Abstract
Protein interaction topologies are critical determinants of biological function. Large-scale or proteome-wide measurements of protein interaction topologies in cells currently pose an unmet challenge that could dramatically improve understanding of complex biological systems. A primary impediment includes direct protein topology and interaction measurements from living systems since interactions that lack biological significance may be introduced during cell lysis. Furthermore, many biologically relevant protein interactions will likely not survive the lysis/sample preparation and may only be measured with in vivo methods. As a step toward meeting this challenge, a new mass spectrometry method called Real-time Analysis for Cross-linked peptide Technology (ReACT) has been developed that enables assignment of cross-linked peptides “on-the-fly”. Using ReACT, 708 unique cross-linked (<5% FDR) peptide pairs were identified from cross-linked E. coli cells. These data allow assembly of the first protein interaction network that also contains topological features of every interaction, as it existed in cells during cross-linker application. Of the identified interprotein cross-linked peptide pairs, 40% are derived from known interactions and provide new topological data that can help visualize how these interactions exist in cells. Other identified cross-linked peptide pairs are from proteins known to be involved within the same complex, but yield newly discovered direct physical interactors. ReACT enables the first view of these interactions inside cells, and the results acquired with this method suggest cross-linking can play a major role in future efforts to map the interactome in cells.

Keywords: Cross-linking, PIR, protein interaction reporter, real-time MS strategy, FTICR MS, protein interaction topologies, in vivo protein interactions, XL-MS
INTRODUCTION
Proteins are the most abundant functional molecules inside cells and perform a bewildering array of biological processes required to support life. The versatility of protein function has its origins in topological shapes and features that these polymeric macromolecules can adopt. Moreover, the crowded intracellular environment profoundly influences their shape such that proteins that appear unstructured in vitro can adopt a more defined conformation inside cells.1,2 These induced topological features occur as a consequence of interaction within cellular compartments that may not be replicated in cell lysates or purified components.3–6 Thus, methods that can reveal information about global protein topology under physiologically relevant conditions, within native interactions with intended partners inside cells could greatly advance understanding of protein function.
Recently, methods based on cross-linking-mass spectrometry (XL-MS) have emerged as viable techniques to investigate protein–protein interactions (PPIs).7–18 These methods involve “fixing” the biological system through covalent chemical modification of amino acid residues and investigating the cross-linked sites using mass spectrometry methods. An attractive aspect of this technology is the potential to identify PPIs and unique topological features and yield large-scale data. Recent efforts have shown feasibility for measurements of PPIs in bacterial cells.19–21 An advantage of cross-linking is the potential to study protein topologies that are resistant to other techniques, such as disordered protein domains and membrane proteins. Unlike X-ray crystallography or NMR structure determination, cross-linking data can provide unique structural insight on many proteins as they exist in their natural cellular environment in a single experiment. XL-MS technologies have the capacity to produce large-scale data sets, although technical limitations have constrained the scope of current methods to the identification of a few (<100) cross-linked peptides in vivo.10,19–21
Real-time Analysis for Cross-linked peptide Technology, or ReACT, was developed to enable large-scale application of cross-linking technology. ReACT analysis of cross-linked peptides from cross-linked E. coli cells yielded data sets approximately 10-fold larger than previously reported. The increased capacity for cross-linked peptide identification achieved with ReACT enabled creation of the first PPI network derived solely from covalent chemical cross-linking on cells. These data provide in vivo topological information on many known interactions, including ribosomal proteins, elongation factor TU, 60 kDa chaperonin, and more. Excitingly, many of the cross-linked sites can be mapped onto existing cocrystal structure data and support the existence of these complex structures inside cells. Furthermore, many cross-linked sites were identified among proteins known to participate in complexes but not known to interact directly. Finally, although the results acquired with ReACT represent only a very small fraction of those possible in cells, the new capabilities presented by ReACT suggest that cross-linking technologies will grow to provide large-scale topological data on protein interactions in cells.
METHODS
ReACT Algorithm
The ReACT algorithm was written in ion trap control language, a native language used with Thermo Electron mass spectrometers. The flowchart in Figure 1 outlines how the algorithm operates. Charge state exclusion alternates between two parameter sets depending on the order, n, of each stage of MSn analysis. The set of parameters allows ions with charge state ≥4+ to be selected from high resolution mass spectral acquisition. This is done to focus instrument capabilities on cross-linked species for subsequent tandem mass spectrometry analyses. All ions generated during high resolution MS2 acquisition for which charge states are assigned are considered during the mass relationship discovery phase of the experiment. By identifying these relationships as the analytes elute from the LC column, ReACT effectively achieves real-time application of previously described analysis strategies for PIR cleavable cross-linkers. 22,23 Any two released and observed peptide masses added to the reporter mass must equal the observed precursor mass within a user definable mass tolerance (eq 1):
| (1) |
where PRECURSOR is the mass of any selected precursor ion, REPORTER is the mass of the reporter ion,14 and PEPTIDEn is the mass of the released peptide n. This equation is applied during real-time data acquisition and requires checking N MS2 high resolution product ions with each other. This amounts to N2/2 calculations where N is equal to the number of detected isotopic distributions in the MS2 pattern. In an effort to make this process more efficient, masses observed in the MS2 spectra are considered only if they satisfy the following statement:
| (2) |
where STUMP is the residual mass modification that remains on lysine residues after CID cleavage. This limits the computational space of the calculation by considering only ions lower in mass than PIR partial cleavage products. Partial cleavage products result from incomplete cleavage of the PIR cross-linked products and are observed when the reporter ion remains covalently linked to one of two peptides involved in the cross-link. These products are not used to determine whether eq 1 has been satisfied but can be used to add further confidence to putative relationships. In the event that two ion masses from the MS2 spectrum satisfy eqs 1 and 2, they are stored for targeted MS3 analysis in the next scan cycle. In this way, no loss of instrument duty cycle occurs during the relationship calculation. Up to two 13C offsets are considered to allow for possible incorrect monoisotopic peak assignment for cross-linked precursors or product ions. A 13C offset is defined as the mass difference in daltons (Da) between 12C and 13C.
Figure 1.
(Left) A flowchart that describes how the REACT algorithm functions during LC–MSn experiments. (Right) An idealized practical diagram of how the algorithm would operate on real data directly corresponding to the flowchart.
Liquid Chromatography–Mass Spectrometry
All samples were analyzed on a custom dual linear RF ion trap Fourier transform ion cyclotron resonance mass spectrometer, hereafter referred to as the Velos-FT. However, it should be noted that in principle, ReACT-based experiments are possible on any mass spectrometry platform that is capable of high resolution MS2 and low resolution MS3. The mass spectrometer is directly coupled with a Waters NanoAcquity UPLC system. Cross-linked peptide samples were loaded onto a trap column (3 cm × 100 μm i.d.) packed with 200 Å Magic-C4AQ (Michrom) using a flow rate of 2 μL/min of 99% solvent A (H2O containing 0.1% formic acid) and 1% solvent B (acetonitrile containing 0.1% formic acid) and washed for a total of 10 min. Peptides were then eluted from the trap column and separated by reversed-phase chromatography over an analytical column (30 cm × 75 μm i.d.) packed with 100 Å Magic-C4AQ at a flow rate of 200 nL/min using a linear gradient from 90% solvent A/10% solvent B to 60% solvent A/40% solvent B over 120 min for a 2 h data acquisition or 240 min for a 4 h data acquisition. The structure of a ReACT method consists of the following mass spectrometry data acquisition parameters. The first acquisition is a high-resolution precursor acquisition (50,000 resolving power (RP) @ 400 m/z). The second is a high resolution MS2 acquisition on ≥4+ charge state isotope distributions. This requires the use of charge state exclusion. Dynamic exclusion is utilized with the following parameters: repeat count = 2, repeat duration = 15 s, dynamic exclusion list size = 500, dynamic exclusion duration = 30 s. FT preview mode and predictive automated gain control (pAGC) were not utilized. Monoisotopic precursor selection was used. A series of four RF ion trap MS3 acquisitions were used to acquire fragmentation spectra of peptides observed in cross-linked relationships. These MS3 events include acquisition on the 1+ and 2+ charge states of the peptides found in PIR relationships. Acquisition of MS3 spectra on two charge states better allows one to address unequal charge state distribution that may result from cleavage of the cross-linked complex.
PIR Cross-Linker Synthesis
PIR synthesis was performed using solid phase peptide synthesis (SPPS) methods.24 The Endeavor 90 (Apptec, Louisville, KY) SPPS unit was used for all PIR synthesis steps with the single exception of the final N-hydroxy ester (NHX, where X = succinimide or phthalimide) ester formation step. Biotin Rink-PIR (BRink)14 and Rink-PIR (2Rink) synthesis has been previously described.12 Briefly, the super acid sensitive resin (SASRIN) with a glycine residue precoupled was utilized (Bachem, Munich, Germany). Synthesis of the cross-linker proceeds through fluorenylmethyloxycarbonyl (Fmoc) N-terminally protected SPPS methods.25 Additions to the resin occur in order and are the following: Fmoc-Lys (biotin), Fmoc-Lys (Fmoc), Fmoc-Rink (all amino acids obtained from Bachem), and succinic anhydride (Sigma-Aldrich, St. Louis, MO). 2Rink is synthesized through the same series of steps with the exception of the addition of Fmoc-Lys (biotin). The activated NHS-ester form of the cross-linker is created in a final esterification step immediately prior to use with TFA-NHS.26 Overall yield for this synthesis is ~90%. Purity was confirmed by direct infusion ESI-MS analysis. Cross-linker is cleaved from the resin using 1% trifluoroacetic acid (TFA) in methylene chloride and purified using a semi-preparative partisil C18 column (Whatmann, United Kingdom) at low pH to prevent hydrolysis of the NHS ester. BRink and 2Rink were dissolved in dimethylsulfoxide to a concentration of 100 mM.
Biotin Aspartate Proline-PIR (BDP) synthesis is also accomplished using Fmoc chemistry. SASRIN-glycine resin was used for the solid support. Amino acid additions to the resin occur in order and are the following: FMOC-Lys (Biotin), FMOC-Lys (FMOC), FMOC-Pro, FMOC-Asp (otBu), and succinic anhydride. The activated NHX form of the cross-linker is created in a final esterification step immediately prior to use with TFA-NHX26 (X = phthalamide or succinimide). Cleavage from the solid support and deprotection of Asp (otBu) was performed simultaneously using 95% TFA 5% methylene chloride. Purification was performed immediately subsequent to Asp deprotection and cleavage via diethyl ether precipitation using 1:15 (cleavage mixture:diethyl ether). Diethyl ether solution was centrifuged at 3400g to pellet precipitate. Diethyl ether was decanted, and pellet was dried to yield ~90–95% pure BDP-ester. Purity was assayed via direct infusion ESI-MS analysis. BDP was dissolved in dimethylsulfoxide to a concentration of 500 mM to form the stock solution.
Purified Protein Sample Preparation
Alcohol dehydrogenase (S. cerevisiae), α-lactalbumin (Bos taurus), carbonic anhydrase (Bos taurus), cytochrome c (Equus caballus), hemoglobin (Homo sapiens), ribonuclease A (Bos taurus), and myoglobin (Equus caballus) were all obtained from Sigma Aldrich (St. Louis, MO) and used as received. Each protein was dissolved at a concentration of 1 mg/mL in phosphate-buffered saline (PBS) buffer, pH 7.4. The cross-linking reaction was performed by adding BDP-NHS at a final concentration of 1 mM and incubating the reaction solution at room temperature for 1 h with constant mixing. A second sample of ribonuclease A was labeled using 2Rink at the same concentration at the same concentration as the BDP RNase A sample analogue. After cross-linking, disulfide bonds were reduced using 5 mM tris(2-carboxyethyl) phosphine (TCEP), and the resulting free thiols were alkylated using 10 mM iodoacetamide (IAA). Digestion was carried out using a 1:200 w/w ratio of sequencing grade modified trypsin (Promega, Madison, WI) to protein and incubating at 37 °C overnight with constant mixing. The samples were desalted using C18 Sep-Pak (Waters Corporation, United Kingdom) and dried in a centrifugal concentrator (Genevac, Gardiner, NY). The cross-linked, digested samples were redissolved in solvent A then stored at −80 °C until LC–MS analysis.
E. coli Sample Preparation
In vivo cross-linking of E. coli was performed as described previously.20 Briefly, E. coli K12 cell suspensions were harvested at OD 0.6–0.8. The cells were pelleted and washed 5 times with 1 mL of PBS before cross-linking. A 150 μL cell pellet was resuspended in 150 mL of PBS, and BDP-NHP was added to the suspension with a final concentration of 10 mM. The reaction was carried out at 4 °C for 1 h. The cells were pelleted, washed, and then lysed by heating to 95 °C in 4% sodium dodecylsulfate (SDS) 1x Tris buffer at pH 8.5. The sample was ultrasonicated to shear DNA, centrifuged at 16000g for 10 min to remove insoluble material, and then added to a 30 kDa molecular weight cutoff (MWCO) filter (Millipore, Billerica, MA) and concentrated by centrifugation at 7500g for 30 min. A protein extract yield of 2.0 mg/mL was determined using a Coomassie Plus assay (Pierce, Rockford, IL). The sample was reduced, alkylated, and digested as described above for the purified protein samples. Strong cation exchange (SCX) fractionation of the sample was performed using Macro SCX Spin Columns (Nest Group Inc., Southborough, MA) and ammonium acetate in 25% acetonitrile/75% water for elution. Fractions were collected at 0, 50, 80, 300, 500, and 1000 mM ammonium acetate. Prior to affinity enrichment each fraction was desalted using C18 Sep-Pak 50 cm3 (Waters Corporation, United Kingdom). The fractions were biotin affinity enriched for BDP cross-linked peptide products using Ultralink Monomeric Avidin (Pierce, Rockford, IL). To each fraction 300 μL of settled avidin resin was added in 500 μL of 100 mM ammonium bicarbonate. Enriched cross-linked peptide samples were stored at −80 °C until LC–MS analysis.
In addition to two biological replicates using the above protocol, a third sample was prepared with 10× greater number of cells by volume (1.5 mL cell pellet) to ascertain whether cross-linking reaction product concentration serves to limit detection by ReACT. This sample was prepared with the same protocol as above with a few minor exceptions. The first exception is SCX separation was done online using a 4.6 mm i.d. × 100 mm SCX column packed in-house with polysulfethyl aspartamide media, identical to that which is used in the spin columns (Nest Group Inc., Southborough, MA) on a standalone liquid chromatograph (Agilent Technologies, Santa Clara, CA). The separation was achieved using an isocratic step gradient at a flow rate of 1.5 mL/min and steps at 0, 50, 100, …, 300 mM with 5 min per step. Solvent A consisted of 25% acetonitrile/75% water, while solvent B consisted of 25% acetonitrile/75% water/1 M ammonium acetate.
Data Interpretation and Sequence Identifications
ReACT provides a list of cross-linked relationships observed during an entire data acquisition. Raw mass spectrometry data is converted to mzXML format using ReAdW (version 4.3.1). MS2 accurate precursor mass and MS3 fragmentation patterns are extracted from the mzXML files and converted to Mascot Generic Format (mgf) for Mascot (version 2.3.1) sequence database searches using MzXML2Search (version 4.4) or mzXML was searched directly using SEQUEST (version UWPR2011.01.1). Mascot searches were conducted with a 10 ppm precursor mass tolerance and 0.8 Da fragment ion tolerance. SEQUEST searches were conducted with 10 ppm precursor mass tolerance and 0.36 Da fragment tolerance (0.11 Da fragment offset). The most probable match for each query was accepted (with an expectation value threshold <0.05) and mapped back to the cross-linked relationship for in vitro experiments with purified proteins. Sequence databases utilized here include those of all proteins (21 sequences including isoforms) and SwissProt E. coli (4178 sequences) (http://www.uniprot.org). False discovery (peptide level) during sequence identification for cross-linking experiments with E. coli cells was estimated using well-described reverse sequence decoy database search methods.27 Relationship discovery in real-time was performed with 20 ppm tolerance between the putative cross-linked precursor and released peptide product and reporter masses. Tolerance of 20 ppm was chosen for relationship discovery as an effective compromise between method sensitivity and false relationship discovery. False relationship discovery was estimated by performing ReACT analysis on a E. coli lysate digest without cross-linker added, and fewer than <5% of all acquired MS2 spectra result in false mass relationships with 20 ppm relationship tolerance. To obtain a direct empirical estimate for ReACT false relationship discovery, non-cross-linked E. coli digest was analyzed utilizing the same ReACT mass relationship tolerance as cross-linked samples. In a series of three technical replicate ReACT experiments of all MS2 spectra (10451) acquired, 31 yielded false mass relationships containing the expected PIR reporter ion (±20 ppm) within. After performing database searches on all falsely discovered mass relationships, none resulted in fully identified cross-linked products, illustrating the stringency of filtering results using 20 ppm on the mass relationship stage and <5% FDR on the peptide identification. At the mass tolerance utilized in ReACT experiments, it is concluded there is negligible contribution of the real-time PIR mass relationship FDR on the overall FDR for reported cross-linked identifications. For comparison, the E. coli cell results presented here comprise 84154 MS2 spectra searched by ReACT from both biological and technical replicates. ReACT identified 3960 cross-linked mass relationships containing reporter ion from these spectra; 2934 fully identified cross-linked relationships were made where both released peptides were identified with <5% sequence assignment FDR. From these results after reducing for redundancy, 708 unique cross-linked sites were identified with <5% false discovery. All discovered relationships were filtered for mass redundancy on the precursor, MS2, and peptide sequence level to curate only the highest confidence set of results. The results are presented in tabular format in the Supporting Information.
Structural Modeling
All models were created and rendered using Pymol (Delano Scientific). E. coli tryptophanase and 30s ribosome structural models were created using coordinates from PDB identifiers 2OQX and 3FIH, respectively.
RESULTS AND DISCUSSION
ReACT is an integrated mass spectrometry analysis platform for real-time identification of cross-linked peptides. This approach relies on measurement and validation of mass relationships that arise from CID cleavable cross-linkers illustrated above in eq 1 (see Methods). The general ReACT strategy is outlined in Figure 1. High resolution MS1 spectra are acquired and deconvoluted to obtain the neutral mass and charge states of all species detected. For any species with charge state 4+ or greater, a high resolution MS2 is acquired in a data-dependent fashion (e.g., selection of the top N most abundant 4+ ionic species). Next the MS2 is deconvolved to obtain the neutral mass and charge state of all species detected. ReACT analysis automatically identifies spectral features that satisfy the mass relationship defined in eq 1 as expected for MS-cleavable cross-linkers. Released peptide ions found to fulfill these relationships are then automatically selected for MS3 analysis, and peptide fragmentation spectra are acquired. Simple real-time informatics strategies for mass spectrometry have recently been explored by others,28,29 successfully increasing specificity and sensitivity of the overall analysis, while minimizing the need for repeated sample analyses or lengthy postanalysis data processing. With ReACT, the final step is to extract the MS3 information and perform a database search with conventional proteome database search tools such as SEQUEST, Mascot, or others. Since ReACT uses mass relationships to direct MS3 events, the number of spectra to be searched scales with the number of relationships found. The selectivity of ReACT decreases demand on instrument duty cycle and yet enables specific targeting of cross-linked peptides that are often observed with lower abundance. These species may be missed by intensity-based data-dependent analyses. The loss of analysis time spent on species that do not meet these criteria is eliminated using ReACT, allowing for improved detection of many more cross-linked peptide species than possible previously. ReACT is a relatively simple algorithm that could be implemented on any mass spectrometer with high mass measurement accuracy capabilities and the capacity to make experiment decisions on-the-fly. Hopefully, these capabilities will be incorporated on future MS operating systems to allow ReACT to be implemented in many laboratories.
Chemical cross-linkers compatible with ReACT must possess a low energy CID cleavage site to facilitate cross-linked peptide relationship recognition and subsequent MS3 peptide fragmentation pattern acquisition. A series of CID cleavable cross-linkers developed in-house, named Protein Interaction Reporter (PIR) cross-linkers, were utilized in this work (Supplemental Figure 1). Since the initial report of PIR molecules with CID cleavable features,14 many cross-linkers have been developed with CID cleavable bonds and are compatible with ReACT. A comprehensive list of cross-linkers with cleavable bonds has been presented in a recent review by Paramelle et al.30 Although these compounds have a variety of structural and chemical properties, each contains the basic features of a mass encoded reporter ion and two low energy CID cleavable bonds. In addition, the BDP and BRink cross-linkers include a biotin moiety, useful for affinity purification of the conjugated reaction products. Among the benefits of using PIR cross-linkers are the engineered fragmentation patterns and the use of a reporter ion as an indicator of labeled species.13 For further information regarding PIR molecules used in this study see Supporting Information.
ReACT has been developed to provide selectivity in LC–MSn analyses to focus on only those ions that are likely cross-linked peptides. This selectivity is illustrated with an example of a cross-linked site identified from E. coli cells (Figure 2). ReACT selectivity for cross-linked species is achieved first on the MS2 precursor stage through exclusion of ions with charge less than 4+, since two tryptic peptides covalently linked will possess on average 4+ charge state or greater.10,31 Many potential analytes are present within the spectrum in Figure 2A; however, the ReACT algorithm selects only those ions with 4+ charge state or higher for MS2 analysis. In fact, the analyte of interest, 718.174 m/z, is the 576th most abundant peak within the spectrum and would likely never have been sampled by conventional intensity-driven data dependent analyses. Requirement of the CID cleavable linker mass relationships to be observed with narrow mass tolerance (±20 ppm) imparts additional specificity in the analysis of the selected high charge state ions. In the example shown, the measurement error between the observed precursor and sum of masses of the relationship (eq 1) is less than 1.5 ppm (Figure 2B). Typically, mass measurement error for observed cross-linked relationships is less than or equal to 5.0 ppm, which significantly reduces false relationship discovery (see Supporting Information). Upon successful relationship detection, ReACT directs MS3 events to automatically acquire fragment ion spectra for sequence identification of the released peptides (1+ and 2+ charge states for each). Both peptides identified in this example belong to tryptophanase (TNAA_ECOLI). The cross-linked sites were mapped onto the existing crystal structure for E. coli tryptophanase (PDB 2OQX), where the red highlighted lysine residues represent the cross-linked sites in this example (gray residues indicate other cross-linking sites found; Figure 2E).
Figure 2.
An example of ReACT data acquired from PIR labeled E. coli cells. (A) High resolution MS1 acquisition for precursor information; inset is an expanded view of the spectrum surrounding the cross-linked peptide precursor, 718.174 m/z. (B) High resolution MS2 acquisition for cross-linked peptide relationship information. (C and D) Low resolution MS3 acquisition for peptide sequence information. (E) Tryptophanase crystal structure (E. coli, PDB 2OQX) with all observed cross-linked sites marked in gray; the cross-link observed in this data is marked in red, while other sites observed in additional cross-linked sites are in gray. To view an animated illustration of cross-linked sites on the molecular structure see: http://brucelab.gs.washington.edu/ReACT_movies.php.
ReACT was initially applied to a set of commercially available purified proteins. The data resultant from this set of experiments is presented in Supplemental Table S1. An unambiguous α–β hemoglobin cross-link was observed, as well as unambiguous homodimeric cross-links supporting protein dimerization of ribonuclease A and carbonic anhydrase. Several cross-linked peptide products were successfully identified in these samples even with a signal-to-noise ratio of ~2, suggesting that the ability to discriminate against lower charge state ions improves the dynamic range over which cross-linked species can be interrogated. The ability of ReACT to extract useful information, even from ions with low signal intensity, is most beneficial for complex samples, as illustrated with in vivo cross-linking samples below. ReACT is customizable for use with any cross-linker that can be cleaved within the mass spectrometer including linkers with mono, bi, or higher order CID cleavage sites. To demonstrate this flexibility, Ribonuclease A (RNase A) was cross-linked with two different PIR molecules, 2Rink and BDP,14,20 and the ReACT approach was applied. For this sample, the respective reporter masses were entered into ReACT so that ions matching either the mass relationship for 2Rink or for BDP would be identified as cross-linked peptide pairs. In either case, ReACT selected the released peptide ions that fulfilled the relationships in eq 1 for MS3 analysis. BDP and 2Rink labeled RNase A digests were mixed in equimolar ratios and four fully identified cross-linked products are discussed next. Of the four, two are obtained from BDP, and two are obtained from 2Rink. All four share a single peptide with a unique second peptide. One pair overlaps between the two linkers (ETAAAKFER-NLTKDR). In Figure 3, this cross-linked site has been identified with both linkers within a single ReACT experiment. These two PIR cross-linkers differ in their engineered cleavage site. In BDP, the proline-aspartate amide bond acts as the low energy cleavage site, whereas, in 2Rink it is the tertiary amine within the Rink core structure. The permanent lysine modification or “stump” mass of these linkers differs (99.032 Da for 2Rink or 197.032 Da for BDP). Therefore, peptides identified with this site have b and y fragment ions with different mass shifts due to the modification (Figure 3A,B). Although this effort is focused on the initial description and application of ReACT, these results demonstrate the capacity of multiple simultaneous cross-linker analyses with ReACT. This feature of ReACT will benefit sample analyses with multiple cross-linker molecules, e.g., with variable structure lengths, reactivity, or physiochemical properties, and may further increase the number of observed cross-linked sites from cells.
Figure 3.
(A) High resolution MS2 spectra acquired on a cross-linked species with two different cross-linkers within the same LC-ReACT experiment. The cross-linked site identified involves the same two peptides from RNase A (ETAAAKFER and NLTKDR). The top contains this site identified with BDP cross-linker (blue), and the bottom contains this site identified with 2Rink cross-linker (red). Low resolution MS3 used to make peptide sequence identification for NLTKDR (B) and ETAAAKFER (C) for both linkers.
PIR technology was used previously to study PPIs and topologies in vivo within E. coli.20 In that study, a total of 65 cross-linked peptide pairs were identified using previously published mass spectrometry analysis methods and informatics tools.22,23 Conclusive identification of these 65 cross-linked pairs was a labor intensive process, requiring multiple LC–MS runs, multiple sample preparations, and significant efforts in data processing and analysis. Since ReACT achieves both PIR relationship and identification of both released peptides within a single run, this approach is at least twice as efficient as previous methods, from sample consumption and instrument time considerations. With cross-linking conditions similar to those of Zhang et al.,20 ReACT analysis of three bioreplicates (one of which was the preparation at 10x scale) resulted in 708 fully identified cross-linked peptide pairs, where both released peptides were identified using SEQUEST with false discovery rate (FDR) below 5% (Supplemental Table S3). These crosslinks peptide pairs are referred to as high confidence. Technical reproducibility of the ReACT method determined from replicate analysis of the same sample twice showed ~70% reproducibility on the basis of uniquely identified linkages (40% reproducibility among bioreplicates). Tabb et al.32 suggest that 35–60% repeatability/reproducibility for technical replicates as an upper limit within standard discovery based proteomics experiments. Both technical and biological reproducibility observed with ReACT are well within this range. For further discussion see Supporting Information. Cross-linked sites where both peptide identities occurred at <5% FDR are referred to as high confidence sites. Because identification of each linked peptide proceeds via independent MS3, it is possible that only a single peptide is identified by MS3 while the other linked peptide fragmentation pattern fails to yield a conclusive assignment at the 5% FDR cutoff. Within E. coli, an additional 657 cross-linked relationships were observed in this category (referred to as low confidence sites). In these cases, accurate released peptide masses and the number of observed matching fragment ions were used to make putative sequence assignments to the peptides above the 5% FDR threshold. Even though the observed SEQUEST score for these ions did not fall within the 5% FDR cutoff, in all cases the accurate peptide mass and the largest number of matching fragment ions search yielded the top scoring SEQUEST candidate. Inclusion of these assignments increased the total number of cross-linked pairs to 1318 cross-linked peptides from E. coli. For the full list of 1318 cross-linked pairs including relationship mass accuracy and peptide sequence FDR (q-values) see Supporting Information. Parallel to ReACT development, a database for storage, visualization, and interpretation of large-scale cross-linking results has been developed (XLink-DB, http://brucelab.gs.washington.edu/crosslinkdbv1).33
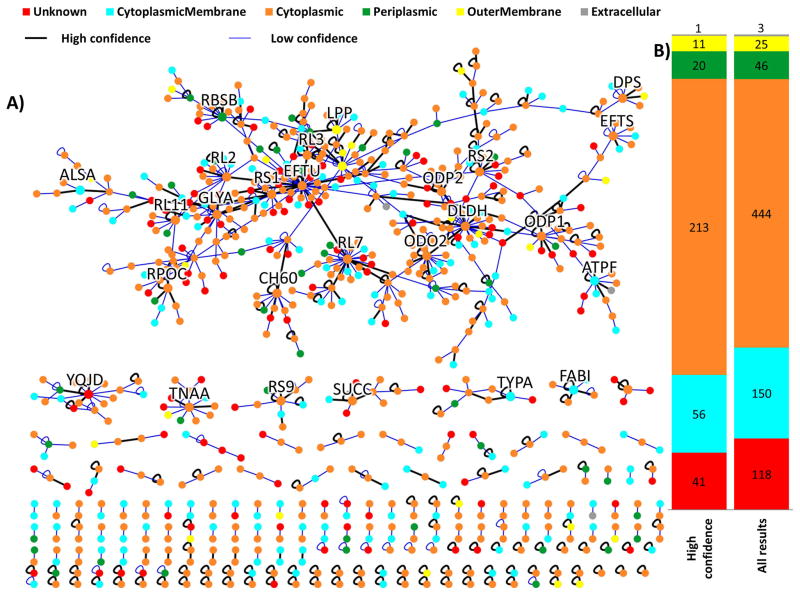
Cross-linked sites from in vivo experiments with E. coli cells were assembled into a protein interaction network (Figure 4A). High confidence cross-linked sites are indicated by a solid black edge, and low confidence cross-linked sites are represented with light blue edges. Major “hubs” for cross-linking have been labeled with their UniProt identifier. Omitted from this network are nodes/proteins that are only represented by a single edge or only contain intramolecular cross-linked sites. Although far from comprehensive, these exciting maps are the first such direct interaction networks derived solely from in vivo cross-linking data acquired from E. coli cells. Figure 4B provides subcellular localization for all proteins identified within cross-linked sites as predicted with PSORTb.34 Approximately 25% of the proteins found within cross-linked sites were found within cell envelope proteins (membrane, cytoplasmic membrane, periplasm). This suggests that cellular cross-linking may be viable for isolating and studying cell envelope proteins and their interactions, which is currently an area not well served by other techniques.
Figure 4.
(A) Interaction network comprising cross-linked information obtained using ReACT on E. coli cells. Node colors represent the subcellular localization for the proteins identified in cross-linked sites. Labeled nodes represent “hubs” for which many cross-links between proteins were detected. (B) Subcellular localization of cross-linked proteins.
ReACT is a shotgun proteomics approach that advances peptide sequence identification for peptides in cross-linked relationships. Identified peptides are used to infer protein identity. However, in contrast to typical shotgun proteomics experiments where identification of many peptides from a single protein supports that protein or protein family’s presence within the sample, a single cross-linked peptide may be the only reactive site identified from an entire protein sequence. It should be noted that this same issue exists for all large-scale cross-linking and post-translational modification studies. To date, this remains a difficult problem to adequately address in large-scale proteomics data sets where modifications are considered. ReACT analysis results in identification of two peptides cross-linked to each other that may or may not belong to the same protein/family. Within the high confidence E. coli cell data presented here, 81% of the cross-linked sites are reported to have both peptides non-redundant (described by a single protein) within the database. Additionally, 12.4% (88 of 708 identified) one of the peptides associated with a cross-linked site are redundant (peptide sequence shared by multiple proteins). Finally, in only 1.5% (11 of 708 identified) of the cases are both peptides redundant in the database. Redundancy values are included for each of the peptides identified in Supplemental Table 3.
For peptides that are redundant among two or more proteins sequences, putative protein identities were inferred through a set of logical criteria derived to address this issue and described here. First, a peptide is preferentially assigned to a single protein from the list if that peptide can be mapped to the same protein as the other peptide in the cross-linked site. This logical assumption is derived from the fact that lysine residues nearby any reacted lysine site will predominantly be within the same protein sequence. Thus, if one of the redundant proteins is the same as the protein that yielded the other non-redundant cross-linked peptide, this entity is chosen. If this step cannot be satisfied, the redundant peptide is preferentially assigned to a protein from the pool of proteins resultant from all non-redundant peptides identified within ReACT data sets. This logical assumption arises from the fact that because the protein was identified as cross-linked on other sites, cross-linker accessibility and reactivity with this protein is demonstrated. If one or more proteins in this pool contain the redundant peptide sequence, the proteins are assigned on the basis of their order of appearance within the database. Finally, if neither of the associations above can be made, a putative protein ID is assigned on the basis of the order of appearance within the entire protein database. With acquisition of larger cross-linking data sets where the number of redundant peptides is likely to become larger, advanced protein assignment methodologies will be implemented. These efforts will rank such assignments on the basis of the frequency of representation of the protein family within the database, relative genomic distance between the two cross-linked proteins (e.g., are the genes for the two proteins within the same operon or under control of a single promoter), established protein interaction databases, or based on proteins uniquely identified in other cross-linked sites (or e-values). 35,36
The primary utility of cross-linking data from cells includes the identification of PPIs and topologies directly from their native physiological environment. These capabilities were specifically illustrated with intact virion capsid proteins in the Potato Leafroll Virus37 and with discovery of OmpA multimer interactions and others in E. coli cells.20,21 Here we focus on the ReACT method and its advanced capacity for identification of structurally informative cross-linked peptides from cells. The size of resultant ReACT data sets presents a significant wealth of structural information derived from cells and precludes full discussion within a single publication. However, key macromolecular interactions within E. coli include the ribosome for which structural data are available and ReACT data on these complexes is discussed below. Nonetheless, the entire data sets of cross-linked peptides from E. coli cells are presented in Supporting Information.
In E. coli, ribosomes have two subunits and are composed of RNA and protein molecules with 56 different protein sequences. Figure 5 illustrates the E. coli ribosome structure (PDB 3FIH) with 3 of 4 interprotein cross-linked pairs identified from cells in this study using ReACT. Visualization of macromolecular complexes such as the ribosome has led to a better understanding of how these complexes function within the cell.38 Here we present measurements from ReACT which for the first time, confirm the protein–protein proximity within cells. In this figure, all interprotein cross-linked sites indicating PPIs are presented where linkage between two different ribosomal protein sequences was observed. For clarity, other ribosomal intraprotein cross-linked pairs (200 cross-linked pairs) that were identified are omitted; however, these crosslinks still provide unique topological information such as proximity and solvent accessibility of lysine residues as they exist in cells. Also omitted are interprotein cross-linked pairs between ribosomal and nonribosomal proteins, e.g., elongation factor TU. Three of four interprotein cross-linked sites within the ribosome assembly were mapped directly to crystallographic data (all cross-link sites are <25 Å). One observed cross-linked pair was not mapped since the available ribosomal crystal structure does not contain these proteins (RL7_ECOLI and RL10_ECOLI). The peptides identified with this cross-linked site are shown to be non-redundant or unique to the protein with which they are associated (Supplemental Table S3). RL10 and RL7 have been cocrystallized with the ribosome in T. thermophilus39 but never resolved. This cross-link between RL7-RL10 illustrates how ReACT can provide new and complementary information on complexes as they exist in cells that have been heavily studied using in vitro techniques. ReACT enables confirmation of many crystallographic measurements of the ribosomal structures with data obtained directly from cells. Many (194 <5% FDR) other nonribosomal interprotein linkages are present within these data which provides new knowledge beyond previously characterized PPIs and topologies. For a summary number of inter- and intraprotein cross-links see Supporting Information.
Figure 5.
E. coli 30s ribosome (PDB 3FIH) with 3 of 4 observed interprotein ribosomal cross-links mapped (RNA has been omitted). To view an animated illustration of cross-linked sites on the molecular structure see: http://brucelab.gs.washington.edu/ReACT_movies.php.
Interprotein cross-links discovered with ReACT provide new information about protein interactions directly from E. coli cells. These data can be broken down into three separate categories: previously observed, likely, and uncharacterized. To do this, the interprotein cross-link results presented in Figure 4 were compared to available protein interaction data from Ecocyc.org (EciD–protein interaction database). From this comparison, 39% of the PPIs presented here have been observed previously through alternative experimental techniques (yeast two hybrid, coIP, etc.). However, even for these known interactions, the data acquired with ReACT provide new topological information on these and help visualize how these proteins interact as they exist inside cells. Moreover, 50% of the PPIs discovered using ReACT were found within one node of a known interacting pair discovered using other experimental techniques. That is, 50% of the PPI discovered in cells with ReACT include proteins that are known to participate in the same complexes, but not previously known to interact directly. For example, protein A interacts with protein B and protein B interacts with protein C, but protein A is not known to interact directly with protein C based on empirical data. Here, these PPI’s are classified as secondary interactors and include for example, N-acetylmuramoyl-L-alanine amidase (AmiA) that has been shown to interact directly with proteins in the 30s (rpsA and rpsO) and the 50s (rplD) ribosome. Although direct cross-linked sites between AmiA and rplD, rpsA, or rpsO were not observed, AmiA was identified as a cross-linked product with rplB (a known direct interaction partner of rplD) of the 50s ribosome with two unique sites. Although this and other interactions appear in existing databases as secondary interactions, in vivo cross-linking results made possible with ReACT illustrate they are present in cells close to one another and can be linked directly together. If these proteins are not directly interacting, the cross-linking data suggests they are at least participating in the same complexes at the same time with nonrandom relative orientation. In summary, 89% of the interactions identified with ReACT are previously known as direct or secondary interactors. Excitingly, ReACT yields new topological data on all these interactions as they exist in cells.
This new topological data is really just the tip of the iceberg. Although ReACT represents a significant breakthrough in terms of number of cross-linked species identified in a single biological system, there is still much improvement to be made before a more comprehensive interactome “view” can be achieved in a single experiment. Two major areas exist where significant technological advancement could improve interactome coverage through cross-linking studies. The first area is with the database search strategies. Postsearch rescoring approaches as described40 are really applicable only to large data sets with the appropriate statistics. A rescoring approach that operates under new models that account for XL-MS specific assumptions, akin to that which was described for noncleavable linkers,35,36 would make further advancement on ReACT data analysis. In addition, application of a so-called stage 1 database restriction as described by Anderson et al.22 also appears to yield a 30% improvement in the number of identified cross-linked species. The second area for improvement is cross-linked sample preparation and purification of cross-linked peptides from complex mixtures. Although significant headway is being made,41 many unidentified and unlabeled tryptic peptide species are observed in the final purified samples. This added complexity hinders ReACT and limits the dynamic range of cross-linked species that can be observed. A multifaceted approach, including orthogonal separation techniques, will be required on the protein and peptide levels to truly achieve samples comprised primary of cross-linked species from cellular, tissue, or otherwise complex cross-linked samples.
CONCLUSIONS
ReACT is a new method for identifying cross-linked peptide pairs using mass spectrometry cleavable cross-linkers that is directly integrated into the mass spectral acquisition. ReACT extends current detection and identification limits of cross-linked peptide pairs by focusing the analysis time and instrument duty cycle on those ions that specifically meet the mass relationships engineered in PIR chemical cross-linkers or similar molecules. Operational time is reduced by not having to perform postacquisition data analysis beyond that of a conventional proteome database search. The ReACT algorithm is compatible for use with any flexible high resolution mass spectrometry platform with up to MS3 capability as well as a wide range of cross-linker chemistries for PPI and topology studies within complex biological systems. ReACT enables the first large-scale identification of cross-linked species from cells, on the order of 100s of cross-linked sites, which represents a 10-fold improvement over any previous report. With further improvements in cross-linked sample preparation methods, cross-linker molecular design and advanced database search strategies optimized for released peptide identification, proteome-wide PPI and topological analyses are a realistic goal.
Supplementary Material
Acknowledgments
The work was supported by the University of Washington Proteomics Resource UWPR95794 and the National Institutes of Health grants 2R01GM086688, 1R01GM097112, 5R01RR023334, and 7S10RR025107.
ABBREVIATIONS
- XL-MS
cross-linking mass spectrometry
- PPI
protein–protein interaction
- FDR
false discovery rate
- NMR
nuclear magnetic resonance
- EIC
extracted ion chromatogram
- TIC
total ion chromatogram
Footnotes
The authors declare no competing financial interest.
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ellis RJ. Macromolecular crowding: obvious but under-appreciated. Trends Biochem Sci. 2001;26(10):597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 2.Dedmon MM, Patel CN, Young GB, Pielak GJ. FlgM gains structure in living cells. Proc Natl Acad Sci USA. 2002;99(20):12681–4. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeya T, Sasaki A, Sakakibara D, Shigemitsu Y, Hamatsu J, Hanashima T, Mishima M, Yoshimasu M, Hayashi N, Mikawa T, Nietlispach D, Walchli M, Smith BO, Shirakawa M, Guntert P, Ito Y. NMR protein structure determination in living E. coli cells using nonlinear sampling. Nat Protoc. 2010;5(6):1051–60. doi: 10.1038/nprot.2010.69. [DOI] [PubMed] [Google Scholar]
- 4.Robinson KE, Reardon PN, Spicer LD. In-cell NMR spectroscopy in Escherichia coli. Methods Mol Biol. 2012;831:261–77. doi: 10.1007/978-1-61779-480-3_15. [DOI] [PubMed] [Google Scholar]
- 5.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447(7147):1021–5. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 6.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33(1):2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Chavez JD, Liu NL, Bruce JE. Quantification of protein-protein interactions with chemical cross-linking and mass spectrometry. J Proteome Res. 2011;10(4):1528–37. doi: 10.1021/pr100898e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9(8):1634–49. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rappsilber J, Siniossoglou S, Hurt EC, Mann M. A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal Chem. 2000;72(2):267–75. doi: 10.1021/ac991081o. [DOI] [PubMed] [Google Scholar]
- 10.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5(4):315–8. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh P, Shaffer SA, Scherl A, Holman C, Pfuetzner RA, Larson Freeman TJ, Miller SI, Hernandez P, Appel RD, Goodlett DR. Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal Chem. 2008;80(22):8799–806. doi: 10.1021/ac801646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang X, Bruce JE. Chemical cross-linking for protein-protein interaction studies. Methods Mol Biol. 2009;492:283–93. doi: 10.1007/978-1-59745-493-3_17. [DOI] [PubMed] [Google Scholar]
- 13.Tang X, Bruce JE. A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. Mol Biosyst. 2011;6(6):939–47. doi: 10.1039/b920876c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X, Munske GR, Siems WF, Bruce JE. Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal Chem. 2005;77(1):311–8. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29(4):717–26. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, Malmstrom L, Aebersold R. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337(6100):1348–52. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 17.Karadzic I, Maupin-Furlow J, Humbard M, Prunetti L, Singh P, Goodlett DR. Chemical cross-linking, mass spectrometry, and in silico modeling of proteasomal 20S core particles of the haloarchaeon Haloferax volcanii. Proteomics. 2012;12(11):1806–14. doi: 10.1002/pmic.201100260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics. 2007;6(12):2200–11. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Tang X, Munske GR, Tolic N, Anderson GA, Bruce JE. Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol Cell Proteomics. 2009;8(3):409–20. doi: 10.1074/mcp.M800232-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng C, Yang L, Hoopmann MR, Eng JK, Tang X, Weisbrod CR, Bruce JE. Cross-linking measurements of in vivo protein complex topologies. Mol Cell Proteomics. 2011;10(10):M110.006841. doi: 10.1074/mcp.M110.006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Zheng C, Weisbrod CR, Tang X, Munske GR, Hoopmann MR, Eng JK, Bruce JE. In vivo application of photocleavable protein interaction reporter technology. J Proteome Res. 2012;11(2):1027–41. doi: 10.1021/pr200775j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson GA, Tolic N, Tang X, Zheng C, Bruce JE. Informatics strategies for large-scale novel cross-linking analysis. J Proteome Res. 2007;6(9):3412–21. doi: 10.1021/pr070035z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoopmann MR, Weisbrod CR, Bruce JE. Improved strategies for rapid identification of chemically cross-linked peptides using protein interaction reporter technology. J Proteome Res. 2010;9(12):6323–33. doi: 10.1021/pr100572u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merrifield RB. Solid-phase peptide synthesis. 3. An improved synthesis of bradykinin. Biochemistry. 1964;3:1385–90. doi: 10.1021/bi00897a032. [DOI] [PubMed] [Google Scholar]
- 25.Sieber P. A new acid-labile anchor group for the solid-phase synthesis of C-terminal peptide amides by the Fmoc method. Tetrahedron Lett. 1987;28(19):2107–2110. [Google Scholar]
- 26.Katritzky AR, Yang B, Qiu G, Zhang Z. ChemInform abstract: A convenient trifluoroacetylation reagent: N-(Trifluoroacetyl)succinimide. ChemInform. 1999;30(19):no–no. [Google Scholar]
- 27.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 28.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5(11):959–64. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graumann J, Scheltema RA, Zhang Y, Cox J, Mann M. A framework for intelligent data acquisition and real-time database searching for shotgun proteomics. Mol Cell Proteomics. 2012;11(3):M111 013185. doi: 10.1074/mcp.M111.013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paramelle D, Miralles G, Subra G, Martinez J. Chemical cross-linkers for protein structure studies by mass spectrometry. Proteomics. 2012;13:438–56. doi: 10.1002/pmic.201200305. [DOI] [PubMed] [Google Scholar]
- 31.Liu F, Wu C, Sweedler JV, Goshe MB. An enhanced protein crosslink identification strategy using CID-cleavable chemical crosslinkers and LC/MS(n) analysis. Proteomics. 2012;12(3):401–5. doi: 10.1002/pmic.201100352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, Carr SA, Clauser KR, Jaffe JD, Kowalski KA, Neubert TA, Regnier FE, Schilling B, Tegeler TJ, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Fisher SJ, Gibson BW, Kinsinger CR, Mesri M, Rodriguez H, Stein SE, Tempst P, Paulovich AG, Liebler DC, Spiegelman C. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010;9(2):761–76. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng C, Weisbrod CR, Chavez JD, Eng JK, Sharma V, Wu X, Bruce JE. CrossLink-DB: Database and software tools for storing and visualizing protein interaction topology data. J Proteome Res. 2012 doi: 10.1021/pr301162j. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–15. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walzthoeni T, Claassen M, Leitner A, Herzog F, Bohn S, Forster F, Beck M, Aebersold R. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9:901–3. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- 36.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–6. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 37.Chavez JD, Cilia M, Weisbrod CR, Ju HJ, Eng JK, Gray SM, Bruce JE. Cross-linking measurements of the Potato leafroll virus reveal protein interaction topologies required for virion stability, aphid transmission, and virus-plant interactions. J Proteome Res. 2012;11:2968–81. doi: 10.1021/pr300041t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 39.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16(5):528–33. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 41.Leitner A, Reischl R, Walzthoeni T, Herzog F, Bohn S, Forster F, Aebersold R. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol Cell Proteomics. 2012;11(3):M111.014126. doi: 10.1074/mcp.M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.