycol) (PEG), poly(vinyl alcohol)(PVA), poly(2-hydroxyethyl methacrylate) (PHEMA), and polyacrylamide (PAM), possess controllable degradation and microstructure, generally show high mechanical properties, but lack biological moieties.[3,7] Due to the distinct properties of each of these hydrogel classes, gels that are based on the combination of natural and synthetic polymers have attracted significant attention for biological and biomedical applications.[8]
Various crosslinking approaches, including chemical and physical, have been employed to create polymer networks and preserve their 3D structures in aqueous environments. In physically crosslinked gels, physical interactions between polymer chains prevent dissociation of the hydrogel, while in chemically crosslinked gels, covalent bonds between polymer chains create stable hydrogels. Physically crosslinked hydrogels are formed through changes in environmental conditions (e.g., pH, temperature, and ionic interactions), hydrogen bonds, and protein interactions. There has been a growing interest in using this class of hydrogels for tissue regeneration as the gelation often occurs in mild conditions and aqueous solution in the absence of chemical crosslinkers.[9] Various injectable hydrogels based on alginate, collagen, agarose, HA, and chitosan have been synthesized by using physical crosslinking approaches for engineering different tissues.[10] These gels can be confined in the damaged site and eliminate the need of invasive surgery. However, low mechanical properties of physically crosslinked hydrogels may limit their tissue engineering applications, particularly in the regeneration of load bearing tissues. Chemically crosslinked gels have been obtained by radical polymerization, chemical reactions, energy irradiation, and enzymatic crosslinking. Some examples of chemically crosslinked gels for tissue engineering applications include PHEMA, glutaraldehyde (GA) crosslinked PVA, elastin, and chitosan, UV crosslinked methacrylated gelatin and elastin, transglutaminases crosslinked fibrinogen hydrogels.[9,11–13] Generally, chemically crosslinked gels have higher mechanical properties compared to their physically crosslinked counterparts, but the residual chemical crosslinkers, organic solvents, and photoinitiator may cause cytotoxicity.
Over the past decade, complex hydrogels have been designed as a result of major breakthroughs in the field of polymer science, microscale technologies, and molecular biology.[4,6] These advances have set the framework to overcome some of the challenges in regenerative medicine by rational design of hydrogels for various medical applications. This review covers the design principles being applied to synthesize advanced hydrogels with enhanced mechanical, biological, chemical and electrical properties. Due to their important biomedical applications, particular emphasis is given to elastomeric, photo-sensitive, hybrid and shape-memory hydrogel systems. In addition, emerging techologies for controlling the micro- and nanoscale architectures of 3D hydrogel constructs and their potential applications are highlighted.
2. Advanced Hydrogels with Tunable Properties
2.1. Elastomeric Materials
Biomaterials have been used as an artificial ECM to support the regeneration of various tissues. Since elasticity is one of the major mechanical characteristics of soft tissues, significant efforts have been made to engineer elastomeric biomaterials, which mimic the ability of native tissues to extend under stress. Mimicking the non-uniform elasticity of innate tissues including skin, blood vessel, lung, cardiac, and muscle is one of the major challenges in tissue engineering. Due to the high stretchability of native tissues, thermoplastic polymers with elongation break of less than 3% fail to replicate the innate tissue elasticity, as they undergo plastic deformation under variable loading.[14] To overcome this limitation, elastomeric hydrogels have been developed for biomedical applications.[15,16] However, one of the challenges associated with these elastomeric systems is their inability to mimic non-uniform elasticity of the native tissue. For example, many of the native tissues display strain stiffening and are responsive to applied strain, which can not be easily obtained by elastomeric systems.[17]
The use of synthetic elastomers for medical devices dates back to 1890s when the rubber industry was developed. Since then, natural and synthetic rubbers, such as silicones, polyolefins, and polydienes, and polyurethanes have been widely used as elastomers to engineer various medical devices due to their biocompatibility, mechanical durability, and low cost.[15]
In the last three decades, the rise of hydrogels as a popular choice of elastomeric materials for a variety of applications has been observed.[18] In this section, we focus on natural- and synthetic-derived elastomeric hydrogels, which are particularly useful for soft tissue engineering applications. We also discuss their limitations and potential applications for engineering biomimetic tissue constructs.
2.1.1. Naturally-Derived Elastin-Based Elastomers
Elastin is one of the main elastomeric proteins in connective tissues that are exposed to repetitive strains such as major vascular vessels, aorta, skin, elastic cartilage, tendon, and lung. Elastin is the essential component that provides elasticity and resilience needed for the proper function of these tissues. For example, the presence of elastin in arterial walls facilitates the blood transfer from the heart, lowers the mechanical work performed by the heart, and preserves the steady flow of oxygen to tissues.[19] In addition, elastin fibers allow blood vessels to withstand continuous cycles of contraction and expansion over the course of a life time.[20] Elastin is also known for being the most persistent and durable protein in the human body, with a half-life of 70 years.[18]
Elastin plays a critical biological role in regulating cellular functions. Various cell-surface proteins including elastin binding protein (EBP),[21] glycosaminoglycans (GAGs),[22] and integrin αvβ3[23] have been identified as receptors for elastin and its derivatives. Binding with these receptors has been shown to facilitate various cellular interactions. For example, it was found that elastin induced the attachment and proliferation of endothelial cells (ECs) and formation of vascular networks.[24] In addition, elastin derivatives could enhance the in vitro proliferation of skin fibroblasts.[23,25] Elastin fibers in the skin were also shown to influence cellular phenotypes during wound healing processes by controlling the differentiation of proliferative dermal fibroblasts into contractile myofibroblasts to help close the wound.[26] The presence of various cell-interactive segments in elastin and its derivatives enable them to modulate cellular functions. For example, VGAPG peptide sequences in elastin facilitate the formation of epidermis layer by inducing the migration and differentiation of epidermal keratinocytes.[27] These unique features demonstrate the potential value of elastin as a biologically active molecule for engineering elastic hydrogels in tissue engineering.
Various techniques have been developed to synthesize and purify elastin molecules from natural sources to engineer elastin-based hydrogels. Elastin can be obtained by partial hydrolysis of decellularized elastin-rich tissues in animals or by expression of recombinant protein.
Decellularized Tissues as Elastin-Based Scaffolds
Natural elastin-containing scaffolds can be generated by tissue decellularization, which removes the cellular component of explant tissues by detergent, enzymatic digestions, and solvent extraction processes. Due to their stability and durability, elastin-based tissues preserve their functions and structure after decellularization. Decellularized elastic scaffolds have been used as suitable replacements of lung, bladder, artery, heart valve, skin, and vascular graft.[28–30] Despite their advantages, decellularized scaffolds have several limitations. For example, the decellularization process involves harsh reaction conditions (e.g., enzymatic, chemical, or physical treatments) that may compromise the biological and mechanical properties of the constructs, particularly when additional steps of tissue purification are used.[31] Other limitations include batch-to-batch variability, risk of pathogen transfer, inability to obtain highly purified elastic tissue, and lack of versatility and uniformity of decellularized elastic tissues.[31]
Elastin Hydrogels Made from Soluble Elastin
Hydrolyzed elastin, soluble in aqueous solvents, has been used to engineer elastic hydrogels. The insolubility of intact elastin fibers in tissues prevents their processing into elastin-based hydrogels. To solve this problem, elastic tissues have been treated with oxalic acid or potassium hydroxide to yield soluble forms of elastin (e.g., α-elastin and K-elastin).[32,33] These hydrolyzed elastin molecules have properties similar to the native tropoelastin, such as ability to coacervate as well as to regulate cell signaling via the elastin receptors. This demonstrates the potential biological value of this class of elastin derivatives for biomedical applications.
Several elastin-based hydrogels have been synthesized from solubilized elastin for engineering different tissues such as skin,[32,34,35] cartilage,[36,37] and blood vessels.[38] For example, α-elastin hydrogels have been fabricated through chemical crosslinking approaches using various types of crosslinking agents.[32,34,40] Highly porous and elastic hydrogels were also engineered by crosslinking α -elastin with glutaraldehyde (GA)[34] and hexamethylene diisocyanate (HMDI)[32] under high pressure CO2. The fabricated hydrogels facilitated the infiltration, attachment, and growth of 3T3 fibroblasts within the 3D structure of the hydrogels.[32,34] In addition, the combination of α-elastin with poly caprolactone (PCL) promoted chondrocyte adhesion and proliferation.[36,39] Regeneration of cartilage tissue has also been achieved by using composite hydrogels containing K-elastin, alginate, and collagen.[37] Chondrocytes isolated from porcine and human were embedded inside the hydrogel composite and subsequently implanted in nude mice. After 12 weeks of implantation, cartilage-specific components including proteoglycans, collagen, and elastin fibers were formed within the engineered tissues which closely mimicked the native articular cartilage.[37] Despite its extensive use in tissue engineering, animal-derived soluble elastin is a heterogeneous mixture of peptides which are partially crosslinked and may not have adequate cell binding sites.[41] In addition, the clinical use of animal-derived proteins is often restricted due to the risk of pathogen transfer and immunological rejection.[42]
Recombinant Elastin-Based Hydrogels
Elastin-based elastomers can be also produced from various recombinant elastin proteins (e.g., recombinant elastin like polypeptides (ELP) and recombinant human tropoelastin). These proteins are obtained via the expression of recombinant DNA in different hosts including plants,[43–45] yeast,[46,47] and Escherichia coli (E. coli).[48]
Recently, human recombinant tropoelastin (rhTE) has been used to generate elastic rhTE-based hydrogels. Previously, rhTE was obtained in very low yield by construction of an expression vector containing the cDNA sequence of an isoform of human tropoelastin.[49] To enhance the production yield, Martin and Weiss developed a 2210-bp synthetic human TEL-encoding gene (SHEL) which contained codons optimized for maximum expression of rhTE in commercial yields.[50] This rhTE has been processed into a variety of promising hydrogels for tissue engineering applications.[27]
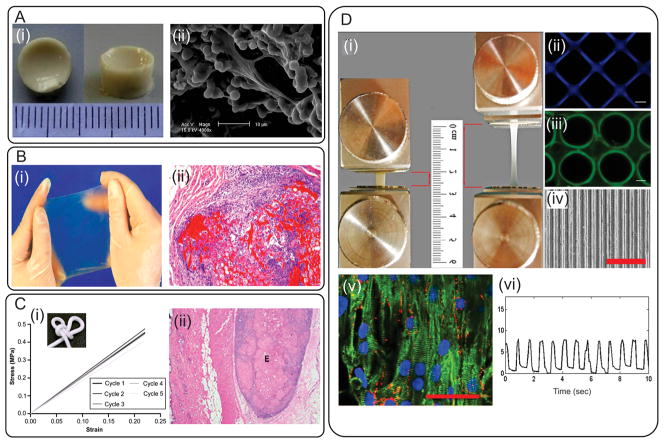
Elastic rhTE-based hydrogels with excellent cell-interactive properties have been created by using various approaches including, enzymatic crosslinking using yeast lysyl oxidase (PPLO),[51] chemical crosslinking,[52,53] using a fungal copper amine oxidase,[54] physical crosslinking,[55] and UV crosslinking.[13,56] For example, rhTE were chemically crosslinked by GA (Figure 1Ai)[52] or bis(sulfosuccinimidyl) suberate (BS3) (Figure 1Bi)[57] to generate hydrogels in various forms such as sheets, sponges, and tubes. The fabricated hydrogels promoted in vitro attachment, proliferation, and growth of dermal fibroblast cells (Figure 1Aii).[52] Furthermore, cellular penetration within the 3D structures of these hydrogels was significantly promoted by increasing the level of porosity and average pore sizes of the gels through the incorporation of GAGs[58] and the use of high pressure CO2.[52] In addition, the BS3 crosslinked rhTE hydrogels, that were implanted subcutaneously in guinea pigs, exhibited high biocompatibility and stability up to 13 weeks of culture (Figure 1Bii).[57] A physical crosslinking approach was also used to generate rhTE hydrogels by increasing the pH of protein solutions, which facilitated the self-assembly of rhTE spherules in a sol-gel transition process.[55] This approach eliminated the use of chemical or enzyme crosslinkers. The resulting hydrogel was highly flexible and elastic with compressive modulus of about 1.7 MPa over 5 cycles (Figure 1Ci). These hydrogels also facilitated the attachment and proliferation of dermal fibroblast in vitro and were stable for two weeks after intradermal injection into rats (Figure 1Cii).[55] Recently, a highly elastic photocrosslinkable hydrogel, methacrylated tropoelastin (MeTro), with tunable physical properties has been synthesized by functionalization of rhTE with methacrylate groups and subsequent UV crosslinking.[13] This approach was used to control the physical properties of resulting hydrogels including swelling behavior, porosity, and mechanical properties by altering the methacrylation degree and MeTro concentration. The fabricated MeTro hydrogels displayed high resilience, reversible deformation with low energy loss following cyclic compressions, and substantial extensibility up to 400% before rupture (Figure 1Di). In addition, in vitro studies showed that MeTro hydrogels supported cellular attachment and growth in both 2D and 3D culture environment.[13] Micropatterns were then created on the surface of these elastic hydrogels with microscale technology to fabricate micropatterned MeTro hydrogels, which were used to align cardiomyocytes (CMs) isolated from rat hearts (Figure 1Dii–iv).[56] The in vitro studies demonstrated that these microfabricated MeTro hydrogels successfully promoted all the characteristics of CMs including attachment, spreading, alignment, phenotype and synchronized beating, which ultimately led to the formation of highly functionalized cardiac tissues (Figure 1Dv–vi).[56] rhTE-based elastomeric hydrogels have shown unique mechanical and biological properties[59] and exhibited potential advantages compared to animal-derived hydrolyzed soluble elastin. First, rhTE is synthesized using a highly reproducible recombinant technology, which eliminates the batch-to-batch variations associated with soluble elastin derived from animal sources. Second, as shown by the animal studies using rhTE-based hydrogels, rhTE carries little risk of immunological rejection upon implantation.[59] Third, the abundance of cell-responsive peptides on rhTE molecules[23] significantly promotes biological properties of rhTE-based biomaterials as compared to soluble elastin.
Figure 1.
Examples of naturally-derived elastin-based hydrogels. A) GA crosslinked rTE/elastin hydrogels produced under high pressure CO2; i) structure of the hydrogel after swelling, ii) SEM image of dermal fibroblast cells penetrated and attached within the 3D structure of the gel. Reproduced with permission.[52] Copyright 2010, Elsevier B.V. B) BS3 crosslinked rTE gel; i) an image from an elastic hydrogel sheet, (ii) hematoxylin and eosin-stained sample explanted after 13 weeks of implantation (hydrogel is shown in bright red). Reproduced with permission.[57] Copyright 2004, Elsevier B.V. C) Physically crosslinked rTE gel; i) representative stress–strain curves over 5 cycles, the resulting gel could be tied in a knot, demonstrating its high flexibility, ii) a hematoxylin and eosin-stained explant showing the injection site (the elastic deposit is marked with an E). Reproduced with permission.[55] Copyright 2009, Elsevier B.V. D) Methacrylated rTE gel; i) image of an elastic MeTro gel before and after stretching, ii–iv) formation of patterns with various geometries on MeTro gel by using different microfabrication techniques, v) immunostaining of CM markers on MeTro gel on day 8 of culture, gel stained for sarcomeric α-actinin (green)/connexin-43 (red)/nuclei (blue) (scale bar = 50 μm), vi) beating behavior of CMs on micropatterned MeTro gel. Reproduced with permission[13] Copyright 2013, Elsevier B.V; Reproduced with permission.[56] Copyright 2013, John Wiley & Sons, Inc.
2.1.2. Synthetic Elastomers
Synthetic degradable elastomers have received significant attention for tissue engineering applications, particularly for soft tissue regeneration. Their unique features include 3D crosslinked networks, which mimic the structure of naturally-derived elastic materials, high elasticity and flexibility, biodegradability, and mechanical properties similar to those of native soft tissues.[16] In addition, the physical properties of these elastomers can be adjusted by changing their processing conditions. The synthesis and preparation of synthetic degradable elastomers have been comprehensively reviewed elsewhere.[15,16] In this section, we introduce several examples of the synthetic elastic materials and composites, which can be used to form elastic hydrogels.
Elastin-like Polypeptides (ELPs)
ELPs containing repetitive amino acids have been synthesized and extensively used to engineer highly elastic hydrogels.[60,61] This class of polymers possesses promising properties for tissue engineering including tunable degradation rates and similarity to native ECM. In addition, their fabrication method allows the incorporation of bioactive peptide moieties within their structures during polymer synthesis.
ELP-based elastomers have been fabricated for the regeneration of blood vessels, cartilage, ocular, and liver tissues.[60,61] For example, cell-laden ELP hydrogels were fabricated by Betre et al. for cartilage repair.[62] To form these injectable cell-laden hydrogels, temperature-triggered coacervation of ELPs was used to encapsulate chondrocytes. The resulting gels facilitated the growth and proliferation as well as the formation of cartilage ECM (e.g., deposition of glycosaminoglycans and collagen).[62] These ELP-based hydrogels also facilitated the in vitro differentiation of human adipose-derived adult stem cells into chondrocytes without the addition of chondrogenic growth factors.[63] Despite the suitable biological properties, these coacervated ELP hydrogels lacked mechanical stability and stiffness, which limited their tissue engineering applications. To fabricate ELP-based gels with higher mechanical properties, researchers have chemically crosslinked a lysine containing ELP by using β-[tris(hydroxymethyl)phosphino]propionic acid (THPP) under physiological conditions.[64–67] The THPP-crosslinked ELP hydrogels supported cell penetration and formation of ECM after injection into an osteochondral defect using a goat model. However, their fast degradation rate was an issue.[67] ELPs containing lysine were also crosslinked using various types of crosslinking agents such as tris-succinimidyl aminotriacetate[66] and bis(sulfosuccinimidyl) suberate.[68]
The unique properties of ELP-based hydrogels, such as their tunable degradation and mechanical properties, and low toxicity, make them a promising class of materials for biomedical applications. However, in vivo biocompatibility of ELP-based hydrogels is still unknown as there are only few in vivo studies on these materials. Therefore, it is crucial to study the immune response against a comprehensive library of ELPs prior to clinical application. It is expected that more systematic approaches for engineering ELPs with controlled biological properties will be developed. Microengineered technologies can also be used to tailor the properties and architectures of ELP-based materials to further advance the potential applications of this class of polymers in regenerative medicine.
Poly(glycerol sebacate) (PGS)
PGS has been synthetized by polycondensation of glycerol and sebacic acid,[69] and used as a promising polyester-based elastomer for soft tissue engineering applications. PGS has been used in various forms including sheets, porous scaffolds, electrospun fibers, and microfabricated constructs for the regeneration of soft tissues such as vascular,[70] cardiac,[71] retinal,[72] cartilage,[73] and neural[74] tissues. Despite having promising properties, PGS has limited water uptake capacity (approximately 2%), which constrains its utility as a hydrogel for soft tissue engineering applications. The hydrophilicity of PGS can be improved by incorporating additional carboxyl groups in PGS backbone[75] or by its copolymerization with PEG.[76–78] Recently, poly(glycerol sebacate citrate) (PGSC) was synthesized by thermally curing citric acid and PGS mixture in a mold.[75] The biodegradation and mechanical properties of elastomeric PGSC scaffolds were controlled by its composition as well as the thermal curing time. The presence of hydroxyl groups in the backbone of PGSC improved the water uptake properties of the elastomer, which can be beneficial for tissue regeneration.[75] Our group has also demonstrated the synthesis of highly elastic PGS-co-PEG copolymers with controlled swelling behaviors.[76] The mechanical properties and degradation of resulting elastomers were finely tuned by changing the water uptake properties of the hydrogels. The elastic modulus of PGS-co-PEG was in the range of 13 kPa to 2.2 MPa, depending on the concentration of PEG incorporated within the copolymer. In addition, the presence of PEG in the polymer network resulted in a 15-fold increase in water uptake capability and a 6-fold increase in elongation as compared with PGS elastomers. The PGS-co-PEG copolymers supported the growth and proliferation of 3T3 fibroblasts over 10 days of culture, demonstrating the suitability of synthesized elastomers for tissue engineering applications.[76]
Due to their tunable physical properties, PGS-based materials are promising candidates for engineering soft tissues, particularly cardiovascular tissues. Despite significant progress in utilizing PGS elastomers for tissue regeneration, there are still some challenges including their fast degradation rate (several weeks), which limits their applications for engineering tissues that require longer time to regenerate (several months to years). In addition, PGS polymers produce acidic degradation products, which causes cytotoxicity in vitro.[74,79] However, in vivo assessment of PGS-based scaffolds showed little to mild inflammation.[73,80,81] It has been shown that increasing the crosslinking density can improve the in vitro cytocompatibility of PGS-based elastomers.[79,82] Other limitations of PGS for soft tissue engineering applications include difficulties in achieving non-linear elastic behavior similar to native soft tissue and their inability to be used as a 3D environment for cellular encapsulation due to their harsh processing conditions (e.g., high temperature).
Polyurethanes (PU)
Since the 1980s, PU materials have been widely used for engineering cardiovascular devices including vascular prostheses, cardiac valves, the total artificial hearts, blood bags, and small diameter grafts for bypass surgeries. Due to their long-term stability, PU-based scaffolds have been utilized for long-term implantation. Tuning biodegradability and durability of PU-based materials is an essential step for their tissue engineering applications. Significant efforts have been made to synthesize a new class of biodegradable PUs for engineering various tissues including vascular grafts,[83] neural tissue,[84] bone,[85] and cardiac muscle.[86,87] These biodegradable PU-based polymers have been synthesized by incorporating chain extenders or soft segments (e.g., caprolactone, lactides, amino acids, and PEG) in PU backbone to induce degradability and in some cases hydrophilicity. For example, Zhang et al. synthesized a series of photocrosslinkable PU hydrogels containing PEG and PCL as the soft segment, lysine diisocyanate (LDI) as the hard segment, and 2-hydroxyethyl methacrylate (HEMA) as the chain terminator.[88] The physical properties of fabricated elastic PU hydrogels were tuned by changing the ratio of PCL/PEG in the soft segment. For example, increasing the amount of PEG enhanced the swelling ratio and degradation rate but reduced the mechanical properties of fabricated PU hydrogels. The fabricated PU hydrogels had swelling ratio in the range of 3.2–66%, elastic modulus ranging from 17–34 MPa, and fracture strain of 5–61% when the ratio of PEG/PCL was changed from 0/100 to 50/50. The PU hydrogels supported mouse chondrocyte attachment and proliferation.[88] Recently, an injectable amine-functionalized PU/PEG block co-hydrogel was synthesized and exhibited its highest elastic modulus at 37 °C.[89] The fabricated composite supported in vitro growth of smooth muscle cells. The results of in vivo test exhibited significant inflammatory response 3 days post-implantation with the presence of recruited ED-1 positive macrophages but the amount of inflammation decreased 4 weeks after implantation.[89] In another recent study, highly elastic PU-based biomaterials, with extensibility of more than 1100%, were fabricated by the solution blending of sodium alginate and an aqueous solution of cationic PU to form cationic dispersions-sodium alginate nanoparticles.[90] Incorporating sodium alginate into PU network improved the mechanical strength as well as the hydrophilicity of the composite network.[90] Similarly, Huang et al. combined a hierarchical PU scaffold with a cell-laden hydrogel composed of gelatin, alginate, and fibrinogen by using a rapid prototyping technique to form 3D vascular constructs.[91] The external PU scaffold provided adequate mechanical support while the internal hydrogel construct supported adipose-derived stem cell growth and proliferation in vitro. The fabricated PU/cell-laden hydrogel was also found to be stable and biocompatible after two weeks of implantation in the abdominal cavity of nude mice.[91]
These results together indicate that biodegradable PU-based composite hydrogels are promising elastic biomaterials for tissue engineering. These highly elastic materials have shown proper durability and good biocompatibility in vitro by supporting cellular adhesion and proliferation during culture. However, the in vivo evaluation of biodegradable PU-based scaffolds demonstrated limited stability and consequently mechanical failures. In addition, the toxic degradation products of PUs (e.g., aromatic diisocyanates) can cause cytotoxicity.[92] To address this problem, aromatic diisocyanates have been replaced with aliphatic ones such as lysine diisocyanates for the synthesis of bioresorbable PU.[93,94] However, more investigation on the in vivo response to biodegradable PU-based materials and their susceptibility to biodegradation is required.
Composite Elastomers
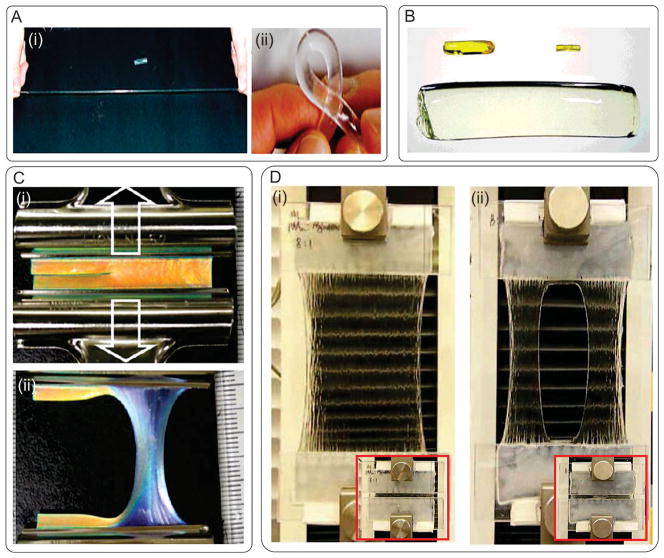
Various types of composite elastomers have been developed including nanocomposite hydrogels (Figure 2A),[95,96] polyrotaxane gel (Figure 2B),[97] double network (DN) gels,[98] hydrophobic bilayers (PDGI)/polyacrylamide (PAAm) (Figure 2C),[99] and PAAm/alginate composite gel[100] (Figure 2D). For example, polymer/clay nanocomposites composed of N-isopropylacrylamide and hectorite clay Laponite XLG were formed by free radical polymerization of the polymer in an aqueous suspension of clay. The resulting hydrogel had tensile modulus in the range of 270–300 kPa and elongation of up to 1300%. The fabricated nanocomposite gels could also withstand high levels of deformation in twisting, bending, and knotting[95] (Figure 2A). In another study, highly elastic hydrogels were formed by ionic crosslinking in combination with physically associated triblock copolymer chains, but these hydrogels could only recover up to 50% of their initial deformation.[101] To solve this problem, Haque et al. incorporated lamellar PDGI in a hydrophilic PAAm matrix as reversible sacrificial bonds to dissociate upon deformation with large energy dissipation.[99] The fabricated hydrogel was highly elastic with a tensile strength of 38 kPa and strain of 2200%. The hydrogel fully recovered its original length within several minutes after stress removal (Figure 2C). Despite their high recovery capability, these PDGI/PAAm gels had lower fracture energy compared to DN gels. Recently, a highly stretchable and tough hydrogel was synthesized by mixing ionically crosslinked alginate, and covalently crosslinked PAAm (Figure 2D).[100] The fabricated hydrogels were able to stretch more than 20 times of their initial length. The hydrogel sheets were also shown to be notch-insensitive and fully recovered after mechanical stretching caused by dropping a metal ball on the hydrogel membrane.[100]
Figure 2.
Examples of composite elastomers. A) N-isopropylacrylamide/clay nanocomposite hydrogel; i) with high level of elongation and ii) torsion. Reproduced with permission.[95] Copyright 2002, John Wiley & Sons, Inc. B) Volume change of a superabsorbent polyrotaxane gel swelled to 45 times the initial weight; before volume change, in dried state, and in swollen state (up to 400% of its dry weight). Reproduced with permission.[97] Copyright 2001, John Wiley & Sons, Inc. C) Crack resistance of a PDGI/PAAm gel; (i) hydrogel with an initial sharp crack along the longitudinal direction, (ii) the hydrogel was stretched perpendicular to the crack direction up to a strain of 3. Reproduced with permission.[99] Copyright 2011, American Chemical Society. D) Highly stretchable alginate/acrylamide gel; i) the gel was glued to two rigid clamps and stretched up to 21 times its initial length, ii) a notch was cut into the gel before stretching to 17 times its initial length. Reproduced with permission.[100] Copyright 2012, Nature Publishing Group.
2.2. Photosensitive Hydrogels
Hydrogels can be crosslinked or degraded by utilizing various approaches such as ionic interactions, pH stimulation, and light exposure.[102] Photosensitive hydrogels have been extensively used for a wide range of tissue engineering applications. In this section, we will discuss about polymer networks, which can be either generated or degraded by UV light exposure.
2.2.1. Photocrosslinkable Gels
To form hydrogels via exposure to light, a photocurable hydrogel precursor is mixed with a photoinitiator and then exposed to light that initiates the crosslinking reaction.[2] Although a range of light wavelengths can be used, ultraviolet (UV) light is most commonly used to induce the photoinitiator to generate free radicals. The activated functional groups then form covalent bonds with free radicals to create crosslinked networks.[103,104] Subsequently, unreacted polymer is washed out upon completion of the crosslinking process. Photocrosslinkable hydrogels offer a number of advantages over other types of crosslinking schemes. For example, they enable controlled spatial crosslinking of the hydrogel to control the microarchitecture of the resulting material,[105–107] which can be used to modulate cellular behavior (e.g., adhesion, migration, and differentiation).[108,109] In addition, photocrosslinking is a simple, rapid, and cost effective technique.[2,6] Despite these attractive features, photocroslinkable hydrogels also demonstrate some drawbacks. For instance, the formation of free radicals upon UV exposure may lead to DNA damage and impair cellular function.[2] In addition, in vivo gelation of photocrosslinkable hydrogel is challenging due to the limited light penetration through the tissues.
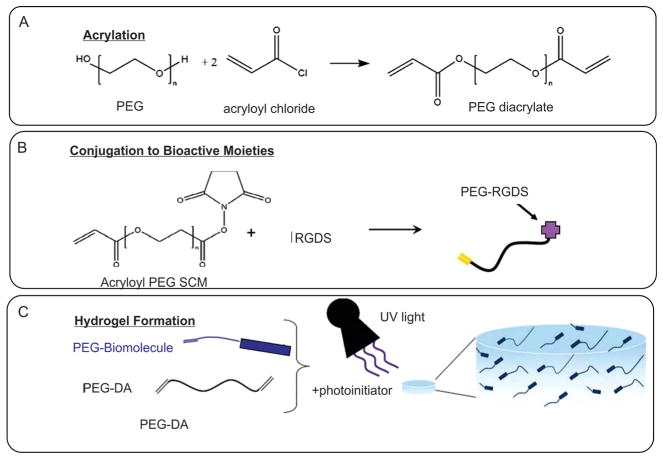
Materials with both synthetic and natural origins have been modified with photocrosslinkable functional groups.[107] For instance, PEG[110–112] and PHEMA[113] were chemically modified by methacrylate groups to synthesize photocrosslinkable hydrogels (Figure 3). Similarly, naturally-derived materials, such as alginate,[114] dextran,[115] agarose,[116] heparin,[117] hyaluronan,[118–121] chitosan,[122] collagen,[123] and gelatin[12,119,124–126] were methacrylated to yield photocurable gels. These photocrosslinkable hydrogels were used as robust 3D environments to engineer biomimetic cell-laden hydrogels for different tissue engineering applications. For instance, 3T3 fibroblast cells were encapsulated within photocurable gelatin hydrogels to test the biocompatibility of the gels.[12,127] Similarly, macrophages,[110] human umbilical vein endothelial cells (HUVECs)[107] and hepatocytes[111] were tested for their cellular response within photocrosslinkable gels based on PEG, gelatin, and HA.
Figure 3.
Schematic for generation of photocrosslinkable hydrogels. A) Modification of PEG polymer with photocrosslinkable acrylate groups. B) Conjugation of biological molecules to photocrosslinkable PEG polymer precursor. C) Formation of hydrogel upon exposure to UV light. Reproduced with permission.[112] Copyright 2011, Wiley Periodical Inc.
Micropatterning using photocrosslinkable gels is a common strategy to modulate cellular behavior. For example, micropatterned gelatin-based hydrogels enabled guidance and alignment of different cell types, such as 3T3 fibroblasts, C2C12 skeletal muscle cells, cardiac side population (CSP) cells, and HUVECs.[128] These technologies will be discussed in details in Section 3. Photocrosslinkable hydrogels allow temporal and spatial control over structural, mechanical, and degradation of the fabricated constructs. For example, in one study methacrylated HA gel was first crosslinked by a Michael-type addition reaction with dithiothreitol (DTT) and then its mechanical stiffness was tuned by additional UV crosslinking.[129] It was shown that the substrate stiffness affected differentiation of MSCs, seeded on the surface of the hydrogel.
Due to their ability to generate micro- and nanostructures, as well as their tunable chemical, biological and mechanical properties, photocrossinkable hydrogels have been extensively used in tissue engineering research. However, the next generation of photocrosslinkable hydrogels could further benefit from novel strategies for in situ crosslinking of these hydrogels within the human body.
2.2.2. Photodegradable Gels
Hydrogel structures can be controlled both spatially and temporally to modulate material properties and therefore their biological response.[130] When photodegradable gels are exposed to irradiation, light-triggered reactions induce dissociation of the crosslinks in the polymer chain and therefore its degradation in the exposed region. This results in either complete degradation or local decrease in the crosslinking density, which influences the physical properties of the gel and subsequently cellular behaviors.[131] For example, the degradation of cell-encapsulated hydrogels can facilitate the deposition of the ECM by the cells as well as guide cellular migration within the hydrogel.
The most commonly used photodegradable functional groups are nitrobenzyl ether,[130] poly(t-butyl acrylate),[132] 4-[4-(1-Hydroxyethyl)-2-methoxy-5-nitrophenoxy] butanoic acid,[133] and bis(4-(dimethylamino)phenyl)(4-vinylphenyl)methyl leluco cyanide.[134] Photodegradable hydrogels from different synthetic sources have been fabricated by the incorporation of the above listed groups to the polymer backbone.[132,133,135–142] For example, the migration of stem cells was guided within photodegradable PEG-based hydrogels.[143] In addition to their bulk degradation ability, photodegradable hydrogels can also be patterned by irradiating light through photomasks to generate specific patterns for modulating or directing cellular migration. In one study, directed migration of fibrosarcoma cells within photodegradable hydrogels was investigated.[130] In another report, DeForest et al. controlled the architecture of PEG-based photodegradable hydrogels, which led to the modulation of the adhesion, spreading, and migration of 3T3 fibroblasts.[144]
Generally, photodegradation enables tuning the hydrogel structure and properties spatially and temporally and offers unique opportunities to control cellular function in 3D environments. But tissue engineering applications of photodegradable hydrogels have been less explored. In addition, similar to their photocrosslinkable counterparts, sensitivity of cells to irradiation may be a concern for cell encapsulation; therefore the process should be optimized.[142] It is expected that photodegradable hydrogels to be utilized to direct cellular behavior dynamically and control local microenvironments for different tissue engineering applications ranging from directed migration of neurons, formation of cardiac fibers, and photorelease of biological molecules.
2.3. Reinforced Composite Hydrogels
Hydrogels are generally made from a single polymer network. But no single polymer holds all the required mechanical and biological properties for tissue engineering applications. Therefore, strategies are being employed to improve the properties of hydrogels, mainly by incorporating different entities to create composite hydrogel matrices.[145–147] These strategies include incorporation of secondary polymers as well as various nanostructures into the main hydrogel. In this section, we will discuss the fabrication and characterization of these composite hydrogels.
2.3.1. Polymer-Composite Hydrogels
The simplest way to create composite materials is by mixing different materials (‘physical blending’). As polymers used to fabricate hydrogels are hydrophilic, they are generally compatible with one another, and miscible in aqueous solutions. For example, alginate hydrogels are widely used in various bioengineering applications.[148] However, alginate alone may not induce the desired cellular response. Therefore, ECM proteins such as collagen and fibronectin, or cell-responsive synthetic polymers such as polylysine are often mixed with alginate to regulate its cell-interactive properties.[149,150] In addition, mechanical properties of the alginate hydrogels were enhanced by incorporating other natural or synthetic polymers (e.g., chitosan, poly(vinyl alcohol) (PVA) and poly(acrylic acid)(PAA)) into hydrogels.[151–153] There are other types of polymer-composite hydrogels utilizing more elaborate strategies to incorporate a secondary polymeric network, such as hybrid networks, interpenetrating polymer networks (IPNs), and semi-IPNs.
Hybrid Hydrogels
One critical drawback of incorporating secondary polymers via physical blending is that only limited amounts of the polymers can be included within the hydrogel network. Often, inadequate inclusion of the secondary polymers within the hydrogels leads to phase separation.[154,155] Therefore, co-polymerization schemes are employed to covalently link the secondary polymers to the hydrogel, and ultimately create fully integrated “hybrid” hydrogels with a wide range of properties. For example, photocrosslinkable hydrogels are commonly prepared by UV-induced radical polymerization of reactive species containing double carbon-carbon bonded functional groups (e.g., vinyl, acrylic and methacrylic).[156,157] Secondary polymers containing those groups can readily undergo copolymerization to form hybrid hydrogels. For example, HA, a polysaccharide component of natural ECM, is made photocrosslinkable by conjugating methacrylated functional groups, and incorporated into PEG hydrogels.[158]
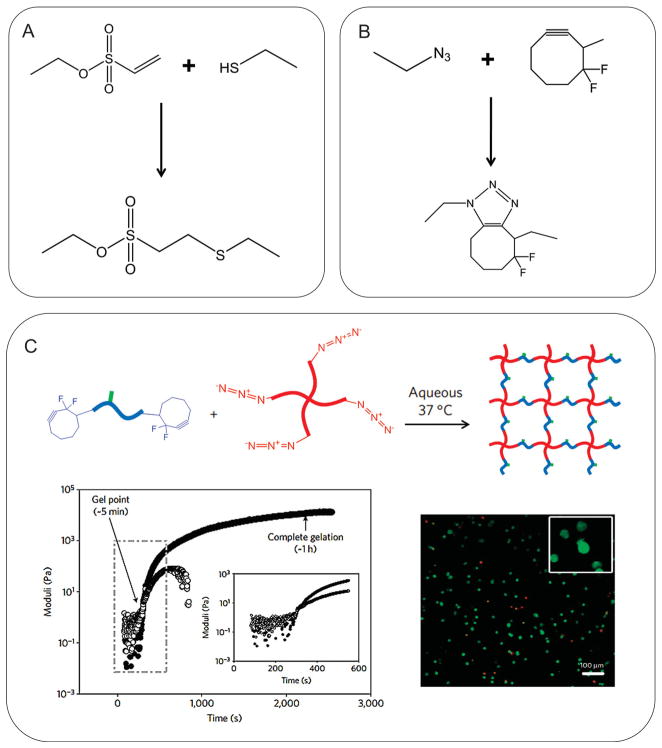
Recently, more biocompatible and selective chemical reactions have been used to create hybrid hydrogels. For example, Lutolf and Hubbell first employed a Michael-type addition reaction to engineer hydrogels using thiol- and vinyl-functionalized PEG molecules (Figure 4A).[159] This thiol-based Michael reaction was especially useful to prepare hydrogels for biological applications as the reaction took place in physiological pH and buffers and in the absence of potentially toxic initiators.[135,160–162] Furthermore, “click” chemistry, which has gained recognition in recent years as a highly bio-orthogonal and facile reaction between alkyne and azide groups, has also been explored to fabricate hydrogels.[162–164] Especially, the biocompatibility of the click chemistry was achieved by the recent development of “copper-free” method (i.e., cycloaddition reaction between cyclooctyne and azide), which circumvented the use of cytotoxic copper catalyst (Figure 4B).[164,165] For example, DeForest et al. utilized the copper-free click chemistry to create PEG-polypeptide hybrid hydrogel (Figure 4C).[166] The reaction between tetraazide-functionalized 4-arm PEG and bis(difluorocyclooctyne)-functionalized polypeptide resulted in hydrogel formation under physiological conditions. The viability of cells encapsulated in the hydrogel was well maintained.[166]
Figure 4.
Biocompatible click-based hydrogels. A) A Michael-type addition between thiol and vinyl sulfone. B) A “copper-free” click chemistry between azide and difluorinated cyclooctyne. Both reactions occurred under physiological conditions. C) Cell-encapsulating hydrogel was fabricated by copper-free click chemistry between 4-arm PEG-tetrazide and bis(difluorocyclooctyne)-polypeptide. Reproduced with permission.[166] Copyright 2009, Nature Publishing Group.
IPN and Semi-IPN Hydrogels
IPNs date back to early 20th century, when Aylsworth first created IPN materials by introducing phenol-formaldehyde resins to natural rubber network.[167] The primary purpose for developing IPN has been to improve the fracture resistance of a material, which otherwise may be too weak or brittle.[168] It is well known that the IPN materials demonstrate enhanced mechanical toughness by efficiently transferring externally applied stress to the secondary polymer network.[98,168] The major distinction of IPN hydrogels from hybrid hydrogels is that the secondary polymers are not covalently conjugated to the hydrogel. Rather, the secondary polymers are placed within the hydrogel already formed, and then allowed to undergo polymerization to form the secondary polymer network. Therefore, the secondary network becomes interlocked within the hydrogel.[169] Semi-IPN hydrogel is another class of polymer-composite hydrogels, in which the secondary polymers are not crosslinked, but only “trapped” within the polymer network.[169]
The strategy of creating IPN hydrogel is continuously explored as a popular mode of structurally reinforcing hydrogels. For example, Gong et al. engineered IPN hydrogel with PAAm and poly(2-acrylamido-2-methylpropanesulfonic acid) (PAMPS), which demonstrated extremely high tensile strength as compared with the single network of PAAM or PAMPS (i.e., 10 to 20 fold increase in fracture stress).[170] Sun et al. have recently reported similar findings, in which by introducing ionically crosslinked alginate network to PAAm network, the resulting IPN hydrogels showed significant stretchability.[100]
There are efforts to create IPNs in order to add different physical characteristics to the hydrogels, rather than focusing purely on the mechanical properties. For example, poly(N-isopropylacrylamide) (PNIPAm) and PAAm networks are often incorporated to engineer IPN hydrogels with temperature- and pH-sensitivity, respectively.[171,172] In addition, Lin et al. introduced electrically conductive polyaniline (PANI) network to non-conductive PAAm hydrogel, and demonstrated that the resulting PANI/PAAm IPN hydrogel showed improved electrical field sensitivity as compared with PAAm hydrogel.[173]
2.3.2. Nanocomposite Hydrogels
With the rapid development of nanotechnologies in recent years, extensive research efforts are being made to develop nanoparticles (NPs) for various biomedical applications.[174] NPs can be engineered from a variety of sources (e.g., polymers, minerals, metals, and semiconductors) and into different shapes (e.g., spheres, rods, shells, wires, and tubes).[174–176] In addition, chemical modification strategies are available to further modulate the properties of NPs.[177] Due to this diverse array of NPs with distinct physical and chemical properties, research efforts are being made to incorporate various types of NPs into hydrogel systems to create reinforced nanocomposite hydrogels.
Mineral NPs
The earliest attempt to reinforce materials with NPs was made by Usuki et al., who incorporated mont-morillonite, a type of natural silicate mineral (“clay”) NPs, into nylon-6.[178] The fabricated nanocomposites showed significant improvement in tensile strength.[178] Since then, various types of clay NPs have been used as prominent composite materials.[179] The inorganic clay NPs are often modified with polymers to render them hydrophilic, and therefore increase solubility and interaction with polymers.[180] The organic clay NPs are also being used to reinforce hydrogels. For example, Kokabi et al. engineered PVA hydrogel reinforced with organic-substituted clay NPs.[181] The resulting PVA-clay nanocomposite hydrogels showed enhanced tensile strength and diminished microbial penetration, while maintaining high water absorbency.[181]
Polymeric NPs
Synthetic polymeric NPs, such as dendrimers and micelles, which are widely used as drug delivery systems, have also been incorporated into hydrogels to utilize their drug releasing capability, as well as enhance their mechanical properties. In one study, Xiao et al. covalently incorporated micelles, made from self-assembly of block copolymers, in PAAm hydrogel.[182] Mechanical properties of the micelle-linked PAAm hydrogel could be controlled by the amount of micelles, in which 2-fold increase in tensile stress and 4-fold increase in Young’s modulus were observed when the micelle concentration was doubled, from 7.5 to 15 mg mL−1. In another study, Desai et al. developed a photocurable PEG hydrogel crosslinked with polyaminoamine dendrimers, and demonstrated cytocompatibility, controlled swelling and degradation by modification of dendrimers (i.e., chain length and charge density).[183]
Metallic NPs
With increasing popularity of hydrogels in various areas of biomedical engineering, there is a growing need to engineer hydrogels with tunable properties (e.g., electrical and optical properties) to meet specific needs. Metallic NPs hold great promise as reinforcing elements to engineer composite hydrogels with unique characteristics, since they possess properties that are not commonly found in polymers or inorganic materials. For example, gold nanoparticles (AuNPs) are actively explored as biosensors, as they possess useful electronic and optical properties (e.g., quantized capacitance and surface plasmon resonance).[184] Pardo-Yisar et al. utilized a “breathing” mechanism to introduce AuNPs into hydrogels, in which PAAm hydrogel was first dehydrated by organic solvent, then followed by swelling in aqueous media containing AuNPs.[185] The resulting AuNP/PAAm composite hydrogels demonstrated significant increase in electrical conductivity. Silver nanoparticles (AgNPs) are also well known for their unique antibacterial activity. Therefore, AgNP-incorporated hydrogels are currently being investigated for their use as antibacterial wound dressing.[186]
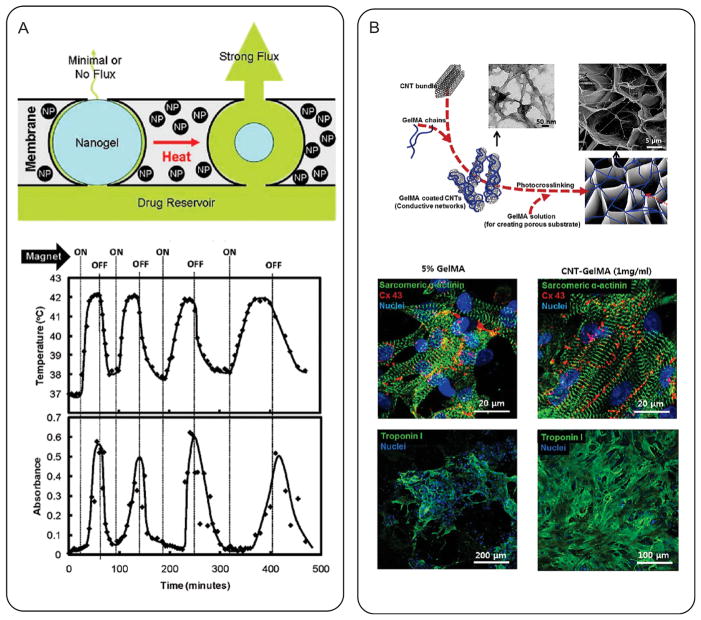
Magnetic nanoparticles (MNPs), such as iron oxide and gadolinium, are the most widely investigated class of metallic NPs for biomedical applications.[187,188] They have been mainly used as contrast agents for magnetic resonance imaging (MRI), with several products already commercially available.[187] Strategies of coating MNPs with hydrogels have been explored to increase their hydrophilicity and biocompatibility, as well as reduce non-specific protein adsorption. For example, Yi et al. utilized a sol-gel reaction to coat iron oxide MNPs with silica hydrogel.[189] Another interesting aspect of MNPs is their ability to produce heat under applied magnetic field (“magnetocaloric effect”).[190] Therefore, MNP-loaded hydrogels are currently being explored in hyperthermia-based cancer therapies (e.g., thermal ablation and temperature-sensitive drug delivery).[188,191,192] For example, Hoare et al. developed a composite film consisting of thermosensitive PNIPAm hydrogel and iron oxide MNPs.[193] They demonstrated that magnetic-induced hyperthermia from iron oxide MNPs caused the volumetric change of PNIPAm hydrogel, thereby inducing the release of cancer drugs (Figure 5A). In another study, Meenach et al. showed that magnetization of PEG-MNP composite hydrogels generated enough heat to kill glioblastoma cells.[194]
Figure 5.
Nanocomposite hydrogels as bioactuators. A) A magnetic triggered composite film consisting of iron oxide MNPs and PNIPAm hydrogels. The magnetic-induced hyperthermia from the iron oxide MNPs caused shrinkage of PNIPAm hydrogel, inducing the release of cancer drugs. Reproduced with permission.[193] Copyright 2009, American Chemical Society. B) CNTs were embedded in GelMA hydrogel to engineer electrically conductive tissue engineering scaffold. CMs on CNT-GelMA composite hydrogels displayed enhanced electrophysiological functions (identified with the expression of various cardiac markers including sarcomeric α-actinin, connexin 43, and troponin I), compared to pure GelMA hydrogels. Reproduced with permission.[195] Copyright 2013, American Chemical Society.
Carbon-based NPs
Carbon-based NPs (CBNs), such as carbon nanotubes (CNTs) and graphene, have gained worldwide popularity in recent years for their excellent multi-functional nature (i.e., mechanical strength, electrical, thermal, and optical conductivity). Therefore, extensive research efforts are under way to utilize their unique and favorable properties to create CBN-based composite materials for a variety of applications, including high-strength materials, nanoscale electronic circuitry, sensors, and actuators.[196–198] CBNs are also being investigated to reinforce hydrogels for biomedical applications. For example, Tong et al. demonstrated a twofold increase in tensile stress of PVA hydrogels by incorporating CNTs (0.05 wt%).[199] Similarly, Zhang et al. reported a 2-fold increase in tensile stress of PVA-graphene oxide (GO) composite hydrogel when GO was increased up to 0.8 wt%.[200]
Hydrogels commonly used in biomedical applications are not conductive towards thermal, optical and electrical stimuli, as they are made from non-conductive polymers. However, there is a growing need for conductive hydrogels to be used as biosensors, actuators and tissue engineering scaffolds. Therefore, CBNs are considered as highly promising materials to engineer conductive stimuli-responsive hydrogels. For example, Zhang et al. developed an electrical- and pH-responsive actuator based on graphene-PAAm composite hydrogel.[201] Shin et al. also demonstrated increased electrical conductivity of methacrylated gelatin (GelMA) hydrogel by incorporating CNTs. The resulting CNT-GelMA composite hydrogel was successfully used as a tissue engineering scaffold to mediate enhanced electrophysiological function of CMs (Figure 5B).[195]
Overall, as hydrogels have become a popular choice of materials used in several areas of bioengineering, strategies of encompassing various components to develop composite hydrogels with tunable properties are expected to be continuously explored. Traditionally, the composite strategies are often employed to engineer hydrogels with enhanced mechanical strength and function. Also, commonly used reinforcing materials have been mostly limited to polymers. However, more extensive research efforts are now being made to impart hydrogels with various properties (e.g., electrical and optical conductivity), with added emphasis on engineering multifunctional hydrogels to tailor to specific needs and complexities often required for present and future challenges. For this purpose, the recent advances in nanotechnology and development of various types of nanomaterials have made it possible to engineer nanocomposite hydrogels, which are likely to attain greater popularity than polymer-based composite hydrogels. This can be due to the fact that the nanomaterials often possess highly specialized and favorable functions that are not commonly found in polymers.
2.4. Shape Memory and Self-Assembled Hydrogels
Shape memory hydrogels (SMHs) are a class of smart biomaterials with multiple shape capability. These hydrogels can vary their shapes when exposed to an external stimulus such as temperature or pH.[202–205] In this section, we review the fundamental mechanisms of shape memory hydrogel formation with their advantages and shortcomings. Particularly, we review the properties of hydrogels with stimuli-responsiveness, self-healing, and shape-memory characteristics. Finally, we point out applications of SMH in regenerative tissue-engineering strategies, particularly of those that are responsive to temperature changes with enhanced control-release and mechanical properties pertinent to biomedical applications and medical devices.[206]
Repeating units such as macrobiomolecules, polysaccharides, and nucleic acids make up the basic building blocks in living organisms. In their native environment, these repeating units are responsible for directing various cellular functions such as morphogenesis, proliferation, migration, and metabolic activity.[207–209] These building blocks are stable at a broad range of temperature gradients but undergo biophysical changes when they are exposed to their thermal transition temperatures (TT), which trigger the reversible transition from a firm state to a softer, rubbery form. This non-linear and specific behavior happens due to cumulative cooperative interactions of the repeating units (e.g., macrobiomolecules, polysaccharides, and nucleic acids). That is, under alteration of temperature, the intermolecular and intramolecular interactions undergo conformational and structural changes. This physiological observation has been introduced artificially in biomaterial design strategies to stimulate a response in natural or synthetic polymers.[210–212]
Among the variables studied to modify the response of polymers and hydrogels, thermal stimulation is the one that causes the largest conformational and structural responses. SMHs are created when these conformational responses are reversible; this reversibility occurs via supramolecular bonds formed by hydrogen bonds, van der Waals interactions, π–π interactions, or metal complexes. These interactions serve to build up network chains from non-covalent interactions between monomers, or non-covalently associate polymer chains together.[213–215] The non-covalent bonds can be combined in different ways to produce SMH networks with adaptable wettability,[216] swelling capability,[217] permeability,[218,219] and sol-gel transition properties.[220] Merging transient, reversible, non-covalent physical bonds with stable chemical bonds is a feature of SMHs that arises during its mechanism of formation.
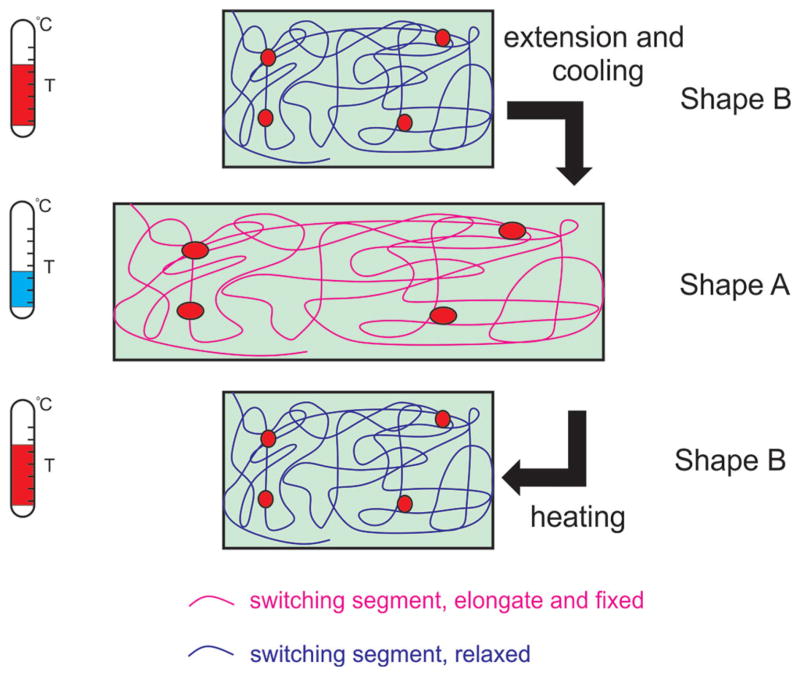
The mechanism of SMH formation can be described as follows. When reversible supramolecular crosslinks and permanent covalent crosslinks are heated to a critical TT, the physical crosslinks dissociate, the network deforms, and only the permanent covalent crosslinks are responsible for an elastic response.[221] Decreasing the temperature below TT causes association of the physical molecular crosslinks; thus, deforming the hydrogel and locking it. Raising the temperature again above TT dissociates the physical molecular crosslinks, releasing stored elastic energy of the permanent crosslinks, and restoring the hydrogel back to its original shape (Figure 6).[222]
Figure 6.
Molecular mechanism of temperature-responsive SMHs. The left panels represent the TT related to the switching of conformational structure. The right panel represents the conformational change of the SMH from shape B to shape A upon cooling, and back from shape A to shape B upon application of heat. The covalent chemical bonds are represented by the red dots and the physical non-covalent bonds by the entangled lines. Reproduced with permission.[222] Copyright 2013, American Chemical Society.
As mentioned above, one important advantage of SMHs is that they rely on the synergistic interplay between networks forming chemical and physical connections.[223] Chemical networks are connected together by covalent chemical bonds; physical networks on the other hand, are connected by transient non-covalent interactions (e.g., hydrophobic or electrostatic interactions). Both chemical and physical crosslinks have advantages and disadvantages. Chemical networks are strong and can be considered permanent. This feature is important for applications that require tough and highly connected networks, but may be detrimental if the hydrogel has to be reprocessed or recycled. On the other hand, the reversibility physical crosslinked networks greatly enhance reprocessing and recycling capabilities. Nonetheless, physical crosslinks are hard to customize, and their applications are specific to polyelectrolyte or biopolymer systems.[213] The combination of these chemical and physical crosslinks offers several advantages to SMHs. First, the reversibility of the physical crosslinked networks in SMHs enable their utilization as sensors,[224] actuators,[225–228] and controlled release platforms.[229] Second, SMHs exhibit self-healing properties;[221] when the network is ruptured, unassociated molecular bonds display a tendency to re-associate if they are brought into close proximity with each other. Third, the SMHs are customizable as they can form strong materials under favorable conditions, but easily de-coupled or de-crosslinked when exposed to temperatures below their TT. Forth, SMHs can be considered as composite hydrogels combining polymers and nanoparticles with specific “memory” characteristics. The detailed chemistry and characteristics of these composite hydrogels have been previously described in Section 2.3.
Temperature-responsive SMHs present promise as delivery vehicles of multiple bioactive molecules or growth factors due to their unique self-healing properties.[230–232] Once the SMH is stimulated, it can modify its shape and selectively attract or release a pre-determined set of biomolecules. For example, Ozadin-Ince et al. developed a coaxial nanofilm with a hydrogel core and a p(tert-butyl acrylate-co-diethylene glycol divinyl ether) shape memory shell to form temperature activated nanotubes using vapor phase deposition.[233] The temperature response of the coaxial nanofilm was then studied through release of encapsulated fluorescent dye. Burst release of fluorescent dye (fluorescein-5-thiosemicarbazide, FTSC) occurred due to the stress applied by the shape memory outer layer when activated at elevated temperatures.[233]
Temperature-responsive SMHs have also been used as smart hydrogels for cell and growth factor encapsulation. For example, Wang et al. developed a biodegradable, partially crosslinked alginate hydrogel with shape-memory properties at body temperature for minimally invasive surgical applications.[234] In this paper, 90% of recombinant insulin-like growth factor-1 (IGF-1) that was encapsulated in these hydrogels was released over several days in vitro, allowing skeletal muscle cell survival, proliferation, and migration within the scaffold over a 28-day period. In another study, a temperature-sensitive hydrogel composed of 2-acrylamido-2-methylpropanosulfonic acid (AMPS), NIPAm, and acrylamide (AAm) was synthesized to selectively bind proteins from serum samples.[224] The AMPS was chosen as a protein complex monomer based on the affinity of sulfonic acid groups to amide groups of bovine serum albumin (BSA). In addition, NIPAm was introduced as a temperature-sensitive material to bind and release BSA upon temperature changes. The shape-memory properties of the fabricated hydrogel combined with charge interactions enabled selectively reorganization and adsorption of BSA from a real serum sample.[224]
A near-infrared light responsive polymer-nanorod composite with a TT in the range of body temperature was also employed for the control-release of anti-cancerous drugs such as doxorubicin.[227] In vitro studies on these composite microspheres demonstrated a ~90% reduction in the activity of cancerous T6–17 cells when the release of doxorubicin was triggered from microspheres exposed to near-infrared light. Due to their high surface area, the microspheres facilitated cumulative release of drug;[227] however their applicability for clinical application such as cancer treatment remains to be proven.
Injectable Smart Hydrogels
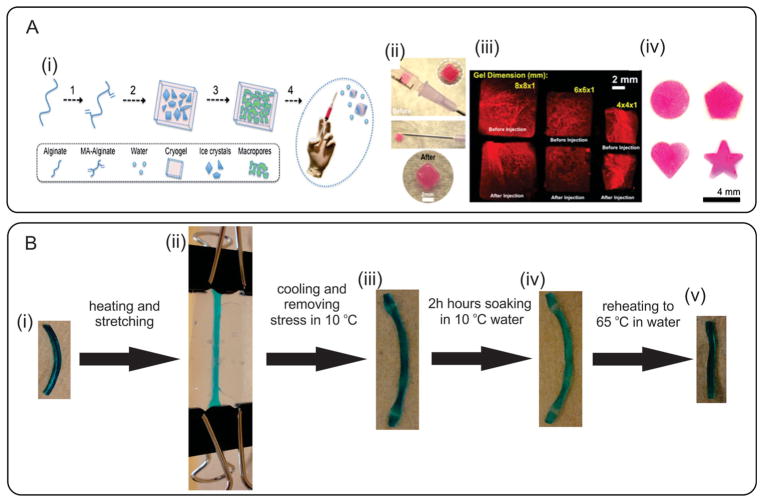
Recently, SMHs with tunable mechanical characteristics have shown promise as injectable hydrogels. For example, Bencherif et al. developed injectable macroporous alginate scaffolds with well-defined shape-memory properties (Figure 7Ai–iv).[235] These injectable hydrogels were highly compressible and could withstand reversible deformations up to 90% strain upon in vivo injection by a using conventional needle–syringe technique. Moreover, the fabricated hydrogels were employed as a delivery vehicle for biomolecules such as bovine serum albumin (BSA) as well as cells. Recently, shear-thinning hydrogels have been also proposed as injectable biomaterials at physiological conditions.[236,237] These hydrogels can be injected by application of shear stress during injection and quickly self-heal after removal of shear. In one study, shear-thinning hydrogels were made from small peptides and used to improve the immune response of H1N1 influenza vaccines.[238]
Figure 7.
Representative examples of SMHs. A) Formation of injectable SMHs: i) cryogelation process: 1) alginate is chemically modified to allow radical polymerization; 2) MA-alginate is added to a chemical initiator at −20 °C to allow ice crystal formation; 3) cryogelation takes place followed by thawing of ice crystals; and 4) conventional needle–syringe injection of preformed cryogels; ii) Photographs showing placement of a cryogel in a syringe (before injection) and hydrogel recovery (after injection); iii) MA-alginate gels with various sizes and shapes. Fluorescent square-shaped gels were syringe injected and showed complete geometric restoration after injection; iv) Cryogels prepared with different geometric shapes. Reproduced with permission.[235] Copyright 2012, National Academy of Sciences. B) Shape-memory behavior of highly stretchable hydrogels: i) The original length of the hydrogel is 26.3 mm; ii) The hydrogel heated in 65 °C water and stretched to 45.2 mm; iii) The shape of the hydrogel immediately after cooling to 10 °C; iv) After 24 h soaking in 10 °C water, the length of hydrogel decreased to 43.0 mm; v) After reheating hydrogel in 65 °C water without any external stress, the length recovered to 26.0 mm. Reproduced with permission.[239] Copyright 2009, American Chemical Society.
SMHs with Enhanced Mechanical Properties
Temperature-responsive SMHs nanocomposites with enhanced mechanical properties have also been investigated. For example, a reinforced SHM was produced by introducing reinforcing networks of cellulose whiskers (CCW) isolated from cotton into poly(vinyl acetate).[240] Upon immersion in artificial cerebrospinal fluid at 37 °C, the nanocomposites showed an adaptable change in elastic modulus, which made them suitable substrates for intracortical electrodes.[240]
In another study, highly stretchable SMHs were produced by covalently crosslinking quad-polymers of N,N-dimethylacrylamide (DMA), 2-(N-ethylperfluoro-octanesulfonamido) ethyl methacrylate (FOSM), hydroxyethyl acrylate, and 2-cinnamoyl-oxyethyl acrylate (Figure 7Bi–v).[239] These hybrid hydrogels, containing physical and covalent crosslinks, exhibited high mechanical strength, elasticity, extensibility, and fracture toughness, with potential applications for ligament or tendon repair.[241] Anthamatten et al. also synthesized copolymers of methyl acrylate (MA), methyl methacrylate (MMA), and iso-bornyl acrylate (IBoA) by adjusting their TT between 28 °C and 55 °C to form highly stretchable SMHs.[242] The developed materials demonstrated fully recoverable strains at 807% for a TT of 28 °C, at 663% for a TT of 37 °C, and at 553% for a TT of 55 °C. Due to their recoverable high-strain, these SMHs may be useful as cardiac patches or synthetic blood vessels.
Various temperature-responsive SMHs have been explored in the medical devices industry with applications in aneurysm treatment,[243] blood clot removal,[244] stents,[245,246] and dialysis needles.[247] Lately, there have been examples of temperature-responsive polymers with promising clinical applications in minimally invasive gastrointestinal surgery.[248] Minimally invasive surgeries can benefit from devices that can change their geometry or shape when placed inside the body. For example, Kratz et al. utilized thermoplastic temperature-responsive polymers for designing intelligent devices, which could be programmed by the clinician to individually adapt their shifting geometry and response temperature to the patients’ needs.[248] In this study, multiblock copolymers were synthesized from biocompatible poly(ω-pentadecalactone), as the hard backbone, and PCL as the crystallizable controlling unit. This polymer network was then used to develop an intelligent temperature-responsive drainage pigtail catheter for gastroenterology applications.[248]
Overall, SMHs hold promise as robust biomedical platforms that can be rationally designed since their structure is controlled by reversible crosslinks. Once created, they can be reversibly disassembled, as their hybrid structures could spontaneously reassemble in one, two or multiple steps. Fundamental research in the area of temperature-responsive SMHs aims to develop materials that can be either light-stimulated remotely or highly compressible to be injected via minimally invasive surgeries. SMHs could also envisioned to be triggered by pH[249] or by shear stress.[250] Therefore, in the future, there may be opportunities for the use of biodegradable and biocompatible medical devices made from SMHs to perform complex movements in the fields of cardiovascular surgery or gastroenterology.
3. Emerging Technologies for Engineering Hydrogel Constructs
Hydrogel constructs with controllable architectural features, at the micro- and nanoscale, have been shown to be useful in directing cell behavior, tissue formation and may be powerful tools for tissue engineering applications. Over the past decades, many research groups have tried to adopt techniques from various engineering fields for the fabrication of hydrogel constructs with controlled architecture.[251–256] However, factors such as low mechanical properties of hydrogels as well as the biocompatibility of the employed chemicals and the fabrication process, should be considered in utilization of these methods. Here, we review the emerging techniques, which can be used to control the geometrical features as well as the distribution of cells and biomolecules within fabricated hydrogel constructs.
3.1. Microfabrication
3.1.1. Photopatterning
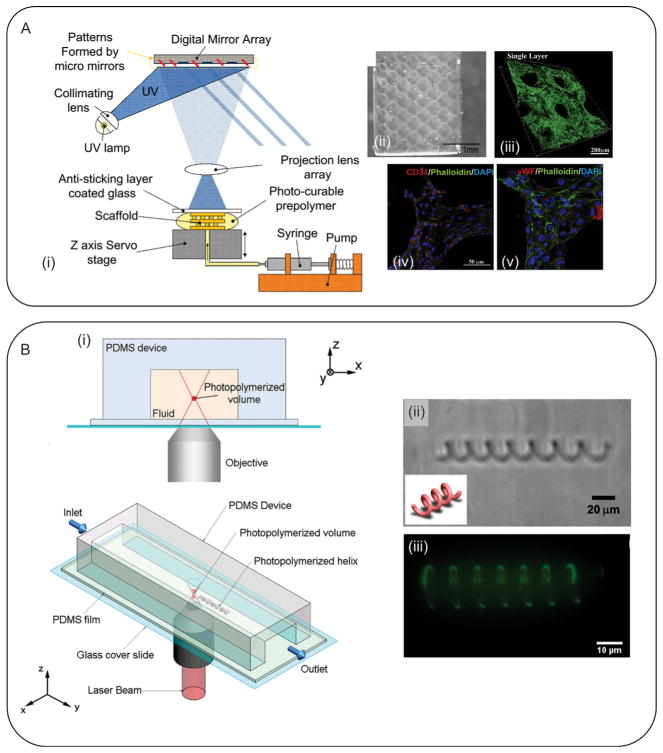
Photolithography, or photopatterning, is one of the techniques that has found popularity in the creation of microfabricated hydrogels.[252] This approach was originally developed for the fabrication of micron-sized features for micro electromechanical systems (MEMS) applications. In photopatterning, a mask is first created, which contains the pattern to be implemented. Certain areas of the mask are kept transparent allowing the light to pass and certain areas are opaque, blocking the light. Upon light irradiation, the hydrogel areas corresponding to the transparent regions of the mask are crosslinked to form the micropatterns, whereas the remaining uncrosslinked parts (under the opaque parts of the mask) are washed out in the washing step.[256] As expected, this photopatterning technique is only applicable to microfabricate photocrosslinkable hydrogels. Recently, researchers have conjugated acrylamide- or acrylate-based groups to prepolymer backbone, which make these hydrogels photocrosslinkable. Some examples of these hydrogels include PEG diacrylate (PEGDA),[257,258] PEG-dimethacrylates (PEGDM),[259] GelMA,[260] gelatin methacrylamid,[261] methacrylated HA,[262] and MeTro.[13,263] To form a polymerized network, a photoinitiator is usually used which forms free radicals upon light irradiation and initiates the polymerization reaction.
During the photopatterning process, the entire thickness of the hydrogel layer is usually crosslinked with no control over the crosslinking depth. The resolution of the features depends on the quality of the employed photomask and the illumination system. However, a resolution down to few microns is readily achievable. Another important property that can limit the dimensions of the microfabricated hydrogel constructs is their aspect ratio (i.e., the ratio between their height and width). Since hydrogels are not usually mechanically strong, high aspect ratio features may collapse.
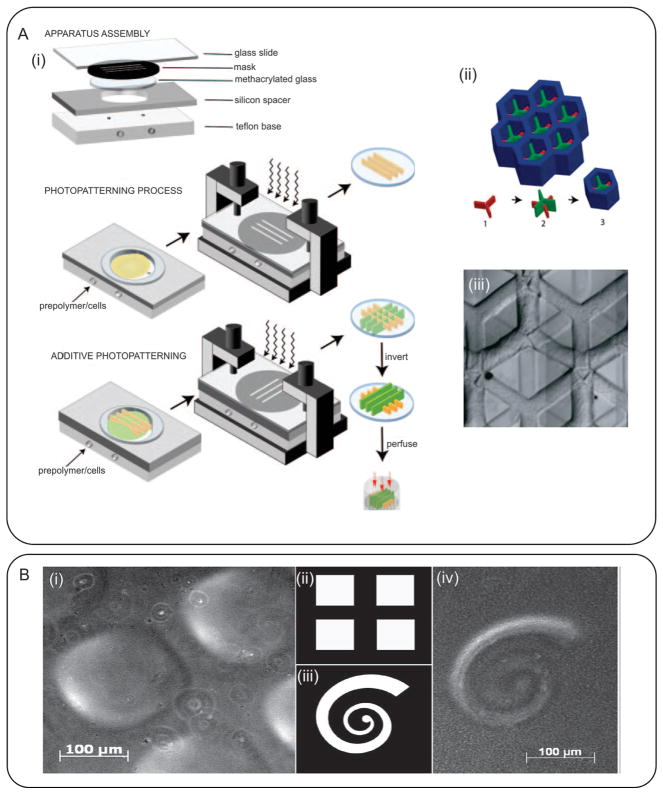
Photopatterning is a versatile technique and allows precise spatial control over the cellular microenvironment. In addition, it allows the fabrication of 3D cell-laden constructs or patterning of different cells through sequential photopatterning of hydrogels containing various cell types. In a notable study, Tsang et al. photopatterned complex 3D structures by successive crosslinking of PEGDA containing different chemicals using three different photomasks (Figure 8A).[257] In addition, they encapsulated primary hepatocytes as a model for liver-on-a-chip research. To control the height of the constructs, the height of the reservoir was changed by means of removable spacers. Cells stayed viable after the fabrication and showed a higher albumin and urea secretion in comparison with unpatterned cells.[257] In another study, Nichol et al. fabricated low aspect ratio HUVEC-laden GelMA constructs through UV crosslinking of the hydrogel and showed high cell viability after the fabrication process.[260] They characterized the hydrogel properties as a function of the UV exposure time and noticed that the mechanical properties were directly related to the UV exposure time and methacrylation degree. In a follow up study, Aubin et al. fabricated lines of cell-laden GelMA hydrogels with different widths and demonstrated that the widths of the patterned lines had a significant impact on the morphology of the cells.[128] Using a similar approach, biomolecules were patterned within photocrosslinkable hydrogels[264] or spatial patterns with different mechanical properties were created.[265] Such patterns can be used to direct cellular activity, migration, and differentiation, which will be further discussed in the following sections.
Figure 8.
Photopatterning of hydrogel constructs. A) Fabrication of cell-laden PEGDA constructs for liver tissue engineering; i,ii) schematic of the additive photopatterning process, in which different cell-laden hydrogels were photocrosslinked sequentially using different photomasks to create a 3D construct, iii) Micrograph image of a typical fabricated cell-laden hydrogel construct. Reproduced with permission.[257] Copyright 2007, The Federation of American Societies for Experimental Biology. B) Typical structures fabricated by degrading the surface of hydrogels; the hydrogel surface under 100 μm (i) squares and (iii) spiral masks were partially degraded and swell to form positive features observed in (i) and (iv). Reproduced with permission.[266] Copyright 2010, American Chemical Society.
Photodegradable hydrogels can also be used for the fabrication of microscale features within hydrogel constructs. For example, Wong et al. formed a photodegradable hydrogel through conjugating photodegradable ortho-nitrobenzyl (o-NB) groups to PEG.[132] They used photomasks to fabricate microchannels and microwells within the fabricated hydrogels. In addition, they could locally adjust the swelling properties of photodegradable hydrogels. As a result, the photodegraded area was swollen and formed a 3D microstructure (Figure 8B). In another study, Chiu et al. sandwiched a layer of photodegradable PEG-co(L-lactide) diacrylate (PEG-PLLA-DA) between layers of PEGDA.[266] A photomask was then used to selectively irradiate UV light to degrade a pattern within PEG-PLLA-DA layer. Upon the exposure of the construct to high pH conditions, the photopatterned regions were dissolved quickly and 3D microchannels were formed.[266] One of the limitations of this technique is the inability to encapsulate cells within the fabricated construct due to the use of a basic environment.
Photopatterning technique is easy-to-use and does not require sophisticated equipment. For photopatterning of hydrogel constructs, one needs a light source, a photo mask, and a photocrosslinkable hydrogel mixed with a photoinitiator. However, a key challenge is determining the suitable UV exposure time. Longer exposure times are found to affect cellular viability and activity or potentially might result in phenotype variation. Another limitation of this method is the fabrication of only planar constructs. Moreover, controlling cell distribution requires the use of multiple photomasks, which is challenging due to the need to align the photopatterns with the masks before each exposure.
3.1.2. Soft Lithography and Molding
Soft lithography and molding have been widely used in the creation of microfabricated hydrogel constructs.[267] Molds can be fabricated from various materials including plastics, polymers, and metals. However, elastomers such as polydimethylsiloxane (PDMS) and polymers such as poly(methyl methacrylate) (PMMA) have found more popularity.[256] This is due to their ease of microfabrication, biocompatibility, and hydrophobic surface property, which helps the detachment of crosslinked hydrogels from the mold. In addition, coating the mold with temperature responsive hydrogels, such as PNIPAm, have been shown to facilitate hydrogel removal from the mold.[268] Molding has been used for the fabrication of both physically and chemically crosslinkable hydrogels. The fabrication of 3D constructs using molding can be achieved by independent fabrication of multiple layers and their subsequent assembly.
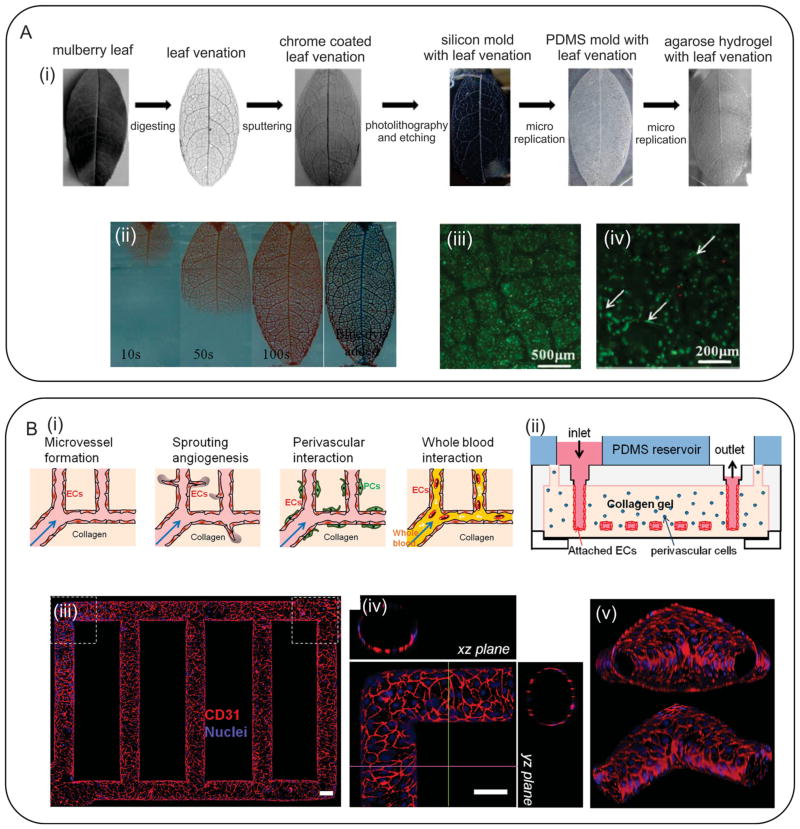
He et al. employed a microreplication method followed by a molding process to fabricate biomimetic perfusable microvascular networks.[269] They digested the soft tissue of a leaf and sputtered its veins with a layer of chrome. The sputtered leaf was used as a photomask in a soft lithography process to fabricate a negative PDMS mold, which was then utilized for the fabrication of agarose hydrogels (Figure 9A). Collagen solution containing ECs was injected within the patterned microchannels. In addition, HepG2 cells were encapsulated within the agarose hydrogel to form a liver-like structure containing perfusable channels. The in vitro results indicated that the presence of the microvascular network resulted in high cellular viability over 3 days of culture.[269] Zheng et al. fabricated microvessel networks within 3D collagen gel by using an injection technique (Figure 9B).[270] They encapsulated human umbilical arterial smooth muscle cells (HUASMCs) as well as human brain vascular pericytes (HBVPCs) within the collagen layer. The channels were also seeded with HUVECs to form functional microvessels. The hydrogel constructs were then cultured for 14 days to study the effects of cell-cell interactions on angiogenesis and sprouting. They observed that HUVECs tend to form cylindrical lumens within the channels. Our groups also used micromolding technique to form microchannels within PDMS samples and used them as molds for fabricating grooved MeTro and GelMA hydrogels.[56] The fabricated constructs were then used to align CMs within the fabricated micropatterns.
Figure 9.
Micromolding of hydrogel constructs. A) Fabrication of a biomimetic microvascular network within agarose hydrogel using a soft lithography process; i) the soft tissue of a leaf was digested and its veins were sputtered with a chrome layer. The sputtered leaf was used as a photomask in soft lithography. The fabricated PDMS mold was used for fabrication of agarose hydrogel containing microvessels; ii) injection of dye within the fabricated agarose samples; iii) HepG2 cell-laden agarose with perfusable microchannels as a model for liver tissue engineering; iv) the HUVECs seeded within the microchannels formed capillaries. Reproduced with permission.[269] Copyright 2013, John Wiley & Sons, Inc. B) Pre-vascularized collagen hydrogel fabricated using micromolding; i) Schematics showing different scenarios that were studied including morphology and barrier function of endothelium, endothelial sprouting, perivascular association, and blood perfusion for fabrication; ii) schematic of the employed microfluidic system; (iii)–(v) confocal images from endothelialized microfluidic, showing the formation of circular vessels. Reproduced with permission.[270] Copyright 2012, National Academy of Sciences.
Another emerging application of the molding techniques is the fabrication of 3D pre-vascularized hydrogel constructs.[271] In this method, a 3D mold of sacrificial fibers is first fabricated and subsequently covered by a hydrogel layer. The sacrificial fibers are then removed from the construct to engineer vascular network.[271] Researchers have used various sacrificial fibers including glass carbohydrate, shellac, gelatin, and metallic needles.[272–275] For example, Golden and Tien formed a gelatin mesh using micromolding and then embedded it as a mold within collagen and fibrin hydrogels.[275] Gelatin mold was then melted away leaving behind perfusable channels, which were seeded with ECs to form functional microvessels.
Micromolding and soft lithography are robust and easy-to-use technologies. Complex structures can be created through layer-by-layer fabrication followed by an assembly step. Molding is also rapid, biocompatible, and scalable. However, these techniques are limited to planar structures and the fabrication of 3D biomimetic geometries such as vessels or veins is challenging. To address this limitation, combination of molding and rapid prototyping strategies could be the next step for the formation of microfabricated complex 3D hydrogels. Moreover, hydrogels that crosslink chemically or through ion transfer such as alginate deform during the molding process; thus strategies that preserve the shape of the hydrogel during its crosslinking are essential.
3.1.3. Rapid Prototyping
Rapid prototyping is an additive-based fabrication technique in which materials or energy is sequentially delivered to form a construct. Rapid prototyping systems are automated and the construct is previously designed using a CAD software or through reconstruction of a geometry from images (MRI, CT, and X-ray).[276] The CAD files are then converted to a standard tessellation language (STL) format, which describes the actual surfaces by raw unstructured triangulated surfaces. The STL files are then converted to sliced models, which are fabricated using the machine through an additive process.[253] In this section, latest advances in the use of laser-based systems will be discussed. The nozzle-based and bioprinting devices will be introduced in the following sections.
In laser-based systems, the beam can only crosslink the hydrogel on the focal plane and the rest of the hydrogel will stay uncrosslinked. As expected, this technique is only applicable to photocrosslinkable hydrogels. The major difference of laser based systems from the mask-based systems is that in the former no physical mask is used and light illumination is not powerful enough to crosslink the complete depth of the hydrogel.[277]
Stereolithography (SLA) is one of the oldest laser-based rapid prototyping strategies.[278] In this approach, 3D constructs are fabricated in a layer-by-layer fashion.[261] The machine follows the sliced model and hydrogel on the focal plane is crosslinked through irradiation in a point by point format. Once all the points on a layer are crosslinked, the focal plane changes and the next layer is fabricated. This method is used to fabricate complex 3D constructs with high resolution. Since the whole area should be swept for each focal plane, SLA is a slow process, which is not favorable for cell encapsulation.
To increase the speed of the fabrication process, researchers have developed a digital light projection (DLP) technology, in which the whole pattern on the focal plane is crosslinked simultaneously.[279] In this method, a digital micro-mirror device or a liquid-crystal display (LCD) can be used to project a dynamic pattern.[280] Gauvin et al. employed a digital micro array system to pattern 3D scaffolds from GelMA in a layer-by-layer fashion (Figure 10A).[281] By programming the system, they were able to control the microstructure of the fabricated scaffolds and consequently tune their mechanical properties. In vitro studies showed that the fabricated scaffolds with 1 mm thickness supported the viability and proliferation of HUVECs during 7 days of culture.
Figure 10.
Fabrication of hydrogel constructs using rapid prototyping. A) SLA with digital mirror device for fabrication of GelMA constructs; i) Schematic representing SLA device with was controlled by a CAD software; (ii) a typical scaffold fabricated using the device; iii) the fabricated scaffold seeded with HUVECs-GFP; iv,v) staining showing the spreading of HUVECs on the surface of the fabricated scaffolds; Reproduced with permission.[281] Copyright 2012, Elsevier B.V. B) Two-photon continuous flow lithography; i) schematic of the employed the device; prepolymer was flown through the channel while a 2PP system was used for crosslinking of prepolymer. ii,iii) bright field and confocal images of a typical fabricated helix-shaped PEGDA. Reproduced with permission.[282] Copyright 2012, John Wiley & Sons, Inc.
Another alternative to SLA is the two photon polymerization (2PP), which is an emerging technology for fabricating 3D hydrogel structures.[283] In 2PP, two laser pulses with high wavelengths are irradiated, which act as a single pulse with a low wavelength at the intersection point to excite the PI and crosslink the hydrogel.[252] As a result, in contrast with SLA systems, which possess a focal plane, 2PP systems have a focal spot. In general, the accuracy of 2PP is higher than SLA.[284] Moreover, 2PP systems can be used to fabricate 3D biomimetic structures such as microvessels, which mimic the vasculature in native tissues. Recently, Laza et al. developed an advanced form of 2PP combined with continuous flow of the prepolymers.[282] Using this approach, they were able to fabricate 3D objects such as helix-shaped constructs from PEGDA gels[282] (Figure 10B). In another study, Ovsianikov et al. demonstrated the capability of 2PP systems for grafting biomolecules to PEG-based hydrogels.[285] The absorption of laser energy by the 2,6-bis(4-azidobenzylidene)-4-methylcyclohexanone (BAC-M) triggered chemical reactions, which resulted in the grafting of the targeted molecule to the backbone polymer. Their technique enabled formation of high resolution (4 μm) complex 3D structures. Moreover, the intensity of the emitted laser and the scanning speed had a direct relationship with the concentration of grafted molecules.[285] A multiphoton system was also used to create patterns of various biomolecules in an aga-rose-based hydrogel modified with coumarin-caged thiols.[264] Neural precursor cells were then seeded on the surface of hydrogels containing gradients of various growth factors to monitor cellular migration over 14 days of culture. It was shown that cells responded to the 3D patterns to a depth of 85 μm.[264]
Laser-based rapid prototyping devices enable the fabrication of 3D constructs in an automated fashion. Moreover, these systems offer high precision and resolution in comparison with photomask-based counterparts. However, the major challenge is their high capital and operating costs, which has limited their use to a few research labs. Moreover, operating systems based on SLA and 2PP are relatively slow, which makes cellular encapsulation challenging.
3.2. Bioprinting
Despite major advances, clinically relevant 3D engineered tissues have not been fabricated, due to our inability to recreate tissue architecture and function.[286,287] Thus, the development of technologies that enable the fabrication of complex multicellular tissue constructs can aid in addressing this challenge.[286] Overall, bioprinting of hydrogels at precise 3D architectural arrangements represents a powerful approach for engineering biomimetic tissue constructs.[288]
Bioprinting techniques originate from additive manufacturing approach, where the sequential deposition of solid layers allows for the precise development of complex structures. In engineering bioprinted tissues, cells can be either embedded within biologically relevant hydrogels or printed free of scaffold support.[288] Bioprinting techniques enable rapid printing of micro- and macro-scale 3D structures with high cell viability.[288–291] A wide variety of bioprinting systems have been developed to engineer and control the design and architectures of 3D hydrogels.[289,290,292,293] In the following sections, we will briefly describe relevant bioprinting methods and highlight potential developments in the future prospects for bioprinting in regenerative medicine.
3.2.1. Inkjet Bioprinting
Efforts to utilize inkjet printers as tools to dispense cells and biological materials were initially reported in the 1980s. In an early study, Klebe used a common Hewlett-Packard desktop printer to deposit both collagen and fibronectin suspensions with cells, thus forming simplified tissue analogs.[294] In inkjet bioprinting, a container, analog to ink cartridges, dispenses drops in the range of 1 to 100 pl via heating and vaporizing, while either a bubble or a piezoelectric actuator forces out the liquid drop.[293] In these systems, pressure waves are generated by a pulse behind an orifice; subsequently, droplet ejection occurs when the wave overcomes the liquid-vapor interfacial tension. These systems were primarily developed to be used with low viscosity and well-dispersed inks, such as cell suspensions. To print high viscosity solutions, such as hydrogels, recently, researchers have intensively attempted to modify inkjet bioprinting systems. One improvement is the development of nozzle-free acoustic ejectors, whereby an X-Y-Z stage is controlled by a computer and synchronized with a pulse generator programmed to build 3D layers.[293] The use of a synchronized opening and closing of a micrometer sized valve orifice in these bioprinters prevents clogging during droplet generation. These systems were successfully utilized to study stem cell genomics, via a method called drop-on-demand single cell isolation.[295] Similarly, an inkjet bioprinting method was used to replace a manual pipetting step for the formation of uniform-sized embryoid bodies.[296] In one study, an array of fibrin gels containing gradients of immobilized growth factors were bioprinted to guide stem cells migration over extended time (>1 day) and length scales (>1 mm).[297] Fibronectin and gelatin gels were also printed by using an inkjet bioprinter to form 3D tissue arrays.[298] In this study, 440 microarrays of simplified 3D structures were fabricated retaining micrometer-sized multilayers with different cell types. This platform was utilized to study the role of co-cultures of HUVECs and liver hepatocellular cells (HepG2) on liver-specific biomarkers, as well as on drug metabolism (Figure 11A). The combination of bioprinting with other widespread fabrication technologies, such as electrospinning, has also attracted significant attention. For example, in a recent report, an inkjet printer set-up was mounted directly adjacent to an electrospinning device, allowing for the alternate fabrication of a layer of electrospun PCL and another layer of chondrocyte-laden fibrin/collagen hydrogel.[299] This integrated platform supported the formation of cartilage tissue both in vivo and in vitro.[299]
Figure 11.
Bioprinting of hydrogels for tissue engineering. A) Inkjet bioprinting approach: i) schematic illustration of a layer-by-layer printing of single cells and proteins; ii) inkjet bioprinted 3D human-tissue chips. Reproduced with permission.[298] Copyright 2011, John Wiley and Sons. B) Laser bioprinting approach: i) Schematic illustration of a laser forward transfer technique (LIFT); ii) A bioprinted cardiac-patch micropatterned with human MSC (PKH26-green) and HUVECs (PECAM-1-red). Reproduced with permission.[87] Copyright 2011, Elsevier. C) Direct-write bioprinting: i) Confocal image of microperiodical pHEMA scaffold fabricated via direct-write bioprinting method; ii) Software reconstruction of bioprinted scaffolds (red) and primary rat hippocampal neurons (green); iii) Software reconstruction of cells alone, demonstrating cell processes guided by scaffold architecture. Reproduced with permission.[300] Copyright 2011, John Wiley and Sons. D) Example of bioprinting integrated with bioelectronics for engineering of whole body parts: i) a bioprinted alginate hydrogel scaffold integrated with an electrically conductive silver nanoparticle (AgNP)-infused inductive coil antenna; ii) Scaffolds seeded with chondrocytes forming an ear-like construct with functional hearing capabilities. Reproduced with permission.[301] Copyright 2011, American Chemical Society.
Generally, inkjet printers are widely used in the fabrication of many devices, from light-emitting diodes to full-color high-resolution flat panel displays.[302] An interesting field that has emerged recently is the utilization of inkjet printers as tools for fabricating biochemical sensing devices. In the fabrication of these biosensors, inkjet printers are utilized to form electrically conducting traces (e.g., electrode, and electric contact) or sensing layers (e.g., polymer film, enzyme or antibody spot, and colorimetric reagent.). All these technologies are profited from developments achieved in printed electronics.[303] The integration of printed electronic with elastomeric stretchable organic fibers can give rise to exciting possibilities in the field of implantable hydrogel-based electronics. We foresee that this may lead to important advances in regenerative medicine towards bionic implants integrating bioprinting, biosensors and advanced biomaterials.[304,305]
3.2.2. Laser-Assisted Bioprinting
In common laser-guided bioprinters, a laser-induced forward-transfer (LIFT) technique transfers a biological material from a source film onto a non-absorbing surface in close proximity to the film.[292,306] Depending on the rheological properties of liquid films and the thickness of the metallic absorbing layer, printing is obtained through a jet formation, occurring above a laser energy limit. The requirement for a high-energy transfer system, however, has been associated with a decrease in cell survival after deposition.[307] One of the recent potential applications of LIFT technique is the fabrication of cardiac-patches made of polyester urethane urea (PEUU) treated with Matrigel.[308] During the fabrication of these patches, a laser printer was utilized to pattern a co-culture of HUVECs and human MSCs at precise regions (Figure 11B). In vitro results demonstrated the possibility of creating vascular micropatterns, which could be arranged in parallel, thus leading to organized cell-cell interactions and formation of a vascular-like network. After implantation in infarcted rats, the bioprinted patches showed significant improvements in cardiac function when compared to a non-printed control.[308] Biological laser printing (BioLP), derived from the LIFT technique, was also utilized to study the formation of microvascular systems in hydrogel treated substrates.[309] In this approach, a laser bioprinter with improved resolution (approximately 30 μm) was utilized to deposit HUVECs on the surface of Matrigel loaded PLGA based biopapers.[309] Results demonstrated a high fidelity of the cells onto the pre-defined patterns. Similarly, in a recent study, 3D multilayered constructs made of fibroblasts and keratinocytes encapsulated in alginate hydrogels were bioprinted. The fabricated constructs demonstrated high structural biomimicry resembling the microarchitecture of human skin.[310,311]
3.2.3. Direct-Write Bioprinting
Direct-write bioprinting has been also widely utilized in the biomedical field given the inexpensive character of manufacturing devices and the relative straightforward principle behind the associated fabrication methods. Direct-write printing generally functions with pressure-driven extrusion methods (via air or mechanical pressure) of pre-cured hydrogels, cell-laden hydrogels or mature cell-aggregate filaments.[290] In principle, a syringe, generally connected to a pump and mounted onto an X-Y-Z stage, allows for controlled deposition of 3D layers or lines containing cells or bioactive materials. To form vasculature, direct-write printers were also integrated with a thin capillary tube with a metallic piston in its center. The piston assisted with the aspiration of a pre-polymer gel and then mechanically extruded it after polymerization.[312] This technique was also employed to bioprint scaffold-free cell aggregates.[313] Due to their ability in prototyping micro- to macro-scale tissue constructs, direct-write printers have been used as a promising alternative to laser and inkjet methods, which tend to dispense much lower volumes of material at a time.
Direct-write 3D printers have also promoted exciting alternatives in tissue engineering. For instance, by controlling the 3D microarchitectures of hydrogels constituted of HEMA absorbed with polylysine, researchers have been able to guide proliferation and alignment of primary rat hippocampal neurons depending on scaffold architecture[300] (Figure 11C). In another example, 3D silk/hydroxyapatite scaffolds were bioprinted with gradient porosity ranging from 200 μm to 750 μm. The scaffolds were then co-cultured with human MSCs and ECs, leading to new tissue formation and bone remodeling.[314] Photopolymerizable thermosensitive p(HPMAm-lactate)-PEG hydrogels were also developed to fabricate complex 3D constructs with tunable mechanical properties.[315]
Direct write printers have also offered exciting prospects in the fabrication of functional and perfusable microvascular networks;[271,316–319] these approaches will be discussed in greater detail in Section 4.1. Similar to the combination of inkjet printing of conductive inks described above, direct write printers have also brought about yet another level of complexity and functionality towards the integration of bionic parts with tissue engineering constructs. This method was recently utilized to fabricate electric antennas on 3D surfaces,[320] optical waveguides,[321] stretchable microelectrodes,[322,323] and conductive grids.[324] Direct write 3D printing technologies has potential to be used for the fabrication of body parts and electronic devices, which brings forth outstanding possibilities in the field of biomedical engineering. Whole 3D body parts integrated with electrically conductive materials have been recently engineered.[301] In this recent work, Mannoor et al. reported the 3D fabrication via direct-write printing of a cell-seeded alginate hydrogel in the anatomic geometry of a human ear integrated to an cochlea shaped electrically silver-nanoparticle infused silicone antenna.[301]
3.3. Microfluidics
Microfluidic technologies are being increasingly applied to various tissue engineering and biological applications. For instance, microfluidic systems have been used for single cell analysis,[325,326] engineering tissue-like structures,[263,327] and fabricating functional tissues for drug screening.[328] Microfluidic technologies offer an attractive strategy, in particular, for creating functional microengineered hydrogels (microgels and microfibers) with 3D morphologies and configurable chemistries. As discussed in the previous sections, these microgels may be used as tissue building blocks, which can be assembled to form complex 3D tissue engineered constructs.[329] Microfluidic technologies have been used to fabricate microgels in the shape of spheres,[330] disks,[331] hemispheres,[332] core–shell structures,[332] Janus-like particles,[333] and fibers[334] by using flow focusing,[335] T-junction geometries,[336] and co-flowing of laminar streams.[337] In addition, microfluidic techniques can be combined with projection-photolithography, which was previously used for particle manipulation,[338,339] to create microgels with more complex geometries and tunable chemical compositions.
This section aims to provide an overview of the current state-of-the-art in the use of various microfluidic systems for the fabrication of engineered microgels and microfibers. We mainly focus on emulsion-based techniques with particular emphasis on co-flow, flow focusing, flow lithography systems, and fiber-spinning techniques. We discuss the advantages and disadvantages of each method and highlight their emerging applications in tissue engineering and regenerative medicine.
3.3.1. Emulsion-Based Systems
Emulsion is a process whereby small particles are formed by dispersing one liquid into an immiscible solution. Microfluidic systems can be used to produce highly monodisperse emulsions by precisely fabricating one drop at a time. The size and chemical composition of the resulting droplets can be manipulated by controlling the flow rates and concentrations of different chemicals in each stream. Microfluidic technologies also enable the fabrication of double, and higher-order emulsions with an exquisite degree of control. Single and multi emulsions are generally produced by co-flow, flow focusing, or a combination of two systems (Figure 12A).[337,340]. These systems can be simply created by using glass capillaries[337] or micromachining techniques such as micromolding in PDMS.[341] By combining hydrodynamics and in situ crosslinking (photo or chemical), microgels can be generated in a continuous and cost-effective manner.
Figure 12.
Emulsion-based systems for creating microengineered hydrogels (microgels) and their applications in tissue engineering. A) Microfluidic methods for creating microgels by using co-flow, flow focusing, and their combination. Reproduced with permission.[342] Copyright 2007, Cambridge University Press. B) Application of cell-loaded microgels for rapid fabrication of tissue constructs. Collagen microgels were fabricated using the flow focusing method and subsequently were seeded with mammalian cells. NIH 3T3-seeded hydrogel beads were tipped into a doll-shaped PDMS mold to a large-scale 3D tissue construct. Reproduced with permission.[343] Copyright 2011, John Wiley and Sons. C) Fabrication of Janus-like particles using a microfluidic flow focusing system. The chemical properties of the particles are adjusted by tuning the flow rate of the two streams (M1 and M2) that merge at the focusing point. Reproduced with permission.[332,344] Copyright 2005, 2008, John Wiley and Sons.
Microgels can be formed from hydrogel materials using emulsion-based microfluidic techniques for tissue engineering applications. In one study, Matsunaga et al. used the flow focusing method to create monodispersed collagen spherical microgels with tunable diameters in the range of 50 to 300 μm.[343] They cultured various types of cells including NIH 3T3 mouse fibroblast cells, HepG2 cells, HUVECs, primary neurons, primary rat hepatocytes, and MIN6 pancreatic cells on the collagen beads and showed that the cells attached to the surface of the bead in less than 2 hours. To form a macroscopic 3D tissue construct, a modular approach was used to induce NIH 3T3-seeded beads to morph into a body-shaped figure inside a PDMS mold (Figure 12B). Results showed that the beads formed strong constructs after 17 hours, indicating that robust tissue constructs can be formed using this rapid fabrication method.[343]
Microfluidic technologies also enable the fabrication of microparticles containing multiple biochemical functions (Figure 12C). For example, Shepherd et al. devised a microfluidic system to form microgels of varying shapes and compositions.[345] They first created droplets with different chemical compositions by focusing two laminar streams in a continuous oil phase sheath-flow. These droplets were subsequently crosslinked in situ to form Janus-like microgels. These particles are of interest in tissue engineering as they are asymmetric and thus can be used to better mimic the mechanical and chemical anisotropy of native tissues.[345]
While creating particles using emulsion-based microfluidics is a promising approach for creating tissue constructs, there are still several challenges that remain to be addressed. Since surface tension effects play a key role in particle formation, only particles with limited shapes can be created. Moreover, using emulsion-based microfluidics requires phase-separating chemistries (immiscible fluids) and stabilization techniques that are generally toxic to the cells. One possible approach could be the formation of particles using coaxial flow of two miscible fluids in microchannels under dripping regime.[346] In this method, droplets are formed due to the break-up of a liquid jet formed by a coaxial stream of core fluid in a sheath flow. These droplets can be subsequently crosslinked downstream of the flow using photopolymerization. Moreover, to prevent the formation of harmful radicals during the crosslinking process, other methods such as radical- and catalyst-free, bio-orthogonal click reactions can be used.[347]
3.3.2. Flow Lithography
Flow lithography combines the advantages of photolithography and microfluidics to create microgels with complex morphologies and chemical compositions (Figure 13).[348] These methods are divided into continuous flow lithography (CFL) and stop flow lithography (SFL). In CFL, microgels are formed by exposing a stream of flowing acrylic oligomer in a PDMS microchannel to pulses of mask-defined UV light. Portions of the oligomer that are exposed to the UV light are crosslinked almost instantaneously to form well-defined solid structures. These structures are then carried away by the continuously flowing polymer precursor liquid that surrounds them.
Figure 13.
Flow lithography for in situ fabrication of cell-laden microgels with complex morphological and chemical properties. (A) Schematic of the flow lithography. Reproduced with permission.[349] Copyright 2008, The Royal Society of Chemistry. B–G) Highly complex microgels fabricated using the flow lithography technique. H) Janus-like hydrogels with spatially controlled chemical properties. Reproduced with permission.[348] Copyright 2006, Nature Publishing Group. I) Cells encapsulated in a microgel, green cells are live and red cells are dead. Reproduced with permission.[349] Copyright 2008, The Royal Society of Chemistry.
CFL has certain advantages over the emulsion-based approach. This process only requires the use of one phase, which eliminates the challenges associated with optimizing device surface chemistry required in two-phase flows. Moreover, Janus-like particles with different chemical properties are fabricated by using polymeric precursors with varying chemical properties. In addition, CFL enables high throughput fabrication of microgels with complex morphologies, which are formed by projecting different shapes with rates as high as 100 particles per second[331] (Figure 13B–H). However, the use of CFL for tissue engineering is limited, as it requires high concentrations of monomer or photoinitiator, which are toxic to cells. Moreover, CFL is restricted to low flow rates because the particles are synthesized in flow and exposed to finite pulses of UV light. To address these limitations, Panda et al. developed a SFL method to synthesize large numbers of cell encapsulated hydrogels in a continuous format (Figure 13A).[349] The SFL process included three steps: i) stopping the liquid flow, ii) polymerizing the patterned solution, and iii) flowing the particles out of the device.[350] Using SFL process, mouse fibroblast cells were encapsulated in PEGDA microgels in about 1 second, which corresponded to 1000 particles per hour.[349] The swelling behavior, mechanical properties, and degradation rate of microgels fabricated by SFL were tailored by adjusting the UV exposure.[351] Similarly, Hwang et al. used SFL to create microgels with tunable size, shape, and degradation profile.[351]
Flow lithography is a powerful technique that enables high precision fabrication of microparticles with excellent control over their geometry, chemical, mechanical, and degradation properties in a high throughput manner. However, fabrication of 3D microgels in one step is still a challenge. Moreover, there are only few examples where the application of flow lithography in the fabrications of cell encapsulated microgels are shown. This may be attributed to the harsh process of particle fabrication, in which high concentrations of photoinitiator or prepolymer are used. Washing the microgels after fabrication and subsequent cell seeding can be an alternative approach to enhance the application of this technique. Flow lithography combined with assembly techniques such as random assembly,[352] manual manipulation,[353] and microfluidic-directed assembly[354] hold great promise to create large tissue constructs with tunable mechanical, biochemical, and morphological properties.
3.3.3. Microfluidic Fiber Spinning
Microfluidics offers unique characteristics such as precise control of flow and chemical compositions in small scale that enables creating hydrogel fibers with controlled geometrical and biological features.[355] Microfluidic fiber spinning includes creating two or more parallel streams of prepolymer solutions and a sheath flow in a microchannel, and the formation of the hydrogel down the stream by chemical, optical, or thermal crosslinking (Figure 14A–C). With this approach, single or multi layer fibers with precise control on their shape, size, cell distribution, and chemical composition can be fabricated by in situ polymerization of the prepolymer solutions.[334] The most attractive polymer used for creating microfibers is alginate, which can be chemically crosslinked by using Ca+2 or Ba+2. Diffusion of Ca+2 ions in the sheath flow into the alginate central stream along the flow direction crosslinks the prepolymer and forms calcium alginate fibers at the exit of the microfluidic device. Other materials including gelatin/hydroxyphenylpropionic acid (Gtn-HPA) and NIPAm were also used to create fibers in a microfluidic platform.[356] The main drawback of these polymers is that they do not contain components of natural ECM, such as RGD moieties for ligand binding, which are essential for cell-cell connections and 3D cellular interactions. To address this challenge, core-shell fibers with ECM proteins in the core and alginate hydrogel in the shell have been fabricated using a double co-axial laminar flow microfluidic device (Figure 14C).[357] With this configuration, a stream of ECM proteins was created in a core flow, which was surrounded by sodium alginate prepolymer and a sheath calcium chloride flow. The calcium alginate shell prevented the ECM proteins to diffuse out during their gelation. The fabricated fibers were shown to reconstitute intrinsic morphologies and functions of living tissues.[357]
Figure 14.
Fiber-based methods for creating functional tissue constructs. A) Creating grooved fibers by using a microfluidic grooved spinneret. Reproduced with permission.[334] Copyright 2011, Nature Publishing Group. B) Microfluidic devices that enables creating fibers with spatially controlled topography and chemical composition. Reproduced with permission.[334] Copyright 2011, Nature Publishing Group. C) Microfluidic fabrication of functional fibers. A core-shell fiber is formed in a coaxial flow microfluidic device. Cells are encapsulated in the core made from ECM proteins and the shell is made from Ca-alginate. Functional fibers can be assembled to form complex tissue constructs using a miniaturized weaving loom. Reproduced with permission.[357] Copyright 2013, Nature Publishing Group.
An important benefit of microfluidic spinning is that the prepolymer stream is confined within the channel and maintains its geometry during the crosslinking process (Figure 14A,B). As a result, the core prepolymer stream preserves the shape of the nozzle and crosslinks in forms of grooved or non-circular fibers. For example, grooved alginate fibers were fabricated by using microfluidic spinning to direct the orientation of neurons.[334] Hollow alginate fibers were also formed by creating three streams of cell medium, sodium alginate, and CaCl2 as core, middle, and sheath flows.[358] These hollow fibers can be potentially used for the vascularization of large tissue constructs.
To create fibers with heterogeneous physiochemical properties, a more sophisticated microfluidic device equipped with controlled pneumatic valves mimicking silk spinning in spiders were designed.[334] With this configuration, microfibers were fabricated with spatially controlled topographies and chemical compositions. In particular, a core shell fiber with hepatocytes in the center and fibroblasts in the outer layer was formed for liver tissue engineering applications.[334]
Fiber fabrication using microfluidic spinning holds a great promise as it enables continuous fabrication of fibers with tunable morphological, structural, and chemical features. Various hydrogels including chemically and optically crosslinkable materials can be synthesized within a microchannel in a coaxial geometry. Moreover, this technology enables the incorporation of cells and chemicals in single- and multi-layer fibers during the manufacturing process. However, the biopolymeric fibers generated by microfluidic spinning are generally weak, which limits their handling and manipulation.[359]
3.4. Wetspinning
Fibers are building blocks of many fibrous constructs used in the fabrication of artificial tissues for regenerative medicine. These fibrous constructs are highly porous and offer high surface area to volume ratio.[360,361] Hydrogel fibers are usually manufactured using wetspinning and microfluidic spinning.[359] Microfluidic spinning has been discussed in Section 3.4. Here, we will highlight recent advancements in the wet-spinning of hydrogel fibers. In tissue engineering, fibers have been assembled into open pore fibrous scaffolds and tissue-like constructs using various techniques such as weaving, knitting, and braiding.[359,362]
In the wetspinning process, hydrogel fibers are formed by injecting a pre-polymer solution into one or multiple coagulation baths. Injection can be performed manually,[363] using a syringe pump,[364] or by using pressurized air.[365] Wetspun fibers have been produced from various biocompatible materials including alginate, collagen/alginate composite,[366] collagen,[363] chitosan,[367] starch/PCL composite,[368] and chitosan/tripolyphosphate composite[369] for engineering various tissues such as bone,[364] neural,[370] and cardiac.[371]
Fibrous scaffolds can be formed from wetspun fibers by random deposition on a substrate,[366] rolling on a spool,[372] patterning in an ordered arrangement using a bioplotter.[372] Mechanical properties of wetspun fibers can be adjusted by adding reinforcing materials such as CNTs and GO.[279,371] For example, adding 4 wt% GO to the sodium alginate prepolymer enhanced the maximum tensile strength and Young’s modulus of the resultant fibers by several folds from 0.3 and 1.9 GPa to 0.6 and 4.3 GPa, respectively.[279] Adding CNTs and GO also improved the electrical conductivity of the fibers.
Wetspinning technique allows cellular encapsulation within the 3D structure of some polymers (e.g., alginate, chitosan). For example, cell-laden Ca-alginate hydrogels were fabricated by using a three-needle pressure-assisted system.[365] However, long exposure of the cells to toxic crosslinkers limits the applicability of wetspinning technique for the fabrication of cell-laden fibers from a wide range of polymers. In general, wet-spinning is an easy-to-use process and does not require sophisticated equipment. However, this method is usually applicable to hydrogels, which are crosslinked chemically or physically through ion transfer. Another limitation of wetspinning is that unlike microfluidic spinning, the prepolymer is not confined. Thus, the geometry of the prepolymer stream may change after its injection into the coagulation bath.
3.5. Electrospinning
Electrospinning is based on creating nanofibers by drawing a viscoelastic polymer from a spinneret and depositing them on a collector plate. This technology has been extensively used to fabricate fibrous scaffolds. Electrospinning is a one step process, in which fibers are drawn and assembled simultaneously. Electrospun scaffolds with randomly distributed nanofibers can be made from hydrogels or their mixture with synthetic polymers for different applications such as cardiac graft,[373] wound dressing,[374] cartilage and bone tissue engineering.[375] To mimic the anisotropic microstructure and mechanical properties of native tissues, scaffolds with aligned fibers have been also fabricated using a rotating disk or mandrel.[376] For example, our group has recently shown that fiber alignment in an electrospun PGS/gelatin scaffold played an important role in the cardiac cells organization, phenotype, and contraction.[373]
Generally, electrospun scaffolds have high fiber packing density and small pore sizes (~10–15 μm).[377] These features limit cell infiltration within the scaffold. Therefore, alternative techniques such as salt/polymer leaching,[378] wet electrospinning using a bath collector[379] or an ice crystal collector,[380] and laser/UV irradiation[381] have been used to create electrospun scaffolds with both large pores and high porosity. Another approach to improve cell infiltration within aligned electrospun sheets is the use of co-electrospinning of a sacrificial component and a polymer.[382] For example, Baker et al. used a custom-built design containing multiple nozzles, which allowed electrospinning of independent networks of PCL and poly(ethylene oxide) simultaneously. In the fabricated structure, poly(ethylene oxide) was degraded spontaneously upon hydration leaving behind large pores which facilitated cellular infiltration.[382] Electrospinning process usually requires harsh environments such as the use of high electric fields and cytotoxic chemicals; thus, encapsulation of cells within the construct during the manufacturing process has always been a challenge.[359]
3.6. Assembly of Microgels
Controlling the distribution of different cell types in tissue engineered scaffolds is a major challenge. In addition, many organs such as liver or kidney have complex 3D structures made from smaller repeating units.[383] As a result, researchers are interested in using directed assembly techniques for combining smaller gel building blocks to mimic the complexity of native tissues. These bottom-up, or modular, approaches use specific microarchitectural structures (building blocks) to engineer macro-size biological tissues. These blocks could be in the shape of planar structures that can be stacked to generate a 3D construct or can be in the form of fibers that are assembled to form a tissue like constructs.[329,384] To create and assemble these building blocks, several methods have been employed such as tissue printing,[385] directed assembly of tissue,[386] and fiber-based techniques.[387] Tissue printing has been discussed in the previous section. Here, we will review the directed assembly of tissue modules and the fiber-based techniques.
3.6.1. Directed Assembly of Tissue Modules
In direct assembly of tissue modules, centimeter scale tissues are made from small modules by manipulating their surface energy in contact with a hydrophobic surface. For example, Du et al. photopatterned PEG-methacrylate (PEGMA) polymer modules in the shapes of rectangles and lock-and-keys.[388] They placed the hydrophilic hydrogels on the surface of a hydrophobic mineral oil. By agitating the liquid, hydrogel modules self assembled to minimize the surface energy. A unified structure was then formed through a secondary crosslinking of the assembled units (Figure 15A). In a follow up study, Du et al. fabricated hollow cell-laden rings.[389] The rings were placed in mineral oil and were assembled into tubular structures. In addition, concentric rings containing HUVECs and smooth muscle cells were fabricated and assembled to form a blood vessel (Figure 15B). The blood vessel was then connected to two needles and perfused.
Figure 15.
Assembly of microgels. A) Schematic diagram of the assembly process; cell-laden modules were immersed inside a hydrophobic oil where they self assembled to minimize the surface energy; Reproduced with permission.[388] Copyright 2008, National Academy of Sciences. B) Fabricated modules and their assembly; i–iii) lock-and-key constructs loaded with FITC-dextran and Nile red; iv,v) rings containing concentric layers of HUVEC- and SMC-laden hydrogel; these rings were assembled to form a microvessel. Reproduced with permission.[384] Copyright 2011, John Wiley & Sons, Inc.
In general, modular approaches allow a precise control over the population and distribution of different cell types within the fabricated tissue-like constructs.[383] However, the application of these methods for fabrication of implantable tissues are limited due to several challenges such as lack of scalability and low mechanical properties of the fabricated constructs.[390]
3.6.2. Fiber-Based Techniques
Fiber-based techniques can either be based on one step or two steps processes. In one step processes such as electrospinning and direct writing,[385,391–394] fibers are fabricated and assembled simultaneously. In two step processes, which encompass textile techniques[395,396] and winding,[387] hydrogel fibers are first fabricated and then assembled into 3D constructs. These fibrous structures are usually highly porous and offer high surface area to volume ratio.[360,361] Recently, Onoe et al. fabricated core-shell fibers from collagen/cell mixture and alginate, after which the fabricated fibers were successfully assembled by using an in-house micro-weaving machine (Figure 14C).[355] The developed woven constructs had low mechanical properties, which made their handling challenging. Thus, the hydrogels were embedded in an agarose layer to enhance their mechanical stability.
Generally, low mechanical properties of alginate fibers prevent their utilization in other textile processes such as braiding and knitting to form complex 3D cell-laden constructs. Recently, Akbari et al. developed composite hydrogel fibers with a strong core material coated with an alginate layer.[397] The developed technology in this study demonstrated the utility of textile techniques in the fabrication of hydrogel constructs for tissue engineering.
Fiber-based techniques can be merged with bioprining and textile-based technologies such as weaving, knitting, and braiding to create 3D vascularized tissue constructs. This will lead to innovative methodologies and devices for both in vitro and in vivo tissue reconstruction.
4. Applications of Engineered Hydrogels
4.1. Pre-Vascularized Hydrogels
Appropriate function of engineered 3D constructs cannot be sustained by diffusion of nutrients into and out of hydrogels alone.[398] Therefore, complex engineered tissues with clinically relevant sizes require a functional vascular network for survival. Host-derived vascularization of implanted constructs, however, is largely limited by the overall difficulty of host cells to invade and form functional capillaries.[399] This limits the therapeutic potential of the implanted constructs due to lack of nutrient delivery and waste removal, which may result in the formation of a necrotic core.[271,400]Creating functional (fully perfusable) biomimetic microvascular systems replicating the structure, biological properties, and biomechanics of native microvascular systems is, therefore, one of the greatest bottlenecks of current tissue engineering approaches. Achieving this goal is expected to promote important steps towards the coveted establishment of tissue regeneration as an effective health care alternative for the increasing lack of transplant organs.
The integration of microfabrication technologies and biologically relevant hydrogels has long held great potential towards addressing the limitations listed above. Therefore, a growing number of reports have aimed at developing complex vascularized networks. In general, these efforts can be divided into two main categories: i) microfabrication (physical formation) of channels and vessel-like structures; ii) biological formation of a vascular system as a result of cellular interactions (Figure 16). In this section, we will describe some of the progresses in the field of hydrogel vascularization. These include the utilization of 3D bioprinting technologies, micropatterning of cell-laden hydrogel substrates, and integration of microfluidic platforms with smart biomaterials.
Figure 16.
Vascularization in tissue engineered constructs. A) Microvascular network fabricated in a microfluidic chip. i) Endothelial cells (CD31-red) interacting with pericytes (alpha-SMA-green) to form perfusable microvascular systems. ii) At higher magnification, the presence of a patent lumen and deposition of ECM (Col IV, pink) are visible from the confocal sections in XZ and YZ. iii–iv) A sprouting assay demonstrates the high-proliferation of mature tubules given the presence of U87MG cancer cells after 2 and 4 days of culture. Reproduced with permission.[414] Copyright 2013, The Royal Society of Chemistry. B) Layer-by-layer assembly of cellular multilayers i) fibronectin-gelatin nanofilms in a co-culture of human umbilical artery smooth-muscle cells and HUVECs. ii) This technique was utilized to form biomimetic multilayered blood vessels with improved selective permeability. Reproduced with permission.[405] Copyright 2007, John Wiley and Sons. C) Photographs of microvascular beds in GelMA hydrogels. i) Bioprinted 500 μm agarose templates surrounded by a GelMA hydrogel replicating 3D branched microvascular structures. ii) After removing the agarose, a patent microvascular network is shown after perfusion with a fluorescent dye (unpublished results). D) Sacrificial glass-carbohydrate as a template for microvascular network fabrication. i) A glass-carbohydrate template bioprinted with a branching architecture to form microvascular systems. ii) 3D lattice architectures of the bioprinted template allowed for the formation of thick cell-laden hydrogel constructs with high cell viability. iii) After sacrificing the glass-carbohydrate, a patent network remained and HUVECs (red) were perfused inside the lumen of a pericyte-laden (10T1/2, green) hydrogel. Reproduced with permission.[271] Copyright 2013, Nature Publishing Group.
Microfabrication or physical formation of vascular networks within hydrogels has been achieved by embedding sacrificial fibers such as PMMA,[401] sugar,[271,402]shellac,[274] gelatin[275] fibers within hydrogel constructs or through printing or molding a pre-vascularized structure. For instance, direct-write printers have been effectively used to create 3D microvascular conduits in hydrogels. Using a fugitive ink composed of an aqueous solution of Pluronic F127, Wu et al. reported the formation of interconnected microchannels in hydrogels via the so-called omnidirectional printing process.[319] Although this method replicated the 3D branching architecture of native vascular networks, the process appeared to yield cytotoxic reaction byproducts.[319] In a notable study, Miller et al., also utilized a 3D printer to extrude a 3D template made from biocompatible glass carbohydrate, which was subsequently fully sacrificed via aqueous dissolution after casting of a cell-laden hydrogel[271] (Figure 16D). The bioprinted microchannels were seeded with ECs, which formed a stable monolayer within a few days of culture. However, the dissolution of the template was shown to result in osmotic damage to encapsulated cells, thus requiring template coating with poly(DL-lactide-co-glycolide) (PDLGA). To address this limitation, an approach was developed whereby agarose hydrogel was utilized as a template material for fabricating of fully perfusable 3D branching microvascular networks in GelMA (Figure 16C). Hollow fibers fabricated with poly(lactide-co-glycolide) (PLGA) and PCL blends showed excellent mechanical properties and allowed for mass transport, thus making them potentially useful for the development of hydrogel-based small-caliber vascular grafts.[403] In another study, to form multilayered microvascular vessels, a layer-by-layer assembly approach was utilized to create bilayered fibronectin/gelatin hydrogel constructs containing human umbilical artery smooth-muscle cells (UASMC) and HUVECs.[404] This approach was originally employed by Matsusaki et al. to create planar microchannels in degradable poly(γ -glutamic acid).[298,405] (Figure 16B)
Biological formation of microvessels generally relies on homotypic or heterotypic cell-cell interactions and their cross-communication with growth factors and the extracellular microenvironment.[398] For instance, it has been shown that ECs, either co-cultured with human MSCs, perivascular lineages or alone, self-assembled to form microvascular networks.[270,398,399,406–409] This tendency becomes more pronounced when these cells are encapsulated in ECM-derived or chemically modified hydrogels following intrinsic homing mechanisms and cross-talk of cell and biomaterial. Recently, our group has developed highly vascularized GelMA hydrogels by encapsulating blood-derived endothelial colony forming cells (ECFCs) and bone marrow-derived MSCs in a hydrogel matrix.[409] Results demonstrated an extensive formation of in vitro capillary-like networks comprised of distinct lumens. Moreover, the organized ECFCs were surrounded by αSMA-expressing MSCs occupying periluminal positions within the network, thus replicating the multicellular architecture of native microcapilaries. Interestingly, the physical properties of the hydrogels had a significant effect in the capillary formation, where an increase in the degree of methacrylation and correlative change in stiffness had an adverse effect on the average capillary branch length, number of branch points and other vascular specific markers.[409]
In addition, the presence of growth factors and signaling markers such as vascular endothelial growth factor (VEGF) can boost the microvessel formation process. D’Andrea et al. developed a synthetic peptide (QK), which replicated the exact region of the VEGF binding interface and activated VEGF receptors. This peptide was then used to induce endothelial proliferation and VEGF signaling mechanisms on a Matrigel-coated substrate.[410] Synthetic peptides were further modified to incorporate acrylate terminal groups which were then utilized with photolabile materials to create micropatterns of vasculogenic regions in PEG based hydrogels via photolithographic techniques.[411] In a similar study, Chiu et al. guided EC proliferation in a microfabricated chitosan-collagen hydrogel rich in Tβ4, an angiogenic and cardio-protective peptide that enhances cardiomyocyte survival.[412,413] This approach enabled capillary-mediated anchorage and anastomosis of an artery and a vein placed front-to-front.[412] In a recent study, Kim et al. integrated a microfluidic chip perfused with a fibrin gel to perform a comprehensive study on the assembly of EC mediated formation of perfusable microcapilaries.[414] The microvessels generated by this approach presented morphological and biochemical markers replicating those of native blood vessels and capillaries, such as strong barrier function and stability[414] (Figure 16A).
Both physical and biological techniques for pre-vascularization of tissues hold a great promise for the fabrication of functional biomimetic vascular networks. However, there are some challenges that have limited the progress of these methods. Challenges associated with the physical fabrication of vascular networks are: i) harsh removal process for majority of sacrificial materials and their incompatibility with cells; ii) inability to form 3D biomimetic networks; and iii) the slow processing time. The challenges associated with the biological systems are: i) the slow generation rate of the vessels; ii) lack of control over the size and spatial distributions of the formed conduits. In general, one can expect that a combination of physical and biological techniques could be employed to fabricate functional biomimetic vascular networks. In this approach the main vessels are formed by use of biocompatible sacrificial fibers and the capillaries can be created biologically through cell-cell and cell-matrix interactions.
4.2. Injectable Fillers for Soft-Tissue Engineering
Soft tissue filler procedures have become extremely popular in the US over the past decade as promising injectable materials due to their capabilities to cure in situ, fill irregular shape defects, be utilized in minimally invasive surgical procedures, and incorporate cells and growth factors.[415,416] As reported by American Society of Plastic Surgeons, the number of soft filler injections has increased by 205% between 2000 and 2012, with approximately 2 million injection procedures in 2012 alone.[9] The most widely used materials to create soft tissue injectable fillers include autologous fat, collagen, HA, and biosynthetic polymers.[417,418] The use of autologous fat as injectable filler for facial defects dates back to 1893 followed by utilization of liquid silicone as soft tissue fillers in the 1960s in the US.[419] Despite their significant success, the use of silicone as injectable filler was not approved by the United States Food and Drug Administration (FDA) for cosmetic purposes due to the development of foreign-body reactions.[420]
The number of soft fillers on the market has significantly increased after approval of the first collagen-based dermal filler, Zyderm I, in 1981.[421] Since then, more than ten types of fillers, with different composition, molecular weight, viscosity, mechanical properties, and longevity, have been approved by FDA for clinical applications.[416,422] (Table 1). Collagen-based fillers were the gold standard injectable materials before the development of HA-based fillers due to their reduced risk of immunological reactions. However, their utilization has declined over the past few years, which can be attributed to their rapid resorption (6 months).[423] Cosmoderm and Zyderm are the most common collagen-based fillers, which have been used for superficial injections.[417]
Table 1.
Injectable fillers developed for soft tissue engineering.
| Product Name | Material Type | Durability | Clinical Applications | Advantages | Disadvantages |
|---|---|---|---|---|---|
| Viable fat | Autologous fat | Months to years | Deep defects | Safe Inexpensive Abundant supply |
Donor morbidity Variable reproducibility Requires complex processing |
| Zyderm 1 | Bovine collagen | 2–4 months | Superficial defects Acne scars Fine lines |
Safe, reliable, contains lidocaine Ease of administration |
Allergic reaction in 1–3% of patients Effective in short-term Requires skin testing prior to use |
| Zyderm 2 | Bovine collagen | 2–6 months | Lip augmentation Moderate defects Deeper acne scars |
Similar to Zyderm 1 | Similar to Zyderm 1 |
| Zyplast | Crosslinked Bovine Collagen | 2–6 months | Lip augmentation Deep defects |
Resistant to degradation Safe, reliable, contains lidocaine Ease of administration |
Skin necrosis when using in glabella Allergic reaction in 3% of patients |
| AlloDerm | Collagen acellular human dermis | 6–12 months | Lip augmentation Deep wrinkles |
Safe No allergy testing is required | Expensive Requires surgical implantation Shrinkage and reformation of wrinkles over time |
| Cymetra | Micronized injectable form of AlloDerm | 3–6 months | Lip augmentation Deep scars |
Safe No allergy testing is required Contains lidocaine |
Skin necrosis when using in glabella Expensive Clumps within the needle |
| Cosmoderm | Cell-cultured Collagen | 3–4 months | Superficial defects Acne scars Shallow wrinkles |
Safe No allergy testing is required Contains lidocaine |
Effective in short-term Side effects in up to 4% of the patients |
| Cosmoplast | Crosslinked Collagen | 3–4 months | Deeper defects Wrinkles Lip augmentation |
Similar to Cosmoderm | Similar to Cosmoderm |
| Hylaform gel | Avian-derived HA | 3–4 months | Moderate defects Lip augmentation |
Safe Reliable No allergy testing is required |
Effective in short-term Immunologic reaction in patients allergic to avian products (eggs) |
| Hylaform Plus | Avian-derived HA | 3–4 months | Deep skin defects Facial wrinkles |
Similar to Hylaform gel | Effective in short-term Immunologic reaction in patients allergic to avian products Skin discoloration due to Hylaform Plus injection |
| Restylane | Bacterial cultured HA | 6–12 months | Skin defects and wrinkles Nasolabial folds Lip augmentation |
Safe Predictable results No allergy testing is required Slow degradation rate compared to collagen |
High risk of bruising Rare immunologic reactions Postprocedure swelling Higher cost compared to collagen injections |
| Perlane | Bacterial cultured HA | 6–12 months | Deeper defects Lip augmentation Shaping facial contours |
Similar to Restylane | Similar to Restylane |
| Captique | Bacterial cultured HA | 3–6 months | Superficial defects Fine wrinkles |
Safe Predictable results No allergy testing is required Slow degradation rate compared to collagen |
Effective in short-term |
| Juvederm | Bacterial cultured HA | 3–6 months | Superficial, moderate and deep defects | Safe Predictable results No allergy testing is required |
Effective in short-term Immunological reactions in some cases |
| Sculptra | Poly-(L-lactic acid) microparticles | 1–2 years | Deep defects | Safe Long term results |
Immunological reactions in some cases |
| Radiesse | Calcium hyodroxyapatite microspheres | 1–2 years | Deep defects Acne and scars Vertical lip lines |
Long-term results No allergy testing is required No inflammatory reactions |
Rarely development of nodules when injected superficially |
| Artecoll/ArteFill | Poly(methyl methacrylate) microspheres in 3.5% bovine collagen and 0.3% lidocaine | Permanent after 50% resorption | Deep defects | Safe Longevity |
Requires allergy testing Not suitable for injection into the lips and areas with thin skin |
| Reviderm Intra | Dextran beads in a hylan gel | Months to years | Lip augmentation Deep skin defects |
Safe Long-term results |
Significant post-injection swelling |
| Silicone/Silikon-1000 | Liquid silicone | Permanent | Lip augmentation Deep defects |
Safe Permanent |
Foreign body reactions |
| Endoplast 50 | Elastin and Collagen | 12 months | Lip augmentation Deep defects |
Long-term results | Allergy testing is required |
| Bio-Alcamid | 96% water, 4% poly(alkyl imide) | Permanent | Deep defects | Long-term results No allergy testing is required |
Inflammatory reactions Infections in the injection site |
| Aquamid | Polyacrylamide hydrogel | Permanent | Lip augmentation Deep defects |
Long-term results | Granuloma formation Infections in the injection site |
Among injectable filler materials, HA is the most commonly used filler, with around 1.3 million injection procedures annually in the US.[422] Due to its high water content, upon injection HA-based hydrogels fill the soft tissue defect by absorbing water to increase their volume injection. Despite significant progress, the use of HA fillers has been limited for clinical applications due to their fast degradation, which classified HA as a temporary filler.[424] Therefore, repeated injections are required to maintain desired outcome when using HA-based fillers.
Various approaches have been used to increase the in vivo longevity of HA fillers such as crosslinking, increasing HA concentration, and fabrication of composite materials.[424–426] For example, Tan et al. recently synthesized a double crosslinked HA filler with tunable degradation and mechanical properties.[425] These HA-based fillers were produced through the crosslinking of an amine- and an aldehyde-functionalized HA via Schiff-based linkage followed by genipin crosslinking as the second crosslinking step.[425] In another study, injectable fillers for adipose tissue engineering were created by covalently crosslinking of HA on the surfaces of PLGA microspheres by using 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride (EDC) crosslinker.[426] To increase the lifetime and persistence of injectable HA filler, recently Hillel et al. produced in situ photocrosslinked PEG/HA composite fillers as soft tissue replacements.[424] After subcutaneous injection, the material blends containing photoinitiator (eosin Y, N-vinyl-2-pyrrolidone (NVP), and triethanolamine) were photocrosslinked into the dermis layer by using a light-emitting diode (LED)[424] (Figure 17). The elasticity and degradation rate of photocrosslinked PEG/HA composites were tuned by changing the ratio of PEG and HA. In addition, the implanted hydrogels demonstrated enhanced biocompatibility and volume retention after injection in both human and rodent. However, an inflammation response and formation of a thin pseudocapsule were observed around the injection sites, which could be attributed to the mismatch between the mechanical properties of the implant and the native tissue. In addition, no neovascularization and adipose tissue formation were noted within the implant.[424]
Figure 17.
Transdermal injection of photocrosslinkable PEG/HA hydrogels. A) The composite blend was injected into the dermis, B) the uncrosslinked mixture was massaged into the desirable shape under the skin, C) the material was then crosslinked by using an array of LEDs emitting light, which penetrated up to 4 mm of tissue depth. Reproduced with permission.[424] Copyright 2011, Advancing Science, Serving Society.
The use of injectable fillers offers several advantages over the prefabricated scaffold for engineering soft tissues. These injectable materials allow for minimally invasive procedures and easy manipulation during the surgery, eliminating the surgical complications. In addition, injectable fillers can be used to fill and repair complex shaped defects and wounds. However, there are some challenges in their clinical applications. These challenges include difficulties in controlling the shape and the position of injected materials, tissue responses due to the differences between the properties of the injectable filler and the native tissue, and the short lifetime of the injected materials.[415] Therefore, it is still required to develop innovative hydrogel-based fillers, which more closely mimic the native soft tissue microenvironment. The future success in the field of injectable filler for soft tissue engineering will likely feature hybrid materials, which combine the properties of various materials, each uniquely contributing to a specific need of soft tissue developments. These properties include adequate biological cues, high elasticity, and in vivo durability.
4.3. Directing Cellular Behavior Using Hydrogels
Over the past decade researchers have put effort to control the biophysical, biochemical, and biological cues in cell-laden hydrogels to closely mimic the ECM.[427–429] These sophisticated hydrogels with precise architecture that can respond dynamically to exogenous extracellular cues, or release growth factors on-demand, may have the ability to recapitulate tissue and organ function in the future.[5,430,431] For example, stroke, traumatic brain injury, Alzheimer’s and Parkinson’s diseases are neurological disorders with patient-specific outcomes, and hydrogel-based regenerative strategies have attracted attention to regenerate the central nervous system (CNS).[432] However, the spatial and temporal variation of physical and biological properties of CNS is a major challenge facing its regeneration. Hydrogel-based biomaterials appear as a suitable candidate, because their mechanical properties align well with those of the CNS.[12,433–435] More importantly, injectable hydrogels have been developed which can encapsulate biological cues and be topographically modified to influence cell behavior. The subject of this section is to highlight the latest strategies to direct cellular behavior by hydrogel microenvironments with the intention to reconstruct normal activity of tissues and organs.
4.3.1. Modification of Cellular Behavior in 3D
Control and optimization of the cellular activity in cell-laden hydrogels is of great importance in many tissue engineering applications. For example, a key factor that affects the success of neural tissue engineering strategies is the ability to guide cells and control their morphology. For instance, thermally crosslinkable xyloglucan and xyloglucan-graft-poly-d-lysine (PDL) hydrogels were injected within the brain of adult rats and their effect on the behavior of neurites and neurons was assessed.[436] The PDL-based hydrogels facilitated the infiltration of axons in a controlled manner as well as repressed a severe inflammatory response after implantation. In addition, the secretion of laminin, a promoter of axonal outgrowth, was upregulated.[437] However, one of the limitations of this approach was directing neuron axonal guidance.[436] In another study, RGD-modified PEGcell-laden hydrogels were produced by incorporating micelles and encapsulating human MSCs to direct cell-hydrogel interactions, and improve cell viability and in vitro gene transfection.[438] These biocompatible PEG-based hydrogels were shown to be suitable candidates for the delivery of hMSCs to a specific repair site.
Tailoring the delivery mechanisms of proteins and growth factors responsible for the growth and survival of cells is an important challenge to overcome during the design of hydrogels in regenerative medicine. In the case of neural tissue engineering, delivering neurothropic factors (NF) is an important strategy for directional guidance of axonal growth in vivo. Controlled release of NF can recreate the innervation of distant brain regions of Parkinson’s patients.[439] Lampe et al. engineered PLGA microparticles entrapped within a PEG-based hydrogel to locally release two types of NF with different release profiles.[439] Microparticle incorporated hydrogels were then analyzed for their abilities to release NF in different regions of the brain. The results indicated coordinated drug delivery ability and reduced localized inflammatory response upon implantation.[439] The knowledge acquired in the modulation of the endogenous immune response to implanted hydrogels may also be useful in the development of intracortical electrodes with biocompatible properties.[440] In another study, cartilage regeneration were also achieved by the controlled spatiotemporal release of growth factors like transforming growth factor-β1 (TGF-β1) from gelatin microspheres.[441] The hMSCs-laden microspheres covalently crosslinked with genipin exhibited important deposition of cartilage matrix composed mainly by glycosaminoglycans (GAGs) and collagen type-II in vitro.[441] It is known that besides matrix stiffness and cell-matrix adhesive interactions, the hydrogel crosslinking mechanism (covalent versus physical) could fundamentally guide cellular behavior such as stem cell differentiation.[442,443] A recent study has elucidated the mechanistic effects underlying stem cell differentiation in covalently crosslinked 3D microenvironments while correlating cell interaction, matrix degradation, and adhesion/ligand interactions at the nanoscale.[444]
Injectable biodegradable hydrogels containing cells provide means to improve transplanted cell viability in the nervous system by physical protection from compaction, shear forces, and acute inflammatory responses. PEG-based hydrogels have shown to be promising candidates for this purpose, as the degradation profile and mechanical properties of the gel can be tailored.[445] The delivery of basic fibroblast growth factor (bFGF-2) was shown to enhance the survival of neural precursor cells in vitro, demonstrating the importance of controlling the chemical microenvironment surrounding cells in 3D hydrogels.[445]
A typical problem with the direct injection of neural-derived stem or progenitor cells to the lesion site is the poor viability and functionality of transplanted cells when in contact with the local tissue environment. To address this challenge, Li et al. engineered an injectable hydrogel system as a supportive niche to provide a permissive microenvironment for transplanted cells to survive, differentiate, and remyelinate CNS lesions.[446] The hydrogels was based on thiol-functionalized HA and thiol-functionalized gelatin. It was found that transplanted neural progenitor cells within the fabricated hydrogels were able to remyelinate axons in lesions in adult spinal cord.[446]
In another study, a hydrogel derived from the ECM of the central nervous system capable of in vivo polymerization and conformation upon injection to fill irregular lesion geometries was developed.[447] It was shown that the fabricated CNS-ECM hydrogels resembled the biochemical compositions, mechanical properties, and neurotrophic potential of the brain and the spinal cord ECM. In addition, these hydrogels promoted 3D neurite outgrowth after 7 days in culture.[447] Injectable alginate-based hydrogels can also be used to promote bone formation as described by Moshaverinia et al.[448] The results showed that the alginate microbeads encapsulating periodontal ligament stem cells (PDLSCs) and gingival mesenchymal stem cells (GMSCs) ensured cell viability and promoted osteogenic differentiation in vitro by the upregulation of Runx-2 and osteocalcin genes. Taken together, these injectable hydrogels open possibilities for implants to readily adapt to differently shaped brain lesions upon implantation.
4.3.2. Cellular Guidance by Hydrogel Topography
The physical contact between a substrate such as the ECM and a biomaterial generally directs cell growth and migration.[449,450] For example, during the development of the nervous system, contact-mediated cues suggest orientation of neuroblasts parallel to glial fibers.[435] To simulate the guidance of neural cells similarly to what occurs during tissue formation, hydrogels containing topographical cues have been developed.[451] Horne et al. demonstrated that 3D nanofibrous PCL scaffolds incorporating immobilized brain-derived neurotrophic factor (BDNF) enhanced neural stem cell (NSC) proliferation and differentiation towards neurons in vitro.[451] Additionally, the effect of non-uniform stiffness of DNA-crosslinked hydrogels on spinal cord cells was evaluated based on neurite outgrowth and neuronal biomarker expression.[452] The results indicated that neuronal cells have mechanosensing capability that allow the cells to respond by changing neurite outhgrowth.[452] These studies provide insight into neurodegenerative and neurophatological diseases where the mechanical properties of the nervous system are compromised. Additionally, micropatterned posts in polyacrylamide hydrogels modified with bound collagen can be used to direct MSC morphology and modify the deposition of ECM in a topography-dependent manner.[453] Vascular sprout formation can also be controlled by varying the matrix mechanical and topographical properties of degradable PEG diacrylate (PEGDA) hydrogels.[454] This in vitro vascularization model indicated sprouting in directions toward the stiff regions of the PEGDA hydrogel, and gave insightful cues to guide neovascularization in engineered hydrogels.[454]
It is likely that in the future, researchers will continue to design hydrogels with fully controllable biochemical and biomechanical properties to understand their effects on cell-cell and cell-biomaterial interactions. The hydrogel composition, the ligand-to-receptor ratio, the hydrogel mechanics and topography will all regulate cell function in a biomimetic 3D microenvironment.[455]
4.4. Organs-on-a-Chip
One of the emerging applications of tissue engineering efforts is in developing functional tissues that can be exploited as disease models or drug screening systems.[456,457] Current drug screening protocols are based on evaluation of drug activity in 2D cultures, animal models, and clinical trials.[458] However, it is now widely recognized that the 2D cultures cannot stimulate cell-cell- and cell-matrix interactions required for normal cellular activity.[456,459] Animal models are also expensive, highly variable, and difficult to control. Moreover, it has been shown that animal models are not always predictive of human physiological responses.[458] All these challenges call for platforms in which human tissues are engineered and employed for drug testing. However, successful in vitro study of cellular activity can only be achieved if the employed platform mimics the in vivo environment.[460] This means that cells should be cultured three dimensionally in a biomaterial containing ECM similar to the physiological environment. Moreover, to eliminate the oxygen and nutrient diffusion limitations, an effective perfusable network of channels should be devised that act as the circulatory system. In addition, cells or tissue-like constructs should be stimulated mechanically and electrically if required to simulate the in vivo conditions and ensure normal cellular activity.
The concept of organs-on-a-chip has emerged in the past decade as in vitro miniaturized models which usually contain human cells and offer structural, biochemical, mechanical, and functional characteristics of living organs.[461–463] These systems can potentially fill the gap between in vitro tests and clinical trials and can be utilized for high throughput screening of drugs in pharmaceutical research with more relevance in comparison to animal models.[464] Researchers have engineered various miniaturized organs such as liver,[465] lung,[328] intestine,[466] kidney,[467] and heart muscle.[468] In one study, a lung-on-a-chip platform that replicated the breathing movements of the alveolus was fabricated. The device comprised of two layers of channels separated by a thin PDMS membrane containing micrometer size pores.[328] The membrane was coated by ECM and seeded with human lung alveolar epithelial cells on one side and human lung capillary ECs on the other side. The side containing ECs was exposed to liquid while a stream of air was passed through the other side of the membrane covered with epithelial cells to create an air-liquid interface. The membrane was deformed by applying cyclic suction to mimic the deformation of the alveolus due to breathing. In another study, Agarwal et al. fabricated alginate hydrogels containing microgrooves to engineer a heart muscle-on-a-chip.[468] The cells seeded on the surface of micropatterned hydrogels offered higher activity in comparison to those seeded on the surface of flat alginate sheets. Recently, Chen et al. introduced a two level microfluidics platform in which primary porcine aortic valvular interstitial cells (VICs) encapsulated within a GelMA layer and valvular ECs were separated by a porous membrane.[469] The platforms allowed studying the interaction between the two cell types as a model for heart valve-on-a-chip. The results suggested that the presence of ECs prevented from differentiation of VICs to myofibroblasts especially when ECs were exposed to shear stress.
As a result of the recent advancements in the fields of microfluidics, MEMS, and biomaterials the ultimate goal of creating a body-on-a-chip to replace animal models in drug screening looks achievable. In such systems, various organs will be cultured on a single chip and will be connected to each other by the vascular system. For testing oral drugs, the drug enters the system and should be absorbed by the intestine.[461] The drug is then taken to various organs by the circulatory system to determine their efficacy, absorptions, transport, metabolism, and clearance.[470] Hydrogels are promising 3D environments for cellular growth and their compositions can be tuned to mimic ECM of native organs. By combining the advanced hydrogels and microfabrication technologies with recent progresses in the area of organ-on-a-chip, the fabrication of functional organs can be realized. These systems can potentially be used as disease models or high throughput drug screening devices.
5. Conclusions and Future Directions
During the past decades, hydrogels have emerged as promising biomaterials supporting cellular viability, proliferation, and differentiation. However, achieving the ultimate goal of tissue engineering, which is the fabrication of biological substitutes for damaged organs, relies on the design and synthesis of advanced hydrogels mimicking the in vivo environment. However, the current state of hydrogels used in tissue engineering applications, in general, mainly focuses on certain aspects of the tissue properties such as physicochemical properties, therefore do not replicates the complexities of the native environment in its entirety, and often lead to unsatisfactory results. Thus, the development of advanced hydrogels with tunable mechanical properties, electrical conductivity, degradation rate, and biological properties are of great importance. In addition, growth factors and suitable cell sources should be incorporated within 3D hydrogels at predefined patterns, thus resembling the native tissue to pave a road towards engineering functional tissues. It is also expected that merging the field of advanced hydrogels with advanced technologies such as Bio-MEMS, microfluidics, 3D printing, textile engineering, and composite fabrication can further the field of tissue engineering and regenerative medicine.
Another bottleneck to overcome is the fabrication of pre-vascularized tissues that can integrate with the host’s blood circulation system upon implantation. Fabrication of effective vascular networks in engineered tissues, however, requires: i) the development of advanced engineering tools to create 3D biomimetic constructs; ii) the establishment of strategies that can utilize cell-cell and cell-environment interactions to control angiogenesis. Rapid prototyping approaches, such as stereolithography, could be utilized to pattern hydrogels to create well-defined microchannels to guide vascularization. In addition, vascularization could be further stimulated by applying external forces (i.e., fluid perfusion, cyclic mechanical stress). One research area, less studied in the past few years, is the fabrication of effective neural networks within engineered tissues, which interact with the native neural system and can transfer electrical impulses. To achieve this goal, fabrication of smart hydrogels containing biomolecules that can direct neural network formation is a key step. Another important application of hydrogels is their use as injectable biomaterials acting as cell and drug carriers. Novel hydrogels should be synthesized, which self-assemble or crosslink upon injection in situ. Injectable hydrogels have potential application in bone, spinal cord, cartilage, and cardiovascular regenerative medicine. Although material selection for clinical applications is currently limited, detailed pharmacological studies to delineate biological responses and ramifications to demonstrate the biocompatibility of hydrogels would likely lead to more clinically approved materials.
Organ-on-a-chip research is another emerging area in which hydrogels play a key role. Recently, researchers have attempted to engineer in vitro systems in the form of miniaturized organs, which mimic the function of native tissues. These platforms can be used as disease models for therapeutic and drug screening applications. Utilization of these organ-on-a-chip platforms can potentially facilitate the translation of therapeutic drugs from the lab to the clinic.
Acknowledgments
This article is part of an ongoing series celebrating the 25 th anniversary of Advanced Materials. N.A. acknowledges the support from the National Health and Medical Research Council. A.K. acknowledges funding from the National Science Foundation CAREER Award (DMR 0847287), the office of Naval Research Young National Investigator Award, the National Institutes of Health (HL092836, DE019024, EB012597, AR057837, DE021468, HL099073, EB008392), and the Presidential Early Career Award for Scientists and Engineers (PECASE). A.T. and M.A. acknowledge the financial support of the Natural Sciences and Engineering Research Council of Canada (NSERC). J.A.U. acknowledges the financial support of the Secretaria Nacional de Educacion Superior, Ciencia, Tecnologia e Innovacion (Senescyt) of Ecuador. N.A.P. acknowledges funding from the National Science Foundation (10-33746), the National Institutes of Health (EB00246, EB012726, U54-CA-143837), the Gates Foundation (1007287) and the Pratt Foundation.
Biographies
 Nasim Annabi is a postdoctoral fellow at the Harvard-MIT Division of Health Sciences Technology, Brigham and Women’s Hospital, and the Wyss Institute for Biologically Inspired Engineering at Harvard University. Her research is based on developing advanced biomaterials and combining them with micro- and nanoscale technologies to control the cellular microenvironment and engineer complex tissue constructs.
Nasim Annabi is a postdoctoral fellow at the Harvard-MIT Division of Health Sciences Technology, Brigham and Women’s Hospital, and the Wyss Institute for Biologically Inspired Engineering at Harvard University. Her research is based on developing advanced biomaterials and combining them with micro- and nanoscale technologies to control the cellular microenvironment and engineer complex tissue constructs.
 Nicholas A. Peppas is the Fletcher S. Pratt Chair in Biomedical Engineering, Chemical Engineering, and Pharmacy, and the Director of the Center for Biomaterials, Drug Delivery and Bionanotechnology at the University of Texas at Austin. He has been elected a member of the National Academy of Engineering (NAE), the Institute of Medicine (IOM) of the National Academies, the Royal National Academy of Spain, and the National Academy of Pharmacy of France. He is the recipient of the 2012 NAE Founders Award for his contributions to biomaterials and hydrogels. He received his Diploma in Engineering (D. Eng.) from NTU of Athens, Greece in 1971 and his Sc.D. from MIT in 1973, both in chemical engineering.
Nicholas A. Peppas is the Fletcher S. Pratt Chair in Biomedical Engineering, Chemical Engineering, and Pharmacy, and the Director of the Center for Biomaterials, Drug Delivery and Bionanotechnology at the University of Texas at Austin. He has been elected a member of the National Academy of Engineering (NAE), the Institute of Medicine (IOM) of the National Academies, the Royal National Academy of Spain, and the National Academy of Pharmacy of France. He is the recipient of the 2012 NAE Founders Award for his contributions to biomaterials and hydrogels. He received his Diploma in Engineering (D. Eng.) from NTU of Athens, Greece in 1971 and his Sc.D. from MIT in 1973, both in chemical engineering.
 Ali Khademhosseini is an associate professor at Harvard University, and holds appointments at the Harvard-MIT Division of Health Sciences Technology, Brigham & Women’s Hospital. In addition, he is on the faculty of the Wyss Institute for Biologically Inspired Engineering at Harvard University and the World Premier International-Advanced Institute for Materials Research (WPI-AIMR) at Tohoku University. He is an internationally recognized bioengineer regarded for his contributions and research in the areas of bioengineering, biomaterial synthesis, microscale technologies, and tissue engineering. His research involves the development of micro- and nanoscale technologies to control cellular behavior, fabrication of microscale biomaterials and engineering systems for tissue engineering, drug discovery and cell-based biosensing.
Ali Khademhosseini is an associate professor at Harvard University, and holds appointments at the Harvard-MIT Division of Health Sciences Technology, Brigham & Women’s Hospital. In addition, he is on the faculty of the Wyss Institute for Biologically Inspired Engineering at Harvard University and the World Premier International-Advanced Institute for Materials Research (WPI-AIMR) at Tohoku University. He is an internationally recognized bioengineer regarded for his contributions and research in the areas of bioengineering, biomaterial synthesis, microscale technologies, and tissue engineering. His research involves the development of micro- and nanoscale technologies to control cellular behavior, fabrication of microscale biomaterials and engineering systems for tissue engineering, drug discovery and cell-based biosensing.
Contributor Information
Dr. Nasim Annabi, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA. Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, 02115, USA
Dr. Ali Tamayol, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA. Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Jorge Alfredo Uquillas, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA. Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Mohsen Akbari, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA. Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, 02115, USA
Dr. Luiz E. Bertassoni, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA. Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Chaenyung Cha, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA. Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Gulden Camci-Unal, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA. Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Dr. Mehmet R. Dokmeci, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA. Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Prof. Nicholas A. Peppas, Email: peppas@che.utexas.edu, Department of Biomedical Engineering, Biomedical Engineering Building 3.110B, The University of Texas at Austin, 1 University Station, C0800, Austin, Texas, 78712–1062, USA
Prof. Ali Khademhosseini, Email: alik@rics.bwh.harvard.edu, Center for Biomedical Engineering, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02139, USA. Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, 02115, USA
References
- 1.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv Mater. 2006;18:1345. [Google Scholar]
- 2.Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Adv Mater. 2009;21:3307. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annabi N, Nichol JW, Zhong X, Ji C, Koshy S, Khademhosseini A, Dehghani F. Tissue Eng, Part B. 2010;16:371. doi: 10.1089/ten.teb.2009.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci USA. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seliktar D. Science. 2012;336:1124. doi: 10.1126/science.1214804. [DOI] [PubMed] [Google Scholar]
- 6.Khademhosseini A, Langer R. Biomaterials. 2007;28:5087. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Rodrigues J, Tomas H. Chem Soc Rev. 2012;41:2193. doi: 10.1039/c1cs15203c. [DOI] [PubMed] [Google Scholar]
- 8.Perez RA, Won JE, Knowles JC, Kim HW. Adv Drug Delivery Rev. 2013;65:471. doi: 10.1016/j.addr.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Hennink WE, van Nostrum CF. Adv Drug Delivery Rev, Suppl. 2012;64:223. [Google Scholar]
- 10.Macaya D, Spector M. Biomed Mater. 2012;7:012001. doi: 10.1088/1748-6041/7/1/012001. [DOI] [PubMed] [Google Scholar]
- 11.Westhaus E, Messersmith PB. Biomaterials. 2001;22:453. doi: 10.1016/s0142-9612(00)00200-3. [DOI] [PubMed] [Google Scholar]
- 12.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Annabi N, Mithieux SM, Zorlutuna P, Camci-Unal G, Weiss AS, Khademhosseini A. Biomaterials. 2013;34:5496. doi: 10.1016/j.biomaterials.2013.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garlotta D. J Polym Environ. 2001;9:63. [Google Scholar]
- 15.Chen Q, Liang S, Thouas GA. Prog Polym Sci. 2013;38:584. [Google Scholar]
- 16.Liu Q, Jiang L, Shi R, Zhang L. Prog Polym Sci. 2012;37:715. [Google Scholar]
- 17.Winer JP, Oake S, Janmey PA. PloS One. 2009;4:e6382. doi: 10.1371/journal.pone.0006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Annabi N, Mithieux SM, Camci-Unal G, Dokmeci MR, Weiss AS, Khademhosseini A. Biochem Eng J. 2013;77:110. doi: 10.1016/j.bej.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bashur CA, Venkataraman L, Ramamurthi A. Tissue Eng. 2012;18:203. doi: 10.1089/ten.teb.2011.0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martyn CN, Greenwald S. Clin Exp Pharmacol Physiol. 2001;28:948. doi: 10.1046/j.1440-1681.2001.03555.x. [DOI] [PubMed] [Google Scholar]
- 21.Rodgers UR, Weiss AS. Biochimie. 2004;86:173. doi: 10.1016/j.biochi.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Broekelmann TJ, Kozel BA, Ishibashi H, Werneck CC, Keeley FW, Zhang L, Mecham RP. J Biol Chem. 2005;280:40939. doi: 10.1074/jbc.M507309200. [DOI] [PubMed] [Google Scholar]
- 23.Bax DV, Rodgers UR, Bilek MM, Weiss AS. J Biol Chem. 2009;284:28616. doi: 10.1074/jbc.M109.017525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinet A, Fahem A, Cauchard JH, Huet E, Vincent L, Lorimier S, Antonicelli F, Soria C, Crepin M, Hornebeck W, Bellon G. J Cell Sci. 2005;118:343. doi: 10.1242/jcs.01613. [DOI] [PubMed] [Google Scholar]
- 25.Kamoun A, Landeau JM, Godeau G, Wallach J, Duchesnay A, Pellat B, Hornebeck W. Cell Adhes Commun. 1995;3:273. doi: 10.3109/15419069509081013. [DOI] [PubMed] [Google Scholar]
- 26.de Vries HJ, Middelkoop E, Mekkes JR, Dutrieux RP, Wildevuur CH, Westerhof H. Wound Repair Regen. 1994;2:37. doi: 10.1046/j.1524-475X.1994.20107.x. [DOI] [PubMed] [Google Scholar]
- 27.Almine JF, Bax DV, Mithieux SM, Nivison-Smith L, Rnjak J, Waterhouse A, Wise SG, Weiss AS. Chem Soc Rev. 2010;39:3371. doi: 10.1039/b919452p. [DOI] [PubMed] [Google Scholar]
- 28.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, Gavrilov K, Yi T, Zhuang ZW, Breuer C, Herzog E, Niklason LE. Science. 2010;329:538. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AP, England KA, Matson AM, Blazar BR, Panoskaltsis-Mortari A. Tissue Eng, Part A. 2010;16:2581. doi: 10.1089/ten.tea.2009.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow JP, Simionescu DT, Warner H, Wang B, Patnaik SS, Liao J, Simionescu A. Biomaterials. 2013;34:685. doi: 10.1016/j.biomaterials.2012.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nivison-Smith L, Rnjak J, Weiss AS. Acta Biomater. 2010;6:354. doi: 10.1016/j.actbio.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Annabi N, Mithieux SM, Boughton EA, Ruys AJ, Weiss AS, Dehghani F. Biomaterials. 2009;30:4550. doi: 10.1016/j.biomaterials.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Leach JB, Wolinsky JB, Stone PJ, Wong JY. Acta Biomater. 2005;1:155. doi: 10.1016/j.actbio.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Annabi N, Mithieux SM, Weiss AS, Dehghani F. Biomaterials. 2009;30:1. doi: 10.1016/j.biomaterials.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 35.Rnjak J, Wise SG, Mithieux SM, Weiss AS. Tissue Eng, Part B Rev. 2011;17:81. doi: 10.1089/ten.TEB.2010.0452. [DOI] [PubMed] [Google Scholar]
- 36.Annabi N, Fathi A, Mithieux SM, Weiss AS, Dehghani F. J Supercrit Fluids. 2011;59:157. [Google Scholar]
- 37.de Chalain T, Phillips JH, Hinek A. J Biomed Mater Res. 1999;44:280. doi: 10.1002/(sici)1097-4636(19990305)44:3<280::aid-jbm6>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 38.Lamprou D, Zhdan P, Labeed F, Lekakou C. J Biomater Appl. 2011;26:209. doi: 10.1177/0885328210364429. [DOI] [PubMed] [Google Scholar]
- 39.Annabi N, Fathi A, Mithieux SM, Martens P, Weiss AS, Dehghani F. Biomaterials. 2011;32:1517. doi: 10.1016/j.biomaterials.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 40.Palumbo FS, Pitarresi G, Fiorica C, Rigogliuso S, Ghersi G, Giammona G. Mater Sci Eng: C. 2013;33:2541. doi: 10.1016/j.msec.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 41.Broekelmann TJ, Ciliberto CH, Shifren A, Mecham RP. Matrix Biol. 2008;27:631. doi: 10.1016/j.matbio.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badylak SF, Gilbert TW. Semin Immunol. 2008;20:109. doi: 10.1016/j.smim.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Floss DM, Mockey M, Zanello G, Brosson D, Diogon M, Frutos R, Bruel T, Rodrigues V, Garzon E, Chevaleyre C, Berri M, Salmon H, Conrad U, Dedieu L. J Biomed Biotechnol. 2010;2010:274346. doi: 10.1155/2010/274346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Floss DM, Sack M, Arcalis E, Stadlmann J, Quendler H, Rademacher T, Stoger E, Scheller J, Fischer R, Conrad U. Plant Biotechnol J. 2009;7:899. doi: 10.1111/j.1467-7652.2009.00452.x. [DOI] [PubMed] [Google Scholar]
- 45.Floss DM, Schallau K, Rose-John S, Conrad U, Scheller J. Trends Biotechnol. 2010;28:37. doi: 10.1016/j.tibtech.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Schipperus R, Teeuwen RL, Werten MW, Eggink G, de Wolf FA. Appl Microbiol Biotechnol. 2009;85:293. doi: 10.1007/s00253-009-2082-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sallach RE, Conticello VP, Chaikof EL. Biotechnol Prog. 2009;25:1810. doi: 10.1002/btpr.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McPherson DT, Xu J, Urry DW. Protein Expression Purif. 1996;7:51. doi: 10.1006/prep.1996.0008. [DOI] [PubMed] [Google Scholar]
- 49.Indik Z, Abrams WR, Kucich U, Gibson CW, Mecham RP, Rosenbloom J. Arch Biochem Biophys. 1990;280:80. doi: 10.1016/0003-9861(90)90521-y. [DOI] [PubMed] [Google Scholar]
- 50.Martin SL, Vrhovski B, Weiss AS. Gene. 1995;154:159. doi: 10.1016/0378-1119(94)00848-m. [DOI] [PubMed] [Google Scholar]
- 51.Mithieux SM, Wise SG, Raftery MJ, Starcher B, Weiss AS. J Struct Biol. 2005;149:282. doi: 10.1016/j.jsb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Annabi N, Mithieux SM, Weiss AS, Dehghani F. Biomaterials. 2010;31:1655. doi: 10.1016/j.biomaterials.2009.11.051. [DOI] [PubMed] [Google Scholar]
- 53.Mithieux SM. PhD dissertation. University of Sydney; Sydeny, Australia: 2003. [Google Scholar]
- 54.McGrath AP, Mithieux SM, Collyer CA, Bakhuis JG, van den Berg M, Sein A, Heinz A, Schmelzer C, Weiss AS, Guss JM. Biochemistry. 2011;50:5718. doi: 10.1021/bi200555c. [DOI] [PubMed] [Google Scholar]
- 55.Mithieux SM, Tu Y, Korkmaz E, Braet F, Weiss AS. Biomaterials. 2009;30:431. doi: 10.1016/j.biomaterials.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 56.Annabi N, Tsang K, Mithieux SM, Nikkhah M, Ameri A, Khademhosseini A, Weiss AS. Adv Funct Mater. 2013 doi: 10.1002/adfm.201300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mithieux SM, Rasko JEJ, Weiss AS. Biomaterials. 2004;25:4921. doi: 10.1016/j.biomaterials.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 58.Tu Y, Mithieux SM, Annabi N, Boughton EA, Weiss AS. J Biomed Mater Res, Part A. 2010;95:1215. doi: 10.1002/jbm.a.32950. [DOI] [PubMed] [Google Scholar]
- 59.Mithieux SM, Wise SG, Weiss AS. Adv Drug Delivery Rev. 2013;65:421. doi: 10.1016/j.addr.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 60.MacEwan SR, Chilkoti A. Biopolymers. 2010;94:60. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- 61.van Eldijk MB, McGann CL, Kiick KL, van Hest JC. Top Curr Chem. 2012;310:71. doi: 10.1007/128_2011_205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Betre H, Setton LA, Meyer DE, Chilkoti A. Biomacromolecules. 2002;3:910. doi: 10.1021/bm0255037. [DOI] [PubMed] [Google Scholar]
- 63.Betre H, Ong SR, Guilak F, Chilkoti A, Fermor B, Setton LA. Biomaterials. 2006;27:91. doi: 10.1016/j.biomaterials.2005.05.071. [DOI] [PubMed] [Google Scholar]
- 64.Lim DW, Nettles DL, Setton LA, Chilkoti A. Biomacromolecules. 2007;8:1463. doi: 10.1021/bm061059m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim DW, Nettles DL, Setton LA, Chilkoti A. Biomacromolecules. 2008;9:222. doi: 10.1021/bm7007982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trabbic-Carlson K, Setton LA, Chilkoti A. Biomacromolecules. 2003;4:572. doi: 10.1021/bm025671z. [DOI] [PubMed] [Google Scholar]
- 67.Nettles DL, Kitaoka K, Hanson NA, Flahiff CM, Mata BA, Hsu EW, Chilkoti A, Setton LA. Tissue Eng, Part A. 2008;14:1133. doi: 10.1089/ten.tea.2007.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McMillan RA, Caran KL, Apkarian RP, Conticello VP. Macromolecules. 1999;32:9067. [Google Scholar]
- 69.Wang Y, Ameer GA, Sheppard BJ, Langer R. Nat Biotechnol. 2002;20:602. doi: 10.1038/nbt0602-602. [DOI] [PubMed] [Google Scholar]
- 70.Gao J, Ensley AE, Nerem RM, Wang Y. J Biomed Mater Res, Part A. 2007;83:1070. doi: 10.1002/jbm.a.31434. [DOI] [PubMed] [Google Scholar]
- 71.Engelmayr GC, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Nat Mater. 2008;7:1003. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Redenti S, Neeley WL, Rompani S, Saigal S, Yang J, Klassen H, Langer R, Young MJ. Biomaterials. 2009;30:3405. doi: 10.1016/j.biomaterials.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kemppainen JM, Hollister SJ. J Biomed Mater Res, Part A. 2010;94A:9. doi: 10.1002/jbm.a.32653. [DOI] [PubMed] [Google Scholar]
- 74.Sundback CA, Shyu JY, Wang Y, Faquin WC, Langer RS, Vacanti JP, Hadlock TA. Biomaterials. 2005;26:5454. doi: 10.1016/j.biomaterials.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 75.Liu QY, Wu SZ, Tan TW, Weng JY, Zhang LQ, Liu L, Tian W, Chen DF. J Biomater Sci, Polym Ed. 2009;20:1567. doi: 10.1163/092050609X12464345064325. [DOI] [PubMed] [Google Scholar]
- 76.Patel A, Gaharwar AK, Iviglia G, Zhang H, Mukundan S, Mihaila SM, Demarchi D, Khademhosseini A. Biomaterials. 2013;34:3970. doi: 10.1016/j.biomaterials.2013.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martina M, Hutmacher DW. Polym Int. 2007;56:145. [Google Scholar]
- 78.Shi R, Chen D, Liu Q, Wu Y, Xu X, Zhang L, Tian W. Int J Mol Sci. 2009;10:4223. doi: 10.3390/ijms10104223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liang SL, Cook WD, Thouas GA, Chen QZ. Biomaterials. 2010;31:8516. doi: 10.1016/j.biomaterials.2010.07.105. [DOI] [PubMed] [Google Scholar]
- 80.Stuckey DJ, Ishii H, Chen QZ, Boccaccini AR, Hansen U, Carr CA, Roether JA, Jawad H, Tyler DJ, Ali NN, Clarke K, Harding SE. Tissue Eng, Part A. 2010;16:3395. doi: 10.1089/ten.TEA.2010.0213. [DOI] [PubMed] [Google Scholar]
- 81.Pomerantseva I, Krebs N, Hart A, Neville CM, Huang AY, Sundback CA. J Biomed Mater Res A. 2009;91:1038. doi: 10.1002/jbm.a.32327. [DOI] [PubMed] [Google Scholar]
- 82.Chen Q, Jin L, Cook WD, Mohn D, Lagerqvist EL, Elliott DA, Haynes JM, Boyd N, Stark WJ, Pouton CW, Stanley EG, Elefanty AG. Soft Matter. 2010;6:4715. [Google Scholar]
- 83.Bergmeister H, Schreiber C, Grasl C, Walter I, Plasenzotti R, Stoiber M, Bernhard D, Schima H. Acta Biomater. 2013;9:6032. doi: 10.1016/j.actbio.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 84.Rao L, Zhou H, Li T, Li C, Duan YY. Acta Biomater. 2012;8:2233. doi: 10.1016/j.actbio.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 85.Page JM, Prieto EM, Dumas JE, Zienkiewicz KJ, Wenke JC, Brown-Baer P, Guelcher SA. Acta Biomater. 2012;8:4405. doi: 10.1016/j.actbio.2012.07.037. [DOI] [PubMed] [Google Scholar]
- 86.Fujimoto KL, Tobita K, Guan J, Hashizume R, Takanari K, Alfieri CM, Yutzey KE, Wagner WR. J Card Failure. 2012;18:585. doi: 10.1016/j.cardfail.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaebel R, Ma N, Liu J, Guan J, Koch L, Klopsch C, Gruene M, Toelk A, Wang W, Mark P, Wang F, Chichkov B, Li W, Steinhoff G. Biomaterials. 2011;32:9218. doi: 10.1016/j.biomaterials.2011.08.071. [DOI] [PubMed] [Google Scholar]
- 88.Zhang C, Zhang N, Wen X. J Biomed Mater Res A. 2007;82:637. doi: 10.1002/jbm.a.30992. [DOI] [PubMed] [Google Scholar]
- 89.Park D, Wu W, Wang Y. Biomaterials. 2011;32:777. doi: 10.1016/j.biomaterials.2010.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daemi H, Barikani M, Barmar M. Carbohydr Polym. 2013;95:630. doi: 10.1016/j.carbpol.2013.03.039. [DOI] [PubMed] [Google Scholar]
- 91.Huang Y, He K, Wang X. Mater Sci Eng: C. 2013;33:3220. doi: 10.1016/j.msec.2013.03.048. [DOI] [PubMed] [Google Scholar]
- 92.Guelcher SA, Gallagher KM, Didier JE, Klinedinst DB, Doctor JS, Goldstein AS, Wilkes GL, Beckman EJ, Hollinger JO. Acta Biomater. 2005;1:471. doi: 10.1016/j.actbio.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 93.Borcan F, Soica CM, Ganta S, Amiji MM, Dehelean CA, Munteanu MF. Chem Cent J. 2012;6:87. doi: 10.1186/1752-153X-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bonzani IC, Adhikari R, Houshyar S, Mayadunne R, Gunatillake P, Stevens MM. Biomaterials. 2007;28:423. doi: 10.1016/j.biomaterials.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 95.Haraguchi K, Takehisa T. Adv Mater. 2002;14:1120. [Google Scholar]
- 96.Djonlagić J, Žugić D, Petrović Z. J Appl Polym Sci. 2012;124:3024. [Google Scholar]
- 97.Okumura Y, Ito K. Adv Mater. 2001;13:485. [Google Scholar]
- 98.Gong JP. Soft Matter. 2010;6:2583. [Google Scholar]
- 99.Haque MA, Kurokawa T, Kamita G, Gong JP. Macromolecules. 2011;44:8916. [Google Scholar]
- 100.Sun JY, Zhao X, Illeperuma WRK, Chaudhuri O, Oh KH, Mooney DJ, Vlassak JJ, Suo Z. Nature. 2012;489:133. doi: 10.1038/nature11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Henderson KJ, Zhou TC, Otim KJ, Shull KR. Macromolecules. 2010;43:6193. [Google Scholar]
- 102.Camci-Unal G, Zorlutuna P, Khademhosseini A. In: Biofabrication. Forgacs G, Sun W, editors. Elsevier Inc; Oxford, UK: 2013. [Google Scholar]
- 103.Corrales T, Catalina F, Peinado C, Allen N. J Photochem Photobiol, A. 2003;159:103. [Google Scholar]
- 104.Ferreira P, Coelho JFJ, Almeida JF, Gil MH. In: Biomedical Engineering -Frontiers and Challenges. Fazel-Rezai R, editor. Vol. 3. InTech; pp. 55–74. [Google Scholar]
- 105.Rivest C, Morrison DWG, Ni B, Rubin J, Yadav V, Mahdavi A, Karp JM, Khademhosseini A. J Mech Mater Struct. 2007;2:1103. [Google Scholar]
- 106.Liu VA, Bhatia SN. Biomed Microdevices. 2002;4:257. [Google Scholar]
- 107.Camci-Unal G, Cuttica D, Annabi N, Demarchi D, Khademhosseini A. Biomacromolecules. 2013;14:1085. doi: 10.1021/bm3019856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gerecht S, Burdick JA, Ferreira LS, Townsend SA, Langer R, Vunjak-Novakovic G. Proc Natl Acad Sci USA. 2007;104:11298. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhong C, Wu J, Reinhart-King CA, Chu CC. Acta Biomater. 2010;6:3908. doi: 10.1016/j.actbio.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 110.Koh WG, Itle LJ, Pishko MV. Anal Chem. 2003;75:5783. doi: 10.1021/ac034773s. [DOI] [PubMed] [Google Scholar]
- 111.Nemir S, Hayenga HN, West JL. Biotechnol Bioeng. 2010;105:636. doi: 10.1002/bit.22574. [DOI] [PubMed] [Google Scholar]
- 112.Stephens-Altus JS, Sundelacruz P, Rowland ML, West JL. J Biomed Mater Res, Part A. 2011;98A:167. doi: 10.1002/jbm.a.33095. [DOI] [PubMed] [Google Scholar]
- 113.Bryant SJ, Cuy JL, Hauch KD, Ratner BD. Biomaterials. 2007;28:2978. doi: 10.1016/j.biomaterials.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chou AI, Akintoye SO, Nicoll SB. Osteoarthritis Cartilage. 2009;17:1377. doi: 10.1016/j.joca.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meyvis T, De Smedt S, Stubbe B, Hennink W, Demeester J. Pharm Res. 2001;18:1593. doi: 10.1023/a:1013038716373. [DOI] [PubMed] [Google Scholar]
- 116.De Paepe I, Declercq H, Cornelissen M, Schacht E. Polym Int. 2002;51:867. [Google Scholar]
- 117.Nilasaroya A, Poole-Warren LA, Whitelock JM, Martens PJ. Biomaterials. 2008;29:4658. doi: 10.1016/j.biomaterials.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 118.Burdick JA, Chung C, Jia XQ, Randolph MA, Langer R. Biomacromolecules. 2005;6:386. doi: 10.1021/bm049508a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Camci-Unal G, Nichol JW, Bae H, Tekin H, Bischoff J, Khademhosseini A. J Tissue Eng Regener Med. 2013;7:337. doi: 10.1002/term.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Camci-Unal G, Aubin H, Ahari AF, Bae H, Nichol JW, Khademhosseini A. Soft Matter. 2010;6:5120. doi: 10.1039/c0sm00508h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hjortnaes J, Camci-Unal G, Kluin J, Schoen FJ, Khademhosseini A, Aikawa E. Cardiovasc Pathol. 2013;22:e38. [Google Scholar]
- 122.Zhou YS, Ma GP, Shi SQ, Yang DZ, Nie J. Int J Biol Macromol. 2011;48:408. doi: 10.1016/j.ijbiomac.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 123.Gaudet ID, Shreiber DI. Biointerphases. 2012:7. doi: 10.1007/s13758-012-0025-y. [DOI] [PMC free article] [PubMed]
- 124.Hancock MJ, Piraino F, Camci-Unal G, Rasponi M, Khademhosseini A. Biomaterials. 2011;32:6493. doi: 10.1016/j.biomaterials.2011.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Piraino F, Camci-Unal G, Hancock MJ, Rasponi M, Khademhosseini A. Lab Chip. 2012;12:659. doi: 10.1039/c2lc20515g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hosseini V, Ahadian S, Ostrovidov S, Camci-Unal G, Chen S, Kaji H, Ramalingam M, Khademhosseini A. Tissue Eng, Part A. 2012;18:2453. doi: 10.1089/ten.tea.2012.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Nat Methods. 2006;3:369. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 128.Aubin H, Nichol JW, Hutson CB, Bae H, Sieminski AL, Cropek DM, Akhyari P, Khademhosseini A. Biomaterials. 2010;31:6941. doi: 10.1016/j.biomaterials.2010.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guvendiren M, Burdick JA. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 130.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jay SM, Saltzman WM. Nat Biotechnol. 2009;27:543. doi: 10.1038/nbt0609-543. [DOI] [PubMed] [Google Scholar]
- 132.Wong DY, Griffin DR, Reed J, Kasko AM. Macromolecules. 2010;43:2824. [Google Scholar]
- 133.Kloxin AM, Tibbitt MW, Anseth KS. Nat Protocols. 2010;5:1867. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mamada A, Tanaka T, Kungwatchakun D, Irie M. Macromolecules. 1990;23:1517. [Google Scholar]
- 135.Peng K, Tomatsu I, van den Broek B, Cui C, Korobko AV, van Noort J, Meijer AH, Spaink HP, Kros A. Soft Matter. 2011;7:4881. [Google Scholar]
- 136.Klinger D, Landfester K. Soft Matter. 2011;7:1426. [Google Scholar]
- 137.Fairbanks BD, Singh SP, Bowman CN, Anseth KS. Macromolecules. 2011;44:2444. doi: 10.1021/ma200202w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Griffin DR, Kasko AM. J Am Chem Soc. 2012;134:13103. doi: 10.1021/ja305280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ercole F, Thissen H, Tsang K, Evans RA, Forsythe JS. Macromolecules. 2012;45:8387. [Google Scholar]
- 140.Griffin DR, Kasko AM. ACS Macro Lett. 2012;1:1330. doi: 10.1021/mz300366s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Griffin DR, Schlosser JL, Lam SF, Nguyen TH, Maynard HD, Kasko AM. Biomacromolecules. 2013;14:1199. doi: 10.1021/bm400169d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Narayanan RP, Melman G, Letourneau NJ, Mendelson NL, Melman A. Biomacromolecules. 2012;13:2465. doi: 10.1021/bm300707a. [DOI] [PubMed] [Google Scholar]
- 143.Kloxin AM, Tibbitt MW, Kasko AM, Fairbairn JA, Anseth KS. Adv Mater. 2010;22:61. doi: 10.1002/adma.200900917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.DeForest CA, Anseth KS. Nat Chem. 2011;3:925. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schexnailder P, Schmidt G. Colloid Polym Sci. 2009;287:1. [Google Scholar]
- 146.Thomas V, Namdeo M, Murali Mohan Y, Bajpai SK, Bajpai M. J Macromol Sci A. 2007;45:107. [Google Scholar]
- 147.Ramakrishna S, Mayer J, Wintermantel E, Leong KW. Compos Sci Technol. 2001;61:1189. [Google Scholar]
- 148.Augst AD, Kong HJ, Mooney DJ. Macromol Biosci. 2006;6:623. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- 149.Hahn MS, Teply BA, Stevens MM, Zeitels SM, Langer R. Biomaterials. 2006;27:1104. doi: 10.1016/j.biomaterials.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 150.Sawhney AS, Pathak CP, Hubbell JA. Biomaterials. 1993;14:1008. doi: 10.1016/0142-9612(93)90194-7. [DOI] [PubMed] [Google Scholar]
- 151.Baruch L, Machluf M. Biopolymers. 2006;82:570. doi: 10.1002/bip.20509. [DOI] [PubMed] [Google Scholar]
- 152.Hua S, Ma H, Li X, Yang H, Wang A. Int J Biol Macromol. 2010;46:517. doi: 10.1016/j.ijbiomac.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 153.Wang Q, Zhang J, Wang A. Carbohyd Polym. 2009;78:731. [Google Scholar]
- 154.Drumheller PD, Hubbell JA. J Biomed Mater Res. 1995;29:207. doi: 10.1002/jbm.820290211. [DOI] [PubMed] [Google Scholar]
- 155.Bauer BJ, Briber RM. The Effect of Crosslink Density of Phase Separation in Interpenetrating Polymer Networks. Technomic; Lancaster, PA, USA: 1994. [Google Scholar]
- 156.Jia X, Kiick KL. Macromol Biosci. 2009;9:140. doi: 10.1002/mabi.200800284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ifkovits JL, Burdick JA. Tissue Eng. 2007;13:2369. doi: 10.1089/ten.2007.0093. [DOI] [PubMed] [Google Scholar]
- 158.Leach JB, Schmidt CE. Biomaterials. 2005;26:125. doi: 10.1016/j.biomaterials.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 159.Lutolf MP, Hubbell JA. Biomacromolecules. 2003;4:713. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 160.Jin R, Moreira Teixeira LS, Krouwels A, Dijkstra PJ, van Blitterswijk CA, Karperien M, Feijen J. Acta Biomater. 2010;6:1968. doi: 10.1016/j.actbio.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 161.Zhang HJ, Xin Y, Yan Q, Zhou LL, Peng L, Yuan JY. Macromol Rapid Commun. 2012;33:1952. doi: 10.1002/marc.201200439. [DOI] [PubMed] [Google Scholar]
- 162.Xu J, Filion TM, Prifti F, Song J. Chem Asian J. 2011;6:2730. doi: 10.1002/asia.201100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ossipov DA, Hilborn J. Macromolecules. 2006;39:1709. [Google Scholar]
- 164.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc Natl Acad Sci USA. 2007;104:16793. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Agard NJ, Prescher JA, Bertozzi CR. J Am Chem Soc. 2004;126:15046. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 166.DeForest CA, Polizzotti BD, Anseth KS. Nat Mater. 2009;8:659. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Aylsworth JW. 1 111 284. US Patent. 1914
- 168.Myung D, Waters D, Wiseman M, Duhamel PE, Noolandi J, Ta CN, Frank CW. Polym Adv Technol. 2008;19:647. doi: 10.1002/pat.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Thomas DA, Sperling LH. Interpenetrating Polymer Networks. Vol. 2. Academic Press; New York: 1978. [Google Scholar]
- 170.Gong JP, Katsuyama Y, Kurokawa T, Osada Y. Adv Mater. 2003;15:1155. [Google Scholar]
- 171.Xing Z, Wang C, Yan J, Zhang L, Li L, Zha L. Soft Matter. 2011;7:7992. [Google Scholar]
- 172.Shin BC, Jhon MS, Lee HB, Yuk SH. Eur Polym J. 1998;34:1675. [Google Scholar]
- 173.Lin J, Tang Q, Hu D, Sun X, Li Q, Wu J. Colloids Surf A. 2009;346:177. [Google Scholar]
- 174.De M, Ghosh PS, Rotello VM. Adv Mater. 2008;20:4225. [Google Scholar]
- 175.Kestell AE, DeLorey GT. Nanoparticles: Properties, Classification, Characterization and Fabrication. Nova Science; New York: 2010. [Google Scholar]
- 176.Rotello V. Nanoparticles: Building Blocks for Nanotechnology. Springer; New York: 2003. [Google Scholar]
- 177.Rong MZ, Zhang MQ, Ruan WH. Mater Sci Technol. 2006;22:787. [Google Scholar]
- 178.Usuki A, Kojima Y, Kawasumi M, Okada A, Fukushima Y, Kurauchi T, Kamigaito O. J Mater Res. 1993;8:1179. [Google Scholar]
- 179.Usuki A, Hasegawa N, Kato M, Kobayashi S. Inorganic Polymeric Nanocomposites and Membranes. Springer; Berlin Heidelberg: 2005. p. 135. [Google Scholar]
- 180.Liu P. Appl Clay Sci. 2007;38:64. [Google Scholar]
- 181.Kokabi M, Sirousazar M, Hassan ZM. Eur Polym J. 2007;43:773. [Google Scholar]
- 182.Xiao L, Liu C, Zhu J, Pochan DJ, Jia X. Soft Matter. 2010;6:5293. doi: 10.1039/C0SM00511H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Desai PN, Yuan Q, Yang H. Biomacromolecules. 2010;11:666. doi: 10.1021/bm901240g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Daniel MC, Astruc D. Chem Rev. 2003;104:293. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 185.Pardo-Yissar V, Gabai R, Shipway AN, Bourenko T, Willner I. Adv Mater. 2001;13:1320. [Google Scholar]
- 186.Mohan YM, Lee K, Premkumar T, Geckeler KE. Polymer. 2007;48:158. [Google Scholar]
- 187.Pankhurst QA, Connolly J, Jones SK, Dobson J. J Phys D Appl Phys. 2003;36:R167. [Google Scholar]
- 188.Mornet S, Vasseur S, Grasset F, Duguet E. J Mater Chem. 2004;14:2161. [Google Scholar]
- 189.Yi DK, Selvan ST, Lee SS, Papaefthymiou GC, Kundaliya D, Ying JY. J Am Chem Soc. 2005;127:4990. doi: 10.1021/ja0428863. [DOI] [PubMed] [Google Scholar]
- 190.Kumar CSSR, Mohammad F. Adv Drug Delivery Rev. 2011;63:789. doi: 10.1016/j.addr.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Rudolf H, Silvio D, Robert M, Matthias Z. J Phys: Condens Matter. 2006;18:S2919. [Google Scholar]
- 192.Ganta S, Devalapally H, Shahiwala A, Amiji M. J Controlled Release. 2008;126:187. doi: 10.1016/j.jconrel.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 193.Hoare T, Santamaria J, Goya GF, Irusta S, Lin D, Lau S, Padera R, Langer R, Kohane DS. Nano Lett. 2009;9:3651. doi: 10.1021/nl9018935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Meenach SA, Hilt JZ, Anderson KW. Acta Biomater. 2010;6:1039. doi: 10.1016/j.actbio.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 195.Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim Sb, Nikkhah M, Khabiry M, Azize M, Kong J, Wan K-t, Palacios T, Dokmeci MR, Bae H, Tang X, Khademhosseini A. ACS Nano. 2013;7:2369. doi: 10.1021/nn305559j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cha C, Shin SR, Annabi N, Dokmeci MR, Khademhosseini A. ACS Nano. 2013;7:2891. doi: 10.1021/nn401196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ajayan PM, Zhou OZ. Carbon Nanotubes: Synthesis, Structure, Properties, and Applications. Springer-Verlag; New York: 2001. p. 391. [Google Scholar]
- 198.Krüger A. Carbon Materials and Nanotechnology. Wiley-VCH; Weinheim, Germany: 2010. [Google Scholar]
- 199.Tong X, Zheng JG, Lu YC, Zhang ZF, Cheng HM. Mater Lett. 2007;61:1704. [Google Scholar]
- 200.Zhang L, Wang Z, Xu C, Li Y, Gao J, Wang W, Liu Y. J Mater Chem. 2011;21:10399. [Google Scholar]
- 201.Zhang N, Li R, Zhang L, Chen H, Wang W, Liu Y, Wu T, Wang X, Wang W, Li Y, Zhao Y, Gao J. Soft Matter. 2011;7:7231. [Google Scholar]
- 202.Peppas NA, Kavimandan NJ. Eur J Pharm Sci. 2006;29:183. doi: 10.1016/j.ejps.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 203.Sanchez-Chavez IY, Martinez-Chapa SO, Peppas NA. AIChE Journal. 2008;54:1901. [Google Scholar]
- 204.Thomas JB, Tingsanchali JH, Rosales AM, Creecy CM, McGinity JW, Peppas NA. Polymer. 2007;48:5042. doi: 10.1016/j.polymer.2007.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schoener CA, Hutson HN, Peppas NA. Polym Int. 2012;61:874. doi: 10.1002/pi.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Caldorera-Moore M, Peppas NA. Adv Drug Delivery Rev. 2009;61:1391. doi: 10.1016/j.addr.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Peyton SR, Ghajar CM, Khatiwala CB, Putnam AJ. Cell Biochem Biophys. 2007;47:300. doi: 10.1007/s12013-007-0004-y. [DOI] [PubMed] [Google Scholar]
- 208.Ghosh K, Pan Z, Guan E, Ge SR, Liu YJ, Nakamura T, Ren XD, Rafailovich M, Clark RAF. Biomaterials. 2007;28:671. doi: 10.1016/j.biomaterials.2006.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Smith HW, Marshall CJ. Nat Rev Mol Cell Biol. 2010;11:23. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 210.Galaev IY, Mattiasson B. Trends Biotechnol. 1999;17:335. doi: 10.1016/s0167-7799(99)01345-1. [DOI] [PubMed] [Google Scholar]
- 211.Jeong B, Gutowska A. Trends Biotechnol. 2002;20:305. doi: 10.1016/s0167-7799(02)01962-5. [DOI] [PubMed] [Google Scholar]
- 212.de Las Heras Alarcon C, Pennadam S, Alexander C. Chem Soc Rev. 2005;34:276. doi: 10.1039/b406727d. [DOI] [PubMed] [Google Scholar]
- 213.Rybtchinski B. ACS Nano. 2011;5:6791. doi: 10.1021/nn2025397. [DOI] [PubMed] [Google Scholar]
- 214.Krieg E, Rybtchinski B. Chem Eur J. 2011;17:9016. doi: 10.1002/chem.201100809. [DOI] [PubMed] [Google Scholar]
- 215.Silva D, Natalello A, Sanii B, Vasita R, Saracino G, Zuckermann RN, Doglia SM, Gelain F. Nanoscale. 2013;5:704. doi: 10.1039/c2nr32656f. [DOI] [PubMed] [Google Scholar]
- 216.Gu J, Xia F, Wu Y, Qu X, Yang Z, Jiang L. J Controlled Release. 2007;117:396. doi: 10.1016/j.jconrel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 217.Kopecek J, Yang JY. Polym Int. 2007;56:1078. [Google Scholar]
- 218.Peng T, Cheng YL. J Appl Polym Sci. 1998;70:2133. [Google Scholar]
- 219.Lue SJ, Hsu JJ, Chen CH, Chen BC. J Membr Sci. 2007;301:142. [Google Scholar]
- 220.Jeong B, Kim SW, Bae YH. Adv Drug Delivery Rev. 2002;54:37. doi: 10.1016/s0169-409x(01)00242-3. [DOI] [PubMed] [Google Scholar]
- 221.Ravichandran R, Sundarrajan S, Venugopal JR, Mukherjee S, Ramakrishna S. Macromol Biosci. 2012;12:286. doi: 10.1002/mabi.201100325. [DOI] [PubMed] [Google Scholar]
- 222.Behl M, Lendlein A. Mater Today. 2007;10:20. [Google Scholar]
- 223.Berger J, Reist M, Mayer JM, Felt O, Peppas NA, Gurny R. Eur J Pharm Biopharm. 2004;57:19. doi: 10.1016/s0939-6411(03)00161-9. [DOI] [PubMed] [Google Scholar]
- 224.Ran D, Wang YZ, Jia XP, Nie C. Anal Chim Acta. 2012;723:45. doi: 10.1016/j.aca.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 225.Bai T, Han YJ, Zhang P, Wang W, Liu WG. Soft Matter. 2012;8:6846. [Google Scholar]
- 226.Zhang XB, Pint CL, Lee MH, Schubert BE, Jamshidi A, Takei K, Ko H, Gillies A, Bardhan R, Urban JJ, Wu M, Fearing R, Javey A. Nano Lett. 2011;11:3239. doi: 10.1021/nl201503e. [DOI] [PubMed] [Google Scholar]
- 227.Ionov L. J Mater Chem. 2010;20:3382. [Google Scholar]
- 228.Fuhrer R, Athanassiou EK, Luechinger NA, Stark WJ. Small. 2009;5:383. doi: 10.1002/smll.200801091. [DOI] [PubMed] [Google Scholar]
- 229.Hribar KC, Lee MH, Lee D, Burdick JA. ACS Nano. 2011;5:2948. doi: 10.1021/nn103575a. [DOI] [PubMed] [Google Scholar]
- 230.Censi R, Vermonden T, van Steenbergen MJ, Deschout H, Braeckmans K, De Smedt SC, van Nostrum CF, di Martino P, Hennink WE. J Controlled Release. 2009;140:230. doi: 10.1016/j.jconrel.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 231.Vermonden T, Jena SS, Barriet D, Censi R, van der Gucht J, Hennink WE, Siegel RA. Macromolecules. 2009;43:782. doi: 10.1021/ma902186e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Censi R, Di Martino P, Vermonden T, Hennink WE. J Controlled Release. 2012;161:680. doi: 10.1016/j.jconrel.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 233.Ozaydin-Ince G, Gleason KK, Demirel MC. Soft Matter. 2011;7:638. [Google Scholar]
- 234.Wang L, Shansky J, Borselli C, Mooney D, Vandenburgh H. Tissue Eng, Part A. 2012;18:2000. doi: 10.1089/ten.tea.2011.0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bencherif SA, Sands RW, Bhatta D, Arany P, Verbeke CS, Edwards DA, Mooney DJ. Proc Natl Acad Sci USA. 2012;109:19590. doi: 10.1073/pnas.1211516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lu HD, Charati MB, Kim IL, Burdick JA. Biomaterials. 2012;33:2145. doi: 10.1016/j.biomaterials.2011.11.076. [DOI] [PubMed] [Google Scholar]
- 237.Glassman MJ, Chan J, Olsen BD. Adv Funct Mater. 2013;23:1182. doi: 10.1002/adfm.201202034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Huang HZ, Shi JS, Laskin J, Liu ZY, McVey DS, Sun XZS. Soft Matter. 2011;7:8905. [Google Scholar]
- 239.Hao JK, Weiss RA. ACS Macro Lett. 2013;2:86. doi: 10.1021/mz3006389. [DOI] [PubMed] [Google Scholar]
- 240.Shanmuganathan K, Capadona JR, Rowan SJ, Weder C. J Mater Chem. 2010;20:180. doi: 10.1021/am9006337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kishore V, Uquillas JA, Dubikovsky A, Alshehabat MA, Snyder PW, Breur GJ, Akkus O. J Biomed Mater Res, Part B. 2012;100B:400. doi: 10.1002/jbm.b.31962. [DOI] [PubMed] [Google Scholar]
- 242.Li JH, Viveros JA, Wrue MH, Anthamatten M. Adv Mater. 2007;19:2851. [Google Scholar]
- 243.Small W, Buckley PR, Wilson TS, Benett WJ, Hartman J, Saloner D, Maitland DJ. IEEE Trans Biomed Eng. 2007;54:1157. doi: 10.1109/TBME.2006.889771. [DOI] [PubMed] [Google Scholar]
- 244.Small W, Metzger MF, Wilson TS, Maitland DJ. IEEE J Sel Top Quantum Electron. 2005;11:892. [Google Scholar]
- 245.Baer G, Wilson T, Maitland D, Matthews D. J Invest Med. 2006;54:S162. [Google Scholar]
- 246.Yakacki CM, Shandas R, Lanning C, Rech B, Eckstein A, Gall K. Biomaterials. 2007;28:2255. doi: 10.1016/j.biomaterials.2007.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ortega JM, Small W, Wilson TS, Benett WJ, Loge JM, Maitland DJ. IEEE Trans Biomed Eng. 2007;54:1722. doi: 10.1109/TBME.2007.892927. [DOI] [PubMed] [Google Scholar]
- 248.Kratz K, Voigt U, Lendlein A. Adv Funct Mater. 2012;22:3057. [Google Scholar]
- 249.Chiu YL, Chen SC, Su CJ, Hsiao CW, Chen YM, Chen HL, Sung HW. Biomaterials. 2009;30:4877. doi: 10.1016/j.biomaterials.2009.05.052. [DOI] [PubMed] [Google Scholar]
- 250.Guvendiren M, Lu HD, Burdick JA. Soft Matter. 2012;8:260. [Google Scholar]
- 251.Tamayol A, Akbari M, Annabi N, Paul A, Khademhosseini A, Juncker D. Biotechnol Adv. 2012;31:669. doi: 10.1016/j.biotechadv.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zorlutuna P, Annabi N, Camci-Unal G, Nikkhah M, Cha JM, Nichol JW, Manbachi A, Bae H, Chen S, Khademhosseini A. Adv Mater. 2012;24:1782. doi: 10.1002/adma.201104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Billiet T, Vandenhaute M, Schelfhout J, Van Vlierberghe S, Dubruel P. Biomaterials. 2012 doi: 10.1016/j.biomaterials.2012.04.050. [DOI] [PubMed] [Google Scholar]
- 254.Melchels FP, Domingos MA, Klein TJ, Malda J, Bartolo PJ, Hutmacher DW. Prog Polym Sci. 2012;37:1079. [Google Scholar]
- 255.Gauvin R, Parenteau-Bareil R, Dokmeci MR, Merryman WD, Khademhosseini A. Wiley Interdiscip Rev: Nanomed Nanobiotechnol. 2012;4:235. doi: 10.1002/wnan.171. [DOI] [PubMed] [Google Scholar]
- 256.Selimović Š, Oh J, Bae H, Dokmeci M, Khademhosseini A. Polymers. 2012;4:1554. doi: 10.3390/polym4031554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Tsang VL, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN. FASEB Journal. 2007;21:790. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- 258.Albrecht DR, Tsang VL, Sah RL, Bhatia SN. Lab Chip. 2005;5:111. doi: 10.1039/b406953f. [DOI] [PubMed] [Google Scholar]
- 259.Lin-Gibson S, Bencherif S, Cooper JA, Wetzel SJ, Antonucci JM, Vogel BM, Horkay F, Washburn NR. Biomacromolecules. 2004;5:1280. doi: 10.1021/bm0498777. [DOI] [PubMed] [Google Scholar]
- 260.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ovsianikov A, Deiwick A, Van Vlierberghe S, Pflaum M, Wilhelmi M, Dubruel P, Chichkov B. Materials. 2011;4:288. doi: 10.3390/ma4010288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Khademhosseini A, Eng G, Yeh J, Fukuda J, Blumling J, Langer R, Burdick JA. J Biomed Mater Res, Part A. 2006;79:522. doi: 10.1002/jbm.a.30821. [DOI] [PubMed] [Google Scholar]
- 263.Annabi N, Selimovic S, Cox JPA, Ribas J, Bakooshli MA, Heintze D, Weiss A, Cropek DM, Khademhosseini A. Lab Chip. 2013 doi: 10.1039/c3lc50252j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wylie RG, Ahsan S, Aizawa Y, Maxwell KL, Morshead CM, Shoichet MS. Nat Mater. 2011;10:799. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- 265.Khetan S, Burdick JA. Biomaterials. 2010;31:8228. doi: 10.1016/j.biomaterials.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 266.Chiu YC, Larson JC, Perez-Luna VH, Brey EM. Chem Mater. 2009;21:1677. [Google Scholar]
- 267.Cuchiara MP, Allen AC, Chen TM, Miller JS, West JL. Biomaterials. 2010;31:5491. doi: 10.1016/j.biomaterials.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 268.Wang J, Chen L, Zhao Y, Guo G, Zhang R. J Mater Sci: Mater Med. 2009;20:583. doi: 10.1007/s10856-008-3593-0. [DOI] [PubMed] [Google Scholar]
- 269.He J, Mao M, Liu Y, Shao J, Jin Z, Li D. Adv Healthcare Mater. 2013;8:1108. doi: 10.1002/adhm.201200404. [DOI] [PubMed] [Google Scholar]
- 270.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA. Proc Natl Acad Sci USA. 2012;109:9342. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen1 DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat Mater. 2012;11:268. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Park JH, Chung BG, Lee WG, Kim J, Brigham MD, Shim J, Lee S, Hwang CM, Durmus NG, Demirci U. Biotechnol Bioeng. 2010;106:138. doi: 10.1002/bit.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Wong KH, Chan JM, Kamm RD, Tien J. Annu Rev Biomed Eng. 2012;14:205. doi: 10.1146/annurev-bioeng-071811-150052. [DOI] [PubMed] [Google Scholar]
- 274.Bellan LM, Pearsall M, Cropek DM, Langer R. Adv Mater. 2012;24:5187. doi: 10.1002/adma.201200810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Golden AP, Tien J. Lab Chip. 2007;7:720. doi: 10.1039/b618409j. [DOI] [PubMed] [Google Scholar]
- 276.Stratakis E, Ranella A, Farsari M, Fotakis C. Prog Quantum Electron. 2009;33:127. [Google Scholar]
- 277.Khripin CY, Brinker CJ, Kaehr B. Soft Matter. 2010;6:2842. [Google Scholar]
- 278.Melchels FP, Feijen J, Grijpma DW. Biomaterials. 2010;31:6121. doi: 10.1016/j.biomaterials.2010.04.050. [DOI] [PubMed] [Google Scholar]
- 279.Lin H, Zhang D, Alexander PG, Yang G, Tan J, Cheng AW-M, Tuan RS. Biomaterials. 2012 doi: 10.1016/j.biomaterials.2012.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Curley JL, Jennings SR, Moore MJ. J Visualized Exp. 2011 doi: 10.3791/2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Gauvin R, Chen YC, Lee JW, Soman P, Zorlutuna P, Nichol JW, Bae H, Chen S, Khademhosseini A. Biomaterials. 2012;33:3824. doi: 10.1016/j.biomaterials.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Laza SC, Polo M, Neves AA, Cingolani R, Camposeo A, Pisignano D. Adv Mater. 2012;24:1304. doi: 10.1002/adma.201103357. [DOI] [PubMed] [Google Scholar]
- 283.Claeyssens F, Hasan EA, Gaidukeviciute A, Achilleos DS, Ranella A, Reinhardt C, Ovsianikov A, Shizhou X, Fotakis C, Vamvakaki M. Langmuir. 2009;25:3219. doi: 10.1021/la803803m. [DOI] [PubMed] [Google Scholar]
- 284.Aprile V, Eaton S, Laganà M, Cerullo G, Raimondi M, Osellame R. presented at SPIE LASE; 2012; San Francisco, CA, USA. [Google Scholar]
- 285.Ovsianikov A, Li Z, Torgersen J, Stampfl J, Liska R. Adv Funct Mater. 2012;22:3429. [Google Scholar]
- 286.Atala A, Kasper FK, Mikos AG. Sci Transl Med. 2012;4:160rv12. doi: 10.1126/scitranslmed.3004890. [DOI] [PubMed] [Google Scholar]
- 287.McAllister TN, Maruszewski M, Garrido SA, Wystrychowski W, Dusserre N, Marini A, Zagalski K, Fiorillo A, Avila H, Manglano X, Antonelli J, Kocher A, Zembala M, Cierpka L, de la Fuente LM, L’Heureux N. Lancet. 2009;373:1440. doi: 10.1016/S0140-6736(09)60248-8. [DOI] [PubMed] [Google Scholar]
- 288.Derby B. Science. 2012;338:921. doi: 10.1126/science.1226340. [DOI] [PubMed] [Google Scholar]
- 289.Billiet T, Vandenhaute M, Schelfhout J, Van Vlierberghe S, Dubruel P. Biomaterials. 2012;33:6020. doi: 10.1016/j.biomaterials.2012.04.050. [DOI] [PubMed] [Google Scholar]
- 290.Chang CC, Boland ED, Williams SK, Hoying JB. J Biomed Mater Res, Part B. 2011;98:160. doi: 10.1002/jbm.b.31831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Fedorovich NE, Alblas J, Hennink WE, Oner FC, Dhert WJ. Trends Biotechnol. 2011;29:601. doi: 10.1016/j.tibtech.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 292.Guillemot F, Souquet A, Catros S, Guillotin B. Nanomedicine (Lond) 2010;5:507. doi: 10.2217/nnm.10.14. [DOI] [PubMed] [Google Scholar]
- 293.Tasoglu S, Demirci U. Trends Biotechnol. 2013;31:10. doi: 10.1016/j.tibtech.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Klebe RJ. Exp Cell Res. 1988;179:362. doi: 10.1016/0014-4827(88)90275-3. [DOI] [PubMed] [Google Scholar]
- 295.Moon S, Kim YG, Dong L, Lombardi M, Haeggstrom E, Jensen RV, Hsiao LL, Demirci U. PloS One. 2011;6:e17455. doi: 10.1371/journal.pone.0017455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Faulkner-Jones A, Greenhough S, King JA, Gardner J, Courtney A, Shu W. Biofabrication. 2013;5:015013. doi: 10.1088/1758-5082/5/1/015013. [DOI] [PubMed] [Google Scholar]
- 297.Miller ED, Li K, Kanade T, Weiss LE, Walker LM, Campbell PG. Biomaterials. 2011;32:2775. doi: 10.1016/j.biomaterials.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Matsusaki M, Sakaue K, Kadowaki K, Akashi M. Adv Healthcare Mater. 2013;2:534. doi: 10.1002/adhm.201200299. [DOI] [PubMed] [Google Scholar]
- 299.Xu T, Binder KW, Albanna MZ, Dice D, Zhao W, Yoo JJ, Atala A. Biofabrication. 2013;5:015001. doi: 10.1088/1758-5082/5/1/015001. [DOI] [PubMed] [Google Scholar]
- 300.Hanson Shepherd JN, Parker ST, Shepherd RF, Gillette MU, Lewis JA, Nuzzo RG. Adv Funct Mater. 2011;21:47. doi: 10.1002/adfm.201001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Mannoor MS, Jiang Z, James T, Kong YL, Malatesta KA, Soboyejo WO, Verma N, Gracias DH, McAlpine MC. Nano Lett. 2013 doi: 10.1021/nl4007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Komuro N, Takaki S, Suzuki K, Citterio D. Anal Bioanal Chem. 2013;405:5785. doi: 10.1007/s00216-013-7013-z. [DOI] [PubMed] [Google Scholar]
- 303.Teichler A, Perelaer J, Schubert US. J Mater Chem. 2013;1:1910. [Google Scholar]
- 304.Park M, Im J, Shin M, Min Y, Park J, Cho H, Park S, Shim MB, Jeon S, Chung DY, Bae J, Park J, Jeong U, Kim K. Nat Nanotechnol. 2012;7:803. doi: 10.1038/nnano.2012.206. [DOI] [PubMed] [Google Scholar]
- 305.Park S, Wang G, Cho B, Kim Y, Song S, Ji Y, Yoon MH, Lee T. Nat Nanotechnol. 2012;7:438. doi: 10.1038/nnano.2012.81. [DOI] [PubMed] [Google Scholar]
- 306.Guillotin B, Souquet A, Catros S, Duocastella M, Pippenger B, Bellance S, Bareille R, Remy M, Bordenave L, Amedee J, Guillemot F. Biomaterials. 2010;31:7250. doi: 10.1016/j.biomaterials.2010.05.055. [DOI] [PubMed] [Google Scholar]
- 307.Catros S, Guillotin B, Bačáková M, Fricain JC, Guillemot F. Appl Surf Sci. 2011;257:5142. [Google Scholar]
- 308.Gaebel R, Ma N, Liu J, Guan J, Koch L, Klopsch C, Gruene M, Toelk A, Wang W, Mark P, Wang F, Chichkov B, Li W, Steinhoff G. Biomaterials. 2011;32:9218. doi: 10.1016/j.biomaterials.2011.08.071. [DOI] [PubMed] [Google Scholar]
- 309.Pirlo RK, Wu P, Liu J, Ringeisen B. Biotechnol Bioeng. 2012;109:262. doi: 10.1002/bit.23295. [DOI] [PubMed] [Google Scholar]
- 310.Michael S, Sorg H, Peck CT, Koch L, Deiwick A, Chichkov B, Vogt PM, Reimers K. PloS One. 2013;8:e57741. doi: 10.1371/journal.pone.0057741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Koch L, Deiwick A, Schlie S, Michael S, Gruene M, Coger V, Zychlinski D, Schambach A, Reimers K, Vogt PM, Chichkov B. Biotechnol Bioeng. 2012;109:1855. doi: 10.1002/bit.24455. [DOI] [PubMed] [Google Scholar]
- 312.Jakab K, Damon B, Neagu A, Kachurin A, Forgacs G. Biorheology. 2006;43:509. [PubMed] [Google Scholar]
- 313.Norotte C, Marga FS, Niklason LE, Forgacs G. Biomaterials. 2009;30:5910. doi: 10.1016/j.biomaterials.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Sun L, Parker ST, Syoji D, Wang X, Lewis JA, Kaplan DL. Adv Healthcare Mater. 2012;1:729. doi: 10.1002/adhm.201200057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Censi R, Schuurman W, Malda J, di Dato G, Burgisser PE, Dhert WJA, van Nostrum CF, di Martino P, Vermonden T, Hennink WE. Adv Funct Mater. 2011;21:1833. [Google Scholar]
- 316.Hansen CJ, Saksena R, Kolesky DB, Vericella JJ, Kranz SJ, Muldowney GP, Christensen KT, Lewis JA. Adv Mater. 2013;25:96. doi: 10.1002/adma.201203321. [DOI] [PubMed] [Google Scholar]
- 317.Lee W, Lee V, Polio S, Keegan P, Lee JH, Fischer K, Park JK, Yoo SS. Biotechnol Bioeng. 2010;105:1178. doi: 10.1002/bit.22613. [DOI] [PubMed] [Google Scholar]
- 318.Toohey KS, Sottos NR, Lewis JA, Moore JS, White SR. Nat Mater. 2007;6:581. doi: 10.1038/nmat1934. [DOI] [PubMed] [Google Scholar]
- 319.Wu W, DeConinck A, Lewis JA. Adv Mater. 2011;23:H178. doi: 10.1002/adma.201004625. [DOI] [PubMed] [Google Scholar]
- 320.Adams JJ, Duoss EB, Malkowski TF, Motala MJ, Ahn BY, Nuzzo RG, Bernhard JT, Lewis JA. Adv Mater. 2011;23:1335. doi: 10.1002/adma.201003734. [DOI] [PubMed] [Google Scholar]
- 321.Lorang DJ, Tanaka D, Spadaccini CM, Rose KA, Cherepy NJ, Lewis JA. Adv Mater. 2011;23:5055. doi: 10.1002/adma.201102411. [DOI] [PubMed] [Google Scholar]
- 322.Ahn BY, Duoss EB, Motala MJ, Guo X, Park SI, Xiong Y, Yoon J, Nuzzo RG, Rogers JA, Lewis JA. Science. 2009;323:1590. doi: 10.1126/science.1168375. [DOI] [PubMed] [Google Scholar]
- 323.Yeo WH, Kim YS, Lee J, Ameen A, Shi L, Li M, Wang S, Ma R, Jin SH, Kang Z, Huang Y, Rogers JA. Adv Mater. 2013;25:2773. doi: 10.1002/adma.201204426. [DOI] [PubMed] [Google Scholar]
- 324.Ahn BY, Lorang DJ, Lewis JA. Nanoscale. 2011;3:2700. doi: 10.1039/c1nr10048c. [DOI] [PubMed] [Google Scholar]
- 325.Takayama S, Ostuni E, LeDuc P, Naruse K, Ingber DE, Whitesides GM. Nature. 2001;411:1016. doi: 10.1038/35082637. [DOI] [PubMed] [Google Scholar]
- 326.Qasaimeh MA, Gervais T, Juncker D. Nat Commun. 2011;2:464. doi: 10.1038/ncomms1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Leng L, McAllister A, Zhang B, Radisic M, Günther A. Adv Mater. 2012;24:3650. doi: 10.1002/adma.201201442. [DOI] [PubMed] [Google Scholar]
- 328.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 2010;328:1662. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Nichol JW, Khademhosseini A. Soft Matter. 2009;5:1312. doi: 10.1039/b814285h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Xu S, Nie Z, Seo M, Lewis P, Kumacheva E, Stone HA, Garstecki P, Weibel DB, Gitlin I, Whitesides GM. Angew Chem. 2005;117:734. doi: 10.1002/anie.200462226. [DOI] [PubMed] [Google Scholar]
- 331.Dendukuri D, Tsoi K, Hatton TA, Doyle PS. Langmuir. 2005;21:2113. doi: 10.1021/la047368k. [DOI] [PubMed] [Google Scholar]
- 332.Nie Z, Xu S, Seo M, Lewis PC, Kumacheva E. J Am Chem Soc. 2005;127:8058. doi: 10.1021/ja042494w. [DOI] [PubMed] [Google Scholar]
- 333.Nisisako T, Torii T, Takahashi T, Takizawa Y. Adv Mater. 2006;18:1152. [Google Scholar]
- 334.Kang E, Jeong GS, Choi YY, Lee KH, Khademhosseini A, Lee SH. Nat Mater. 2011;10:877. doi: 10.1038/nmat3108. [DOI] [PubMed] [Google Scholar]
- 335.Anna SL, Bontoux N, Stone HA. Appl Phys Lett. 2003;82:364. [Google Scholar]
- 336.Kawakatsu T, Kikuchi Y, Nakajima M. J Am Oil Chem Soc. 1997;74:317. [Google Scholar]
- 337.Utada A, Lorenceau E, Link D, Kaplan P, Stone H, Weitz D. Science. 2005;308:537. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- 338.Chiou PY, Ohta AT, Wu MC. Nature. 2005;436:370. doi: 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- 339.Akbari M, Bahrami M, Sinton D. Microfluid Nanofluid. 2012;12:221. [Google Scholar]
- 340.Duncanson WJ, Lin T, Abate AR, Seiffert S, Shah RK, Weitz DA. Lab Chip. 2012;12:2135. doi: 10.1039/c2lc21164e. [DOI] [PubMed] [Google Scholar]
- 341.Romanowsky MB, Abate AR, Rotem A, Holtze C, Weitz DA. Lab Chip. 2012;12:802. doi: 10.1039/c2lc21033a. [DOI] [PubMed] [Google Scholar]
- 342.Utada A, Chu LY, Fernandez-Nieves A, Link D, Holtze C, Weitz D. MRS Bull. 2007;32:702. [Google Scholar]
- 343.Matsunaga YT, Morimoto Y, Takeuchi S. Adv Mater. 2011;23:H90. doi: 10.1002/adma.201004375. [DOI] [PubMed] [Google Scholar]
- 344.Kim SH, Jeon SJ, Jeong WC, Park HS, Yang SM. Adv Mater. 2008;20:4129. [Google Scholar]
- 345.Shepherd RF, Conrad JC, Rhodes SK, Link DR, Marquez M, Weitz DA, Lewis JA. Langmuir. 2006;22:8618. doi: 10.1021/la060759+. [DOI] [PubMed] [Google Scholar]
- 346.Oh HJ, Kim SH, Baek JY, Seong GH, Lee SH. J Micromech Microeng. 2006;16:285. [Google Scholar]
- 347.Rossow T, Heyman JA, Ehrlicher AJ, Langhoff A, Weitz DA, Haag R, Seiffert S. J Am Chem Soc. 2012;134:4983. doi: 10.1021/ja300460p. [DOI] [PubMed] [Google Scholar]
- 348.Dendukuri D, Pregibon DC, Collins J, Hatton TA, Doyle PS. Nat Mater. 2006;5:365. doi: 10.1038/nmat1617. [DOI] [PubMed] [Google Scholar]
- 349.Panda P, Ali S, Lo E, Chung BG, Hatton TA, Khademhosseini A, Doyle PS. Lab Chip. 2008;8:1056. doi: 10.1039/b804234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Dendukuri D, Gu SS, Pregibon DC, Hatton TA, Doyle PS. Lab Chip. 2007;7:818. doi: 10.1039/b703457a. [DOI] [PubMed] [Google Scholar]
- 351.Hwang DK, Oakey J, Toner M, Arthur JA, Anseth KS, Lee S, Zeiger A, Van Vliet KJ, Doyle PS. J Am Chem Soc. 2009;131:4499. doi: 10.1021/ja809256d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.McGuigan AP, Sefton MV. Proc Natl Acad Sci USA. 2006;103:11461. doi: 10.1073/pnas.0602740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Yeh J, Ling Y, Karp JM, Gantz J, Chandawarkar A, Eng G, Blumling J, III, Langer R, Khademhosseini A. Biomaterials. 2006;27:5391. doi: 10.1016/j.biomaterials.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 354.Chung SE, Park W, Shin S, Lee SA, Kwon S. Nat Mater. 2008;7:581. doi: 10.1038/nmat2208. [DOI] [PubMed] [Google Scholar]
- 355.Onoe H, Okitsu T, Itou A, Kato-Negishi M, Gojo R, Kiriya D, Sato K, Miura S, Iwanaga S, Kuribayashi-Shigetomi K, Matsunaga YT, Shimoyama Y, Takeuchi S. Nat Mater. 2013;12:584. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- 356.Hu M, Deng R, Schumacher KM, Kurisawa M, Ye H, Purnamawati K, Ying JY. Biomaterials. 2010;31:863. doi: 10.1016/j.biomaterials.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 357.Onoe H, Okitsu T, Itou A, Kato-Negishi M, Gojo R, Kiriya D, Sato K, Miura S, Iwanaga S, Kuribayashi-Shigetomi K, Matsunaga YT, Shimoyama Y, Takeuchi S. Nat Mater. 2013;12:584. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- 358.Lee KH, Shin SJ, Park Y, Lee SH. Small. 2009;5:1264. doi: 10.1002/smll.200801667. [DOI] [PubMed] [Google Scholar]
- 359.Tamayol A, Akbari M, Annabi N, Paul A, Khademhosseini A, Juncker D. Biotechnol Adv. 2012;31:669. doi: 10.1016/j.biotechadv.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Tamayol A, Bahrami M. Phys Rev. 2011;83:046314. doi: 10.1103/PhysRevE.83.046314. [DOI] [PubMed] [Google Scholar]
- 361.Tamayol A, Wong K, Bahrami M. Phys Rev. 2012;85:026318. doi: 10.1103/PhysRevE.85.026318. [DOI] [PubMed] [Google Scholar]
- 362.Moutos FT, Freed LE, Guilak F. Nat Mater. 2007;6:162. doi: 10.1038/nmat1822. [DOI] [PubMed] [Google Scholar]
- 363.Enea D, Henson F, Kew S, Wardale J, Getgood A, Brooks R, Rushton N. J Mater Sci: Mater Med. 2011:1. doi: 10.1007/s10856-011-4336-1. [DOI] [PubMed] [Google Scholar]
- 364.Puppi D, Dinucci D, Bartoli C, Mota C, Migone C, Dini F, Barsotti G, Carlucci F, Chiellini F. J Bioact Compat Polym. 2011;26:478. [Google Scholar]
- 365.Jayasinghe SN, Suter N. Biomicrofluidics. 2010;4:014106. doi: 10.1063/1.3328092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Lee BR, Lee KH, Kang E, Kim DS, Lee SH. Biomicrofluidics. 2011;5:022208. doi: 10.1063/1.3576903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Tuzlakoglu K, Alves CM, Mano JF, Reis RL. Macromol Biosci. 2004;4:811. doi: 10.1002/mabi.200300100. [DOI] [PubMed] [Google Scholar]
- 368.Leonor IB, Rodrigues MT, Gomes ME, Reis RL. J Tissue Eng Regener Med. 2011;5:104. doi: 10.1002/term.294. [DOI] [PubMed] [Google Scholar]
- 369.Pati F, Adhikari B, Dhara S. J Mater Sci: Mater Med. 2012:1. doi: 10.1007/s10856-012-4559-9. [DOI] [PubMed] [Google Scholar]
- 370.DeRosa K, Siriwardane M, Pfister B. IEEE 37th Annual Northeast Bioengineering Conf. (NEBEC); Troy, NY, USA. 2011. [Google Scholar]
- 371.Spinks GM, Shin SR, Wallace GG, Whitten PG, Kim SI, Kim SJ. Sens Actuators, B. 2006;115:678. [Google Scholar]
- 372.Landers R, Pfister A, Hübner U, John H, Schmelzeisen R, Mülhaupt R. Asian J Mater Sci. 2002;37:3107. [Google Scholar]
- 373.Kharaziha M, Nikkhah M, Shin S, Annabi N, Masoumi N, Gaharwar A, Unal G, Khademhosseini A. Biomaterials. 2013;34:6355. doi: 10.1016/j.biomaterials.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Ignatova M, Manolova N, Markova N, Rashkov I. Macromol Biosci. 2009;9:102. doi: 10.1002/mabi.200800189. [DOI] [PubMed] [Google Scholar]
- 375.Alves da Silva M, Martins A, Costa-Pinto A, Costa P, Faria S, Gomes M, Reis R, Neves N. Biomacromolecules. 2010;11:3228. doi: 10.1021/bm100476r. [DOI] [PubMed] [Google Scholar]
- 376.Bellan LM, Pearsall M, Cropek DM, Langer R. Adv Mater. 2012 doi: 10.1002/adma.201200810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Leong MF, Chan WY, Chian KS, Rasheed MZ, Anderson JM. J Biomed Mater Res, Part A. 2010;94:1141. doi: 10.1002/jbm.a.32795. [DOI] [PubMed] [Google Scholar]
- 378.Lee YH, Lee JH, An IG, Kim C, Lee DS, Lee YK, Nam JD. Biomaterials. 2005;26:3165. doi: 10.1016/j.biomaterials.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 379.Yokoyama Y, Hattori S, Yoshikawa C, Yasuda Y, Koyama H, Takato T, Kobayashi H. Mater Lett. 2009;63:754. [Google Scholar]
- 380.Simonet M, Schneider OD, Neuenschwander P, Stark WJ. Polym Eng Sci. 2007;47:2020. [Google Scholar]
- 381.woon Choi H, Johnson JK, Nam J, Farson DF, Lannutti J. J Laser Appl. 2007;19:225. [Google Scholar]
- 382.Baker BM, Shah RP, Silverstein AM, Esterhai JL, Burdick JA, Mauck RL. Proc Natl Acad Sci USA. 2012;109:14176. doi: 10.1073/pnas.1206962109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Elbert DL. Curr Opin Biotechnol. 2011;22:674. doi: 10.1016/j.copbio.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Luo H, Chen M, Wang X, Mei Y, Ye Z, Zhou Y, Tan WS. J Tissue Eng Regener Med. 2012 doi: 10.1002/term.1554. [DOI] [PubMed] [Google Scholar]
- 385.Fedorovich NE, Moroni L, Malda J, Alblas J, Blitterswijk CA, Dhert AWJ. In: Cell and Organ Printing. Ringeisen BR, Spargo BJ, Wu PK, editors. Springer; The Netherlands: 2010. p. 225. [Google Scholar]
- 386.Zamanian B, Masaeli M, Nichol JW, Khabiry M, Hancock MJ, Bae H, Khademhosseini A. Small. 2010;6:937. doi: 10.1002/smll.200902326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Liberski AR, Delaney JT, Schäfer H, Perelaer J, Schubert US. Macromol Biosci. 2011;11:1491. doi: 10.1002/mabi.201100086. [DOI] [PubMed] [Google Scholar]
- 388.Du Y, Lo E, Ali S, Khademhosseini A. Proc Natl Acad Sci USA. 2008;105:9522. doi: 10.1073/pnas.0801866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Du Y, Ghodousi M, Qi H, Haas N, Xiao W, Khademhosseini A. Biotechnol Bioeng. 2011;108:1693. doi: 10.1002/bit.23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Kachouie NN, Du Y, Bae H, Khabiry M, Ahari AF, Zamanian B, Fukuda J, Khademhosseini A. Organogenesis. 2010;6:234. doi: 10.4161/org.6.4.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Ghorbanian S. Masters of Engineering. McGill University; Montreal: 2010. [Google Scholar]
- 392.Fedorovich NE, De Wijn JR, Verbout AJ, Alblas J, Dhert WJA. Tissue Eng, Part A. 2008;14:127. doi: 10.1089/ten.a.2007.0158. [DOI] [PubMed] [Google Scholar]
- 393.Cohen DL, Malone E, Lipson H, Bonassar LJ. Tissue Eng. 2006;12:1325. doi: 10.1089/ten.2006.12.1325. [DOI] [PubMed] [Google Scholar]
- 394.Gaetani R, Doevendans PA, Metz CHG, Alblas J, Messina E, Giacomello A, Sluijter JPG. Biomaterials. 2012;33:1782. doi: 10.1016/j.biomaterials.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 395.Onoe H, Gojo R, Tsuda Y, Kiriya D, Takeuchi S. IEEE 23rd International Conference on Micro Electro Mechanical Systems (MEMS); Wanchai, Hong Kong. 2010. [Google Scholar]
- 396.Onoe H, Gojo R, Matsunaga Y, Kiriya D, Kato-Negishi M, Kuribayashi-Shigetomi K, Shimoyama Y, Takeuchi S. IEEE MEMS; Cancun, Mexico. 2011. p. 908. [Google Scholar]
- 397.Akbari M, Tamayol A, Laforte V, Annabi N, Khademhosseini A, Juncker D. Proc. microTAS; Freiburg, Germany. 2013. [Google Scholar]
- 398.Bae H, Puranik AS, Gauvin R, Edalat F, Carrillo-Conde B, Peppas NA, Khademhosseini A. Sci Transl Med. 2012;4:160ps23. doi: 10.1126/scitranslmed.3003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Cuchiara MP, Gould DJ, McHale MK, Dickinson ME, West JL. Adv Funct Mater. 2012;22:4511. doi: 10.1002/adfm.201200976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Malda J, van den Brink P, Meeuwse P, Grojec M, Martens DE, Tramper J, Riesle J, van Blitterswijk CA. Tissue Eng. 2004;10:987. doi: 10.1089/ten.2004.10.987. [DOI] [PubMed] [Google Scholar]
- 401.Takei T, Kishihara N, Ijima H, Kawakami K. Artif Cells, Blood Substitutes, Biotechnol. 2012;40:66. doi: 10.3109/10731199.2011.592492. [DOI] [PubMed] [Google Scholar]
- 402.Bellan LM, Singh SP, Henderson PW, Porri TJ, Craighead HG, Spector JA. Soft Matter. 2009;5:1354. [Google Scholar]
- 403.Diban N, Haimi S, Bolhuis-Versteeg L, Teixeira S, Miettinen S, Poot A, Grijpma D, Stamatialis D. Acta Biomater. 2013;9:6450. doi: 10.1016/j.actbio.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 404.Yoshida H, Matsusaki M, Akashi M. Adv Funct Mater. 2013:1736. [Google Scholar]
- 405.Matsusaki M, Kadowaki K, Nakahara Y, Akashi M. Angew Chem Int Ed Engl. 2007;46:4689. doi: 10.1002/anie.200701089. [DOI] [PubMed] [Google Scholar]
- 406.Baranski JD, Chaturvedi RR, Stevens KR, Eyckmans J, Carvalho B, Solorzano RD, Yang MT, Miller JS, Bhatia SN, Chen CS. Proc Natl Acad Sci USA. 2013;110:7586. doi: 10.1073/pnas.1217796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Nguyen DH, Stapleton SC, Yang MT, Cha SS, Choi CK, Galie PA, Chen CS. Proc Natl Acad Sci USA. 2013;110:6712. doi: 10.1073/pnas.1221526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Rouwkema J, Rivron NC, van Blitterswijk CA. Trends Biotechnol. 2008;26:434. doi: 10.1016/j.tibtech.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 409.Chen YC, Lin RZ, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A. Adv Funct Mater. 2012;22:2027. doi: 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.D’Andrea LD, Iaccarino G, Fattorusso R, Sorriento D, Carannante C, Capasso D, Trimarco B, Pedone C. Proc Natl Acad Sci USA. 2005;102:14215. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Leslie-Barbick JE, Saik JE, Gould DJ, Dickinson ME, West JL. Biomaterials. 2011;32:5782. doi: 10.1016/j.biomaterials.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 412.Chiu LL, Montgomery M, Liang Y, Liu H, Radisic M. Proc Natl Acad Sci USA. 2012;109:E3414. doi: 10.1073/pnas.1210580109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR, Riley PR. Nature. 2007;445:177. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 414.Kim S, Lee H, Chung M, Jeon NL. Lab Chip. 2013;13:1489. doi: 10.1039/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- 415.Hilborn J. Wiley Interdiscip Rev: Nanomed Nanobiotechnol. 2011 doi: 10.1002/wnan.91. [DOI] [PubMed] [Google Scholar]
- 416.Page JM, Harmata AJ, Guelcher SA. J Biomed Mater Res A. 2013 doi: 10.1002/jbm.a.34665. [DOI] [PubMed] [Google Scholar]
- 417.Buck DW, 2nd, Alam M, Kim JY. J Plast Reconstructive Aesthetic Surg. 2009;62:11. doi: 10.1016/j.bjps.2008.06.036. [DOI] [PubMed] [Google Scholar]
- 418.Overstreet DJ, Dutta D, Stabenfeldt SE, Vernon BL. J Polym Sci, Part B: Polym Phys. 2012;50:881. [Google Scholar]
- 419.Narins RS, Beer K. Plast Reconstructive Surg. 2006;118:77S. doi: 10.1097/01.prs.0000234919.25096.67. [DOI] [PubMed] [Google Scholar]
- 420.Rapaport MJ, Vinnik C, Zarem H. Aesthetic Plast Surg. 1996;20:267. doi: 10.1007/s002669900032. [DOI] [PubMed] [Google Scholar]
- 421.Eppley BL, Dadvand B. Plast Reconstr Surg. 2006;118:98e. doi: 10.1097/01.prs.0000232436.91409.30. [DOI] [PubMed] [Google Scholar]
- 422.Hillel AT, Nahas Z, Unterman S, Reid B, Axelman J, Sutton D, Matheson C, Petsche J, Elisseeff JH. Dermatol Surg. 2012;38:471. doi: 10.1111/j.1524-4725.2011.02273.x. [DOI] [PubMed] [Google Scholar]
- 423.Young DA, Christman KL. Biomed Mater. 2012;7:024104. doi: 10.1088/1748-6041/7/2/024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Hillel AT, Unterman S, Nahas Z, Reid B, Coburn JM, Axelman J, Chae JJ, Guo Q, Trow R, Thomas A, Hou Z, Lichtsteiner S, Sutton D, Matheson C, Walker P, David N, Mori S, Taube JM, Elisseeff JH. Sci Transl Med. 2011;3:93ra67. doi: 10.1126/scitranslmed.3002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Tan H, Li H, Rubin JP, Marra KG. J Tissue Eng Regener Med. 2011;5:790. doi: 10.1002/term.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Chung HJ, Jung JS, Park TG. J Biomater Sci, Polym Ed. 2010 doi: 10.1163/092050609X12580983495681. [DOI] [PubMed] [Google Scholar]
- 427.Brandl F, Sommer F, Goepferich A. Biomaterials. 2007;28:134. doi: 10.1016/j.biomaterials.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 428.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Luhmann T, Hall H. Materials. 2009;2:1058. [Google Scholar]
- 430.Tibbitt MW, Anseth KS. Sci Transl Med. 2012:4. doi: 10.1126/scitranslmed.3004804. [DOI] [PubMed] [Google Scholar]
- 431.Santos E, Hernandez RM, Pedraz JL, Orive G. Trends Biotechnol. 2012;30:331. doi: 10.1016/j.tibtech.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 432.Aurand ER, Lampe KJ, Bjugstad KB. Neurosci Res. 2012;72:199. doi: 10.1016/j.neures.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Shin SR, Bae H, Cha JM, Mun JY, Chen YC, Tekin H, Shin H, Farshchi S, Dokmeci MR, Tang S, Khademhosseini A. ACS Nano. 2012;6:362. doi: 10.1021/nn203711s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Clarke EC, Cheng S, Bilston LE. J Biomech. 2009;42:1397. doi: 10.1016/j.jbiomech.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 435.Gumera C, Rauck B, Wang YD. J Mater Chem. 2011;21:7033. [Google Scholar]
- 436.Nisbet DR, Rodda AE, Horne MK, Forsythe JS, Finkelstein DI. Tissue Eng, Part A. 2010;16:2833. doi: 10.1089/ten.TEA.2009.0677. [DOI] [PubMed] [Google Scholar]
- 437.Turney SG, Bridgman PC. Nat Neurosci. 2005;8:717. doi: 10.1038/nn1466. [DOI] [PubMed] [Google Scholar]
- 438.Li Y, Yang C, Khan M, Liu SQ, Hedrick JL, Yang YY, Ee PLR. Biomaterials. 2012;33:6533. doi: 10.1016/j.biomaterials.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 439.Lampe KJ, Kern DS, Mahoney MJ, Bjugstad KB. J Biomed Mater Res, Part A. 2011;96A:595. doi: 10.1002/jbm.a.33011. [DOI] [PubMed] [Google Scholar]
- 440.Marin C, Fernandez E. Front Neuroeng. 2010;3:8. doi: 10.3389/fneng.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Solorio LD, Dhami CD, Dang PN, Vieregge EL, Alsberg E. Stem Cells Transl Med. 2012;1:632. doi: 10.5966/sctm.2012-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 443.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat Mater. 2010;9:518. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Nat Mater. 2013;12:458. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Mahoney MJ, Anseth KS. J Biomed Mater Res, Part A. 2007;81A:269. doi: 10.1002/jbm.a.30970. [DOI] [PubMed] [Google Scholar]
- 446.Li X, Liu X, Cui L, Brunson C, Zhao W, Bhat NR, Zhang N, Wen X. FASEB J. 2013;27:1127. doi: 10.1096/fj.12-211151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Medberry CJ, Crapo PM, Siu BF, Carruthers CA, Wolf MT, Nagarkar SP, Agrawal V, Jones KE, Kelly J, Johnson SA, Velankar SS, Watkins SC, Modo M, Badylak SF. Biomaterials. 2013;34:1033. doi: 10.1016/j.biomaterials.2012.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Moshaverinia A, Chen C, Akiyama K, Ansari S, Xu XT, Chee WW, Schricker SR, Shi ST. J Mater Sci Mater Med. 2012;23:3041. doi: 10.1007/s10856-012-4759-3. [DOI] [PubMed] [Google Scholar]
- 449.Myers JP, Santiago-Medina M, Gomez TM. Dev Neurobiol. 2011;71:901. doi: 10.1002/dneu.20931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.di Summa PG, Kalbermatten DF, Raffoul W, Terenghi G, Kingham PJ. Tissue Eng, Part A. 2013;19:368. doi: 10.1089/ten.tea.2012.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Horne MK, Nisbet DR, Forsythe JS, Parish CL. Stem Cells Dev. 2010;19:843. doi: 10.1089/scd.2009.0158. [DOI] [PubMed] [Google Scholar]
- 452.Jiang FX, Yurke B, Schloss RS, Firestein BL, Langrana NA. Tissue Eng, Part A. 2010;16:1873. doi: 10.1089/ten.TEA.2009.0574. [DOI] [PubMed] [Google Scholar]
- 453.Poellmann MJ, Harrell PA, King WP, Johnson AJW. Acta Biomater. 2010;6:3514. doi: 10.1016/j.actbio.2010.03.041. [DOI] [PubMed] [Google Scholar]
- 454.Turturro MV, Christenson MC, Larson JC, Young DA, Brey EM, Papavasiliou G. PloS One. 2013:8. doi: 10.1371/journal.pone.0058897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.DeForest CA, Anseth KS. Annu Rev Chem Biomol Eng. 2012;3:421. doi: 10.1146/annurev-chembioeng-062011-080945. [DOI] [PubMed] [Google Scholar]
- 456.Ghaemmaghami AM, Hancock MJ, Harrington H, Kaji H, Khademhosseini A. Drug Discovery Today. 2012;17:173. doi: 10.1016/j.drudis.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Sung JH, Esch MB, Shuler ML. Expert Opin Drug Metab Toxicol. 2010;6:1063. doi: 10.1517/17425255.2010.496251. [DOI] [PubMed] [Google Scholar]
- 458.Baker M. Nature. 2011;471:661. doi: 10.1038/471661a. [DOI] [PubMed] [Google Scholar]
- 459.Sung JH, Kam C, Shuler ML. Lab Chip. 2010;10:446. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 460.Sung J, Shuler M. Ann Biomed Eng. 2012;40:1289. doi: 10.1007/s10439-011-0491-2. [DOI] [PubMed] [Google Scholar]
- 461.Huh D, Hamilton GA, Ingber DE. Trends Cell Biol. 2011;21:745. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Sonntag F, Schilling N, Mader K, Gruchow M, Klotzbach U, Lindner G, Horland R, Wagner I, Lauster R, Howitz S. J Biotechnol. 2010;148:70z. doi: 10.1016/j.jbiotec.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 463.Williamson A, Singh S, Fernekorn U, Schober A. Lab Chip. 2013 doi: 10.1039/c3lc50237f. [DOI] [PubMed] [Google Scholar]
- 464.Neuži P, Giselbrecht S, Länge K, Huang TJ, Manz A. Nat Rev Drug Discovery. 2012;11:620. doi: 10.1038/nrd3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Ho C-T, Lin R-Z, Chen R-J, Chin C-K, Gong S-E, Chang H-Y, Peng H-L, Hsu L, Yew T-R, Chang S-F. Lab Chip. 2013 doi: 10.1039/c3lc50402f. [DOI] [PubMed] [Google Scholar]
- 466.Kim HJ, Huh D, Hamilton G, Ingber DE. Lab Chip. 2012;12:2165. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 467.Jang K-J, Mehr AP, Hamilton G, McPartlin L, Chung S, Suh K-Y, Ingber D. Integr Biol. 2013 doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 468.Agarwal A, Farouz Y, Nesmith AP, Deravi LF, McCain ML, Parker KK. Adv Funct Mater. 2013;23:3738. doi: 10.1002/adfm.201203319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Chen MB, Srigunapalan S, Wheeler AR, Simmons CA. Lab Chip. 2013;13:2591. doi: 10.1039/c3lc00051f. [DOI] [PubMed] [Google Scholar]
- 470.Esch M, King T, Shuler M. Annu Rev Biomed Eng. 2011;13:55. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]