SUMMARY
Malaria parasites scavenge nutrients from their host but also harbor enzymatic pathways for de novo macromolecule synthesis. One such pathway is apicoplast-targeted type II fatty acid synthesis, which is essential for late liver stage development in rodent malaria. It is likely that fatty acids synthesized in the apicoplast are ultimately incorporated into membrane phospholipids necessary for exoerythrocytic merozoite formation. We hypothesized that these synthesized fatty acids are being utilized for apicoplast-targeted phosphatidic acid synthesis, the phospholipid precursor. Phosphatidic acid is typically synthesized in a three-step reaction utilizing three enzymes: glycerol 3-phosphate dehydrogenase, glycerol 3-phosphate acyltransferase and lysophosphatidic acid acyltransferase. The Plasmodium genome is predicted to harbor genes for both apicoplast- and cytosol/endoplasmic reticulum-targeted phosphatidic synthesis. Our research shows that apicoplast-targeted P. yoelii glycerol 3-phosphate dehydrogenase and glycerol 3-phosphate acyltransferase are expressed only during liver stage development and deletion of the encoding genes resulted in late liver stage growth arrest and lack of merozoite differentiation. However, the predicted apicoplast-targeted lysophosphatidic acid acyltransferase gene was refractory to deletion and was expressed solely in the endoplasmic reticulum throughout the parasite lifecycle.
Our results suggest that P. yoelii has an incomplete apicoplast-targeted phosphatidic acid synthesis pathway that is essential for liver stage maturation.
Keywords: Plasmodium, fatty acid, phosphatidic acid, liver stage, apicoplast
INTRODUCTION
Malaria continues to be a global health disaster. The disease, caused by Plasmodium species, was contracted by upwards of 219 million people in 2010 leading to 660,000 deaths (WHO, 2012). Although global malaria mortality declined between 2004 and 2010 (Murray et al., 2012), the threat of insecticide resistance as well as evidence for P. falciparum resistance to artemisinin combination therapies (Takala-Harrison et al., 2013) dictates continued research into novel control mechanisms to keep the parasite at bay. Plasmodium parasites harbor an apicoplast, an essential non-photosynthetic plastid of cyanobacterial origin (Funes et al., 2002). Consequently, the apicoplast’s biochemical pathways are attractive targets for antimalarial drugs as they are often fundamentally different from the eukaryotic host’s pathways. Indeed, in silico identification of proteins that likely target to the apicoplast along with ongoing research have uncovered a number of biochemical pathways, including isoprenoid-, fatty acid- and heme biosynthesis as attractive antimalarial drug targets (Ralph et al., 2004).
The apicoplast harbors the parasite’s sole fatty acid biosynthetic pathway, type II fatty acid synthesis (FAS II), which differs in many respects from mammalian FAS I, and thus is an attractive antimalarial drug target. It was initially assumed that FAS II would be an ideal target for blood stage antimalarial therapy (Waller et al., 2003, Gornicki, 2003) but more recently it has been shown, using gene knockouts, that P. falciparum FAS II is not required for in vitro asexual blood stage replication (Vaughan et al., 2009, Yu et al., 2008). Further studies on the rodent malaria models, P. yoelii and P. berghei showed that FAS II was necessary only for late liver stage development and maturation of infectious merozoites (Vaughan et al., 2009, Yu et al., 2008). Indeed, P. yoelii parasites lacking Fab B/F, one of the key enzymes involved in the elongation of the fatty acid carbon backbone, fail to complete the final phases of liver stage development and thus are completely attenuated at this life cycle stage (Vaughan et al., 2009). These studies demonstrated the need for genetic validation, which can prove that a prospective apicoplast drug target is indeed active in the life cycle stage in which the drug study is undertaken. Although it has been shown that FAS II is essential for rodent malaria liver stage maturation, the fate and further role of the synthesized fatty acids is unclear.
Fatty acids are a major component of the phospholipids that make up all cellular membranes and one hypothesis would be that the fatty acids are required for the enormous amount of membrane biosynthesis necessary late in liver stage development for the formation of tens of thousands of exoerythrocytic merozoites (Tarun et al., 2009). The precursor of phospholipid biosynthesis is phosphatidic acid. Phosphatidic acid is formed in a three-step enzymatic reaction (Yao et al., 2013), the first step of which is the conversion of dihydroxyacetone phosphate (DHAP) to glycerol 3-phosphate by glycerol 3-phosphate dehydrogenase (G3PDH). Two fatty acid side chains are then transferred to glycerol 3-phosphate, firstly by glycerol 3-phosphate acyltransferase (G3PAT) to form lysophosphatidic acid and then lysophosphatidic acid acyltranferase (LPAAT) to form phosphatidic acid. In plants, there are two pathways of phosphatidic acid biosynthesis, one is prokaryotic in origin and targeted to the plastid and the second is of eukaryotic origin, is extraplastidial and typically localizes to the endoplasmic reticulum (ER) (Ohlrogge et al., 1995). Furthermore, separate gene families code for the plastidial and extraplastidial enzymes. Analysis of the Plasmodium genome has also uncovered two sets of genes for phosphatidic acid biosynthesis and one set is predicted to target to the apicoplast (Ralph et al., 2004). We hypothesized that apicoplast-targeted phosphatidic acid biosynthesis would be an essential downstream event following fatty acid synthesis by FAS II in the liver stage. Indeed, our research in the rodent malaria parasite P. yoelii demonstrates that G3PDH and G3PAT are localized to the apicoplast only during liver stage development, where they prove to be essential. Unexpectedly, we also show that there appears to be no specific apicoplast-targeted LPAAT. Our results suggest that liver stage FAS II biosynthesis provides fatty acids essential for atypical downstream phosphatidic acid synthesis, likely required for de novo phospholipid creation for exoerythrocytic merozoite formation.
RESULTS
Apicoplast-targeted G3PDH and G3PAT are expressed only during liver stage development
G3PDH and G3PAT are the first two enzymes involved in the biosynthesis of phosphatidic acid and to test for the presence of apicoplast-targeting enzymes involved in phosphatidic acid biosynthesis, we created transgenic P. yoelii XNL parasites that express a 4× myc epitope tag fused to the C-terminus of G3PDH (PY00789, PlasmoDB.org, Py apiG3PDHmyc) or the apicoplast-targeted G3PAT (Py apiG3PATmyc), each of which exhibited a predicted apicoplast-targeting sequence. However, the current annotation of the P. yoelii 17XNL genome is incomplete and no apiG3PAT ortholog was present. Thus, based on the predicted cDNA sequences of the P. berghei apiG3PAT, we created primers to amplify the gene and cDNA from P. yoelii genomic DNA and liver stage cDNA respectively. A complete open reading frame for apiG3PAT was obtained, as well as a gene sequence. More recently, a mostly complete annotation of the P. yoelii YM strain genome has been deposited in PlasmoDB.org and the YM apiG3PAT sequence (PYYM_1420200) is in agreement with the sequence we generated for P. yoelii XNL.
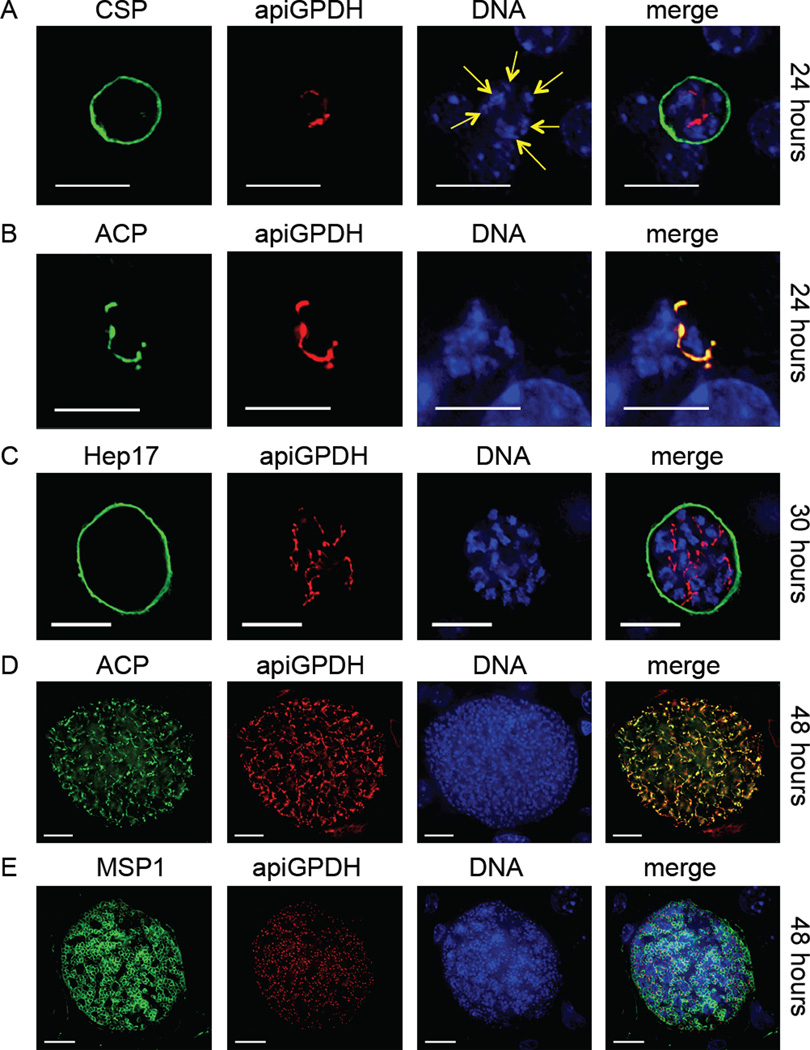
The transgenic myc-epitope expressing parasites were created by gene replacement (Lindner et al., 2013) and thus contained a single epitope-tagged copy of the gene under its endogenous promoter (Fig. S1A, B). We used immunofluorescence assay (IFA) to determine the spatial and temporal expression of apiG3PDH throughout the parasite’s life cycle. Expression was not observed in blood stages (Fig. S2), or salivary gland sporozoites by IFA (Fig. S3) or by Western blot in the case of blood stages (Fig. S4). Furthermore, the number of salivary gland sporozoites per mosquito recovered after infectious blood meal was similar to wildtype (Fig. S5) and we assumed that the creation of the myc-tagged parasite was not affecting the fitness of the parasite. As we predicted, apiG3PDH was expressed in the apicoplast of liver stage parasites. The apicoplast branches extensively during liver stage development until the formation of exoerythrocytic merozoites, by which time, each merozoite contains a single spherical apicoplast (Stanway et al., 2009). Mice injected with apiG3PDH sporozoites were used to assay liver stage development of Py apiG3PDHmyc by IFA. Using an antibody to the plasma membrane protein circumsporozoite protein (CSP) and an antibody to the myc epitope, apiG3PDH expression was clearly seen at 24 hours (Fig. 1A) after sporozoite infection and was reminiscent of that seen for apicoplast-targeted proteins of FAS II (Vaughan et al., 2009). To confirm that apiG3PDH was indeed expressed in the apicoplast, an antibody to the apicoplast lumen protein acyl carrier protein (ACP) was used to demonstrate co-localization at 24 hours (Fig. 1B). Liver stage parasites were further analyzed at 30 hours using the parasitophorous vacuole membrane (PVM) protein Hep17 to delineate the parasite. As expected, the liver stage parasites are larger, the apicoplast structure is more complex and DNA replication has increased from the 24 hour time point (Fig. 1C). A highly branched apicoplast was present at 48 hours of liver stage development, as seen by ACP and myc co-localization (Fig. 1D). At 48 hours of development, some wildtype liver stage parasites have fully matured and contain individual merozoites. This was observed for Py apiG3PDHmyc also and IFA using antibody to merozoite surface protein 1 (MSP1) demonstrated the presence of merozoites, each of which contained an individual spherical apicoplast, based on myc expression (Fig. 1E). Py apiG3PDHmyc parasites completed liver stage development and transitioned to blood stage patency, at which time, apiG3PDH expression was absent (Fig. S2).
Figure 1.
The Plasmodium glycerol 3-phosphate dehydrogenase (apiG3PDH) predicted to target to the apicoplast is expressed only during liver stages and co-localizes with the apicoplast lumen-targeted acyl carrier protein (ACP). The expression of apiG3PDH was assessed with the transgenic epitope-tagged Py apiG3PDHmyc parasite and identified by IFA. At 24 hours (A), nuclear replication was apparent (yellow arrows point to multiple nuclei centers) and apiG3PDH expression was in internal structures reminiscent of the apicoplast. Antibody to circumsporozoite protein (CSP) was used to delineate the parasite plasma membrane (PPM). It was also apparent at 24 hours (B), that apiG3PDH perfectly co-localized with the apicoplast lumen protein acyl carrier protein (ACP). At 30 hours (C), liver stages had increased in size, as had the complexity of apiG3PDH localization, which was contained within the parasitophorous vacuole membrane (PVM) marker Hep17. At 48 hours (D), co-localization of apiG3PDH with ACP clearly shows that apiG3PDH is localized to the extensively branched apicoplast in a late-liver stage parasite. Fully mature liver stages at 48 hours showed the presence of merozoites, localized with the PPM marker merozoite surface protein 1 (MSP1) and each merozoite, as expected, appeared to contain a single apicoplast, based on apiG3PDH expression (E). DNA was stained with 4',6-diamidino-2-phenylindole. Scale bar: 10 µm.
Due to the fact that apiG3PDH was expressed only during liver stage development, we assumed that apiG3PAT would also be expressed in a similar manner. Indeed, phenotypic analysis of Py apiG3PATmyc by IFA showed that, as predicted, apiG3PAT was expressed within the liver stage parasites only and localized to the apicoplast (Fig. S6). Expression was not observed in blood stages (Fig. S2), or salivary gland sporozoites by IFA (Fig. S3) or by Western blot in the case of blood stages (Fig. S4). Furthermore, the number of salivary gland sporozoites per mosquito recovered after infectious blood meal was similar to wildtype (Fig. S5) and we thus assumed that as for the Py apiG3PDHmyc parasite, the creation of the myc-tagged Py apiG3PATmyc parasite was not affecting the fitness of the parasite. As we predicted, apiG3PAT was expressed in the apicoplast of liver stage parasites. This result shows that the predicted apiG3PAT is indeed an apicoplast-targeted enzyme and is expressed exclusively in liver stage parasites. Thus, the P. yoelii genome encodes apicoplast-targeted enzymes that are likely involved in the biosynthesis of phosphatidic acid.
Apicoplast-targeted G3PDH and G3PAT are essential for liver stage development
Deletions in genes of FAS II from P. yoelii lead to complete attenuation of the parasite late in liver stage development in BALB/cJ mice (Vaughan et al., 2009), as do deletions in genes that code for the apicoplast-resident pyruvate dehydrogenase complex (Pei et al., 2010), which provides the acetyl CoA necessary for FAS II activity. On the other hand, deletions of both FAS II and pyruvate dehydrogenase complex genes in the P. berghei rodent malaria parasite result in a severe but not complete attenuation in C57BL/6 mice (Nagel et al., 2013, Yu et al., 2008). Based on this research and on the fact that both P. yoelii apiG3PDH and apiG3PAT are expressed exclusively in liver stages, we hypothesized that gene knockout would be possible and result in the creation of knockout parasites that would affect liver stage development. Previous gene knockouts of P. yoelii FAS II genes using standard methodology have been successful (Vaughan et al., 2009) and we used this methodology to knockout both apiG3PDH and apiG3PAT in P. yoelii blood stage parasites and by limiting dilution isolated knockout parasite clones [Py apig3pdh(−) and Py apig3pat(−)] from two independent transfections (Fig. S7A, B). Blood stage replication (Fig. S8) and salivary gland sporozoite production (Fig. S5) were not affected in the knockout parasites, suggesting that the gene knockouts are having no effect on these stages of the life cycle. However, when salivary gland sporozoites were isolated from knockout parasites and used to infect Swiss Webster mice (50,000 sporozoites were injected intravenously into groups of five mice on three separate occasions), blood stage patency was not seen (Table 1). Conversely, mice injected with wildtype sporozoites all became blood stage patent three days after sporozoite inoculation (Table 1). This result suggested that both Py apig3pdh(−) and Py apig3pat(−) liver stages were unable to complete liver stage development, resulting in liver stage arrest before the maturation of exoerythrocytic merozoites.
Table 1.
Py apig3pdh(−) and Py apig3pat(−) sporozoites fail to complete liver stage development and transition to blood stages.
| Parasite Type | # Sporozoites Injecteda |
Repeat #1 # SW Mice Infected |
Repeat #1 # SW Mice Patent (Day of Patency)b |
Repeat #2 # SW Mice Infected |
Repeat #2 # SW Mice Patent (Day of Patency)b |
|---|---|---|---|---|---|
| Py 17XNL (WT control) | 50,000 | 5 | 5 (day 3) | 5 | 5 (day 3) |
| Py apig3pdh (−) (Clone 1) | 50,000 | 5 | 0 | 5 | 0 |
| (Clone 2) | 50,000 | 5 | 0 | 5 | 0 |
| Py apig3pat (−) (Clone 1) | 50,000 | 5 | 0 | 5 | 0 |
| (Clone 2) | 50,000 | 5 | 0 | 5 | 0 |
50,000 salivary gland sporozoites were injected intravenously into female Swiss Webster (SW) mice and Giemsa-stained thin blood smears were analyzed for patency from days three to fourteen after injection.
No mice injected with knockout parasites became patent.
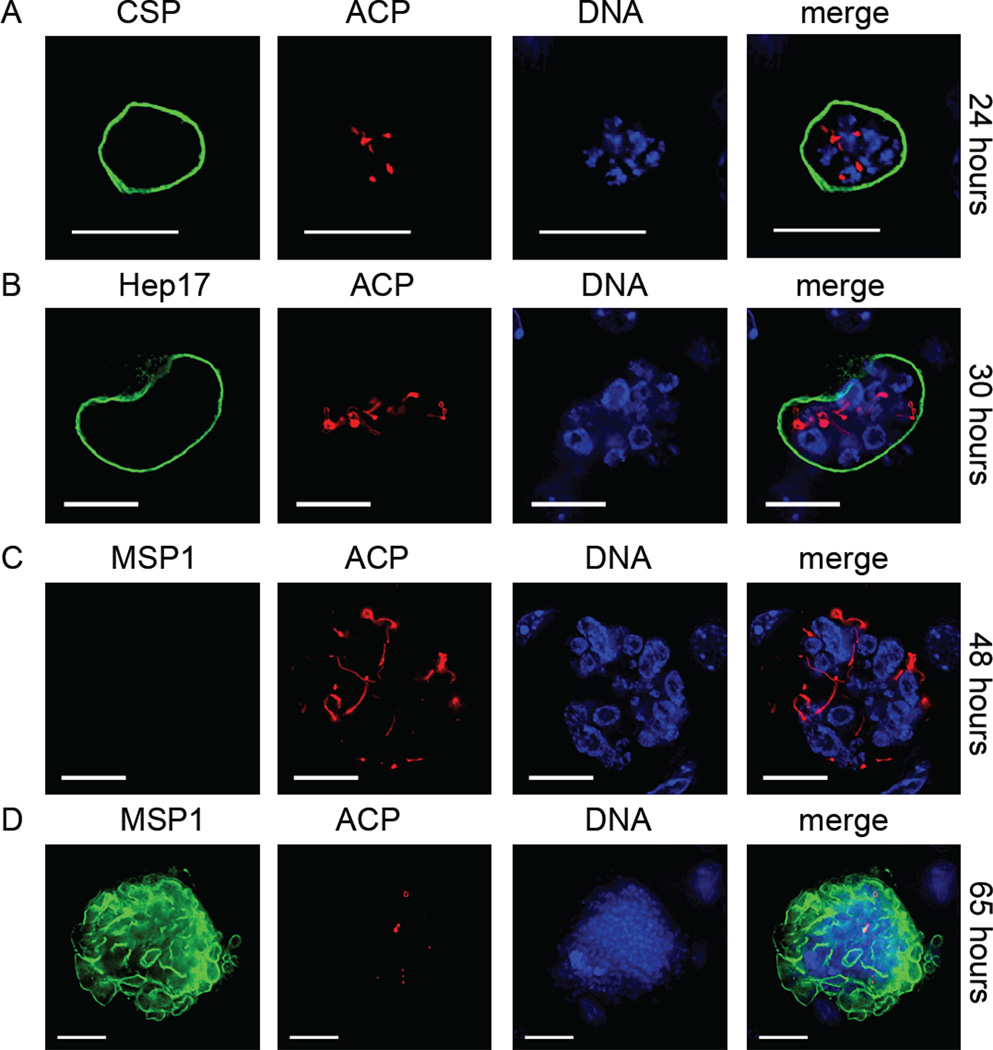
To determine the phenotype associated with the Py apig3pdh(−) and Py apig3pat(−) parasite lines, gene knockout and wildtype sporozoites were injected intravenously into BALB/cAnN mice and their livers removed at increasing time points after injection. IFA was used to determine the effect of gene knockout on liver stage development. Liver stage development of the Py apig3pdh(−) parasite was comparable to Py apiG3PDHmyc at 24 hours and the architecture of the apicoplast was similar (Fig. 2A and refer to ACP expression in Fig. 1B). However, by 30 hours, the Py apig3pdh(−) apicoplast showed signs of aberrant development, and the replication of DNA was also less pronounced when compared to Py apiG3PDHmyc (Fig. 2B and refer to Fig. 1C). By 48 hours only 25% (total number of liver stages examined = 88) of Py apig3pdh(−) liver stages showed any sign of MSP1 expression, and those that did express MSP1 had only a weak signal (Fig. 2C). Most liver stage parasites expressed no MSP1, had aberrant apicoplast-replication when compared to Py apiG3PDHmyc liver stages and were also smaller than Py apiG3PDHmyc 48 hour liver stages (Fig. 2C). At 65 hours after sporozoite inoculation, no liver stages were detectable in the liver of mice inoculated with Py apiG3PDHmyc sporozoites. Conversely, at this time point, Py apig3pdh(−) liver stage parasites were still detectable in the liver, the majority of which (95%, total number of liver stages examined = 48) were expressing MSP1 (Fig. 2D). Late in schizogony, MSP1 is expressed on the parasite plasma membrane (PPM) of the developing cytomeres - multiple invaginations of the PPM that eventually lead to individual merozoite membranes. The MSP1 expression at 65 hours (Fig. 2D) is reminiscent of the start of cytomere formation. Additionally, further nuclear division had occurred in the 65 hour Py apig3pdh(−) liver stages when compared to the 48 hour liver stage although ACP expression was either very weak or lacking in all parasites examined (Fig. 2D). The results suggest that the Py apig3pdh(−) liver stage continues to develop beyond 48 hours and MSP1 expression is turned on but the functionality of the apicoplast is lost. No Py apig3pdh(−) liver stage parasites were detected 96 hours after sporozoite injection by IFA, suggesting that they had been eliminated by the murine immune system.
Figure 2.
Gene knockout of apicoplast-targeted glycerol 3-phosphate dehydrogenase (apiG3PDH) results in arrested liver stage development. Loss of apiG3PDH was assessed with the Py apig3pdh(−) parasite by IFA. At 24 hours (A), liver stage development appeared normal, based on expression of the parasite plasma membrane (PPM)-localized circumsporozoite protein (CSP) and the apicoplast-targeted acyl carrier protein (ACP) and parasite size (compare to Fig. 1A). At 30 hours (B), nuclear replication and apicoplast development were retarded compared to wildtype parasites although the parasitophorous vacuole membrane (PVM), based on Hep17 localization, appeared intact (compare to Fig. 1C). At 48 hours (C), little or no MSP1 expression was seen in Py apig3pdh(−) liver stages (compare to Fig. 1E). Additionally, the liver stages were smaller and both nuclear replication and apicoplast complexity were stunted. At 65 hours (D), Py apig3pdh(−) liver stages were detected, the majority of which were expressing MSP1 but not ACP, suggesting that apicoplast integrity was compromised. DNA was stained with 4',6-diamidino-2-phenylindole. Scale bar: 10 µm.
Due to the fact that we have shown P. yoelii apiG3PDH and apiG3PAT expression only in the apicoplast of liver stage parasites, we hypothesized that the Py apig3pat(−) parasite, like the Py apig3pdh(−) parasites, would attenuate in the liver stage. Indeed, we found that the Py apig3pat(−) liver stage parasites were attenuated during liver stage development and shared a similar phenotype to that seen for Py apig3pdh(−) liver stage progression (Fig. S9). The results show that the knockout of both apiG3PDH and apiG3PAT give similar phenotypes and we hypothesize that the two genes both play a role in the synthesis of apicoplast-generated lysophosphatidic acid.
To determine if the Py apig3pdh(−) and Py apig3pat(−) liver stage parasites could act as an experimental vaccine (Khan et al., 2012), groups of five BALB/cJ mice were immunized with 10,000 knockout sporozoites (from two individual clones) and boosted six weeks later with the same dose. This experimental strategy was carried out twice. Six weeks after the boost, mice were challenged with 10,000 wildtype sporozoites. Control mice, immunized with the salivary gland extract from sporozoite-free mosquitoes became blood stage patent three days after challenge whereas the Py apig3pdh(−) and Py apig3pat(−) sporozoite-immunized mice were completely protected and never became blood stage patent during a two week analysis of Giemsa-stained blood smears (Table 2). Thus, as for Py fabb/f(−) sporozoites (Vaughan et al., 2009), the Py apig3pdh(−) and Py apig3pat(−) sporozoites act as powerful immunogens.
Table 2.
Py apig3pdh(−) and Py apig3pat(−) sporozoites act as immunogens, protecting mice from a wildtype sporozoite challenge.
| Parasite Typea | Primeb | Boost (6 weeks)b |
Challenge (6 weeks after boost)c |
Repeat #1 #BALB/cJ mice patentd |
Repeat #2 #BALB/cJ mice patentd |
Repeat #3 #BALB/cJ mice patentd |
|---|---|---|---|---|---|---|
| Py 17XNL (WT control) | n/a | n/a | 10,000 | 5/5 | 5/5 | 5/5 |
| Py apig3pdh(−) (Clone 1) | 10,000 | 10,000 | 10,000 | 0/5 | 0/5 | 0/5 |
| (Clone 2) | 10,000 | 10,000 | 10,000 | 0/5 | 0/5 | 0/5 |
| Py apig3pat(−) (Clone 1) | 10,000 | 10,000 | 10,000 | 0/5 | 0/5 | 0/5 |
| (Clone 2) | 10,000 | 10,000 | 10,000 | 0/5 | 0/5 | 0/5 |
Control mice were injected with salivary gland lysate from uninfected mosquitoes for the prime and boost.
10,000 salivary gland sporozoites were injected intravenously for both the prime and boost. For controls, mice were injected with an equivalent amount of mosquito salivary gland extract from mosquitoes that received and uninfectious blood meal.
10,000 Py 17XNL wildtype salivary gland sporozoites were injected intravenously for the challenge.
Giemsa-stained thin blood smears were analyzed for patency from days three to fourteen after challenge. No mice immunized with Py apig3pdh(−) and Py apig3pat(−) sporozoites became patent.
Apicoplast-targeted G3PAT rescues G3PAT deficient Escherichia coli
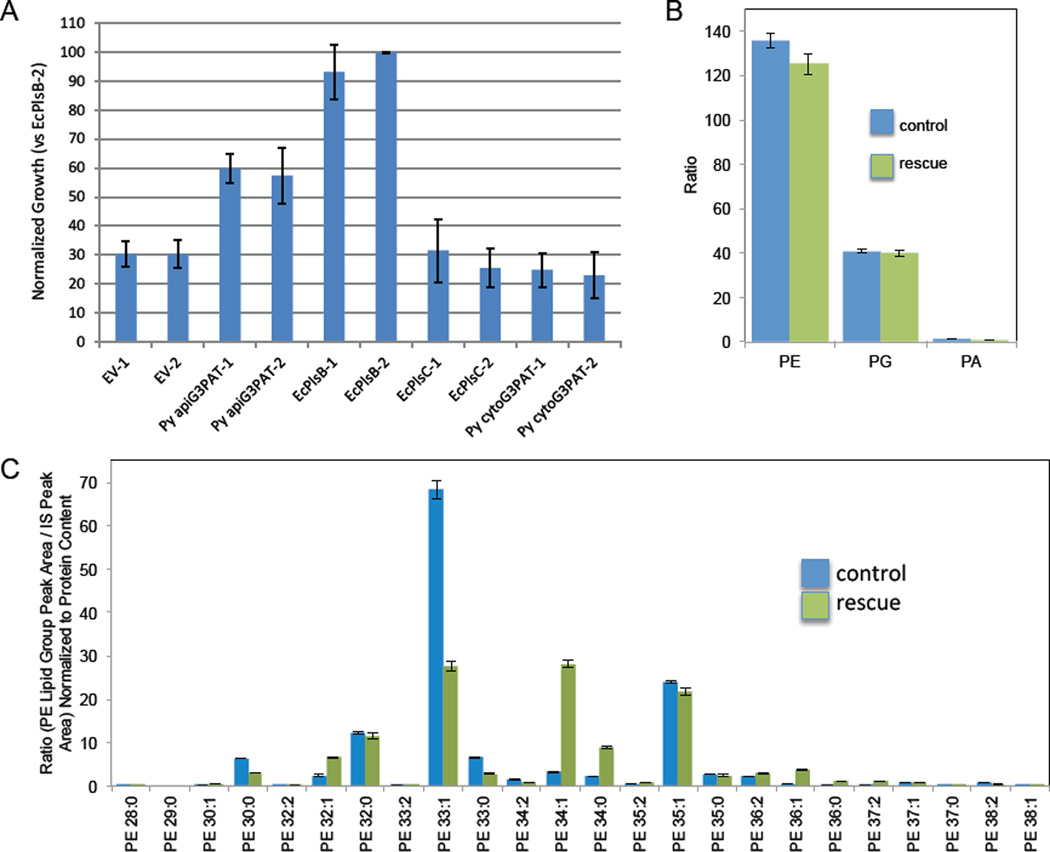
As we observed that P. yoelii apiG3PAT plays a critical role in the development of the parasite, we sought to further determine its enzymatic function. As Py apiG3PAT is predicted to be a G3PAT of prokaryotic origin (due to the algal provenance of the apicoplast (Kohler et al., 1997)), we utilized complementation of the G3PAT-deficient E. coli strain BB26-36 to confirm this activity in vivo (McIntyre et al., 1977). Recombinant P. yoelii apiG3PAT (minus the bipartite leader sequence), the extraplastidial P. yoelii G3PAT (PY06015) and E. coli PlsB (native E. coli G3PAT as a positive control), E. coli PlsC (native E. coli LPAAT, a negative control) or an empty vector product (a further negative control) were expressed from a plasmid utilizing a strong, constitutive Lac promoter. Bacteria harboring one of each of these plasmids were cloned, and two clones of each type were grown in triplicate in permissive or non-permissive conditions (Fig. 3A). Bacteria expressing the native E. coli G3PAT (EcPlsB) grew well in both permissive and non-permissive conditions, whereas those expressing the empty vector product or the native E. coli LPAAT (EcPlsC), that does not exhibit G3PAT activity, did not grow well in non-permissive conditions, although some growth was seen due to the fact that BB26-36 itself can grow poorly in non-permissive conditions. Expression of P. yoelii apiG3PAT allowed bacterial growth to approximately 66% of the levels permitted by EcPlsB, thus indicating that P. yoelii apiG3PAT indeed functions as a G3PAT, as it can partially rescue the growth-arrested phenotype. In contrast, expression of the extraplastidial P. yoelii G3PAT (predicted to be a G3PAT of eukaryotic origin) failed to complement growth deficiency, likely because the enzyme relies upon an acyl-CoA derivative for catalysis rather than the prokaryotic acyl-ACP derivative (Murata et al., 1997).
Figure 3.
The predicted apicoplast-targeted glycerol 3-phosphate acyltransferase (apiG3PAT) functionally rescues the G3PAT-deficient Escherichia coli strain BB26-36. (A) BB26-36 was transformed with plasmids that express an empty vector (EV), P. yoelii apiG3PAT (Py apiG3PAT), the native E. coli G3PAT, PlsB (EcPlsB), the native E. coli lysophosphatidic acid acyltransferase (LPAAT), PlsC (EcPlsC), or the extraplastidial P. yoelii G3PAT (Py cytoG3PAT). Independent clones were grown in triplicate and growth was determined spectrophotometrically. Values were normalized to the amount of growth of EcPlsB clone 2 (EcPlsB-2). Error bars represent the standard deviation across three technical replicates. Results show that only Py apiG3PAT and E. coli PlsB rescue G3PAT deficiency. (B) Mass spectrometric analysis of the phosphatidyl ethanolamine (PE), phosphatidyl glycerol (PG) and phosphatidic acid (PA) content from three technical replicates of BB26-36 transformed with empty vector and grown in minimal media with glycerol (control) and Py apiG3PAT-rescued BB26-36 grown in minimal media (rescue). The ratio is based on the summed phospholipid species peak area in relation to the internal standard peak area, normalized to protein content. Error bars represent the mean ± SEM (from three independent culture replicates). The results show that control and rescued bacteria have similar phospholipid content. (C) PE species were compared between the control and rescue cells. Carbon chain lengths analyzed ranged from C28:0 through C38:1. Lipid quantity is displayed as a ratio (phospholipid species peak area / internal standard peak area) normalized to protein content. Error bars represent the mean ± SEM (three independent culture replicates). When comparing the control with the rescue, the most striking difference was the decrease of the most common control species, C33:1 coupled with a substantial increase in C34:1, in the rescue.
To determine lipid content of the P. yoelii apiG3PAT-rescues BB26-36, we performed targeted mass spectrometry-based lipidomic analysis (Supplemental Table S1) of the major phospholipid classes of the apiG3PAT-rescued BB26-36 cells and compared this analysis of the parental E. coli strain grown in supplemented minimal media (Feng et al., 2011, McIntyre et al., 1977). The phospholipid content of the rescued BB26-36 cells was similar to the control (Fig. 3B) and the majority of all phospholipid was phosphatidyl ethanolamine, as has previously been documented (Oursel et al., 2007). Moreover, the sensitivity of the analysis also enabled detection of phosphatidyl glycerol and phosphatidic acid, the levels of which were comparable between the control and rescued BB26-36 cells (Fig. 3B). Further analysis was undertaken to determine the fatty acid makeup of the phospholipids (Supplemental Table S1). A wide range of fatty acids compositions were observed and significant differences between the control and rescued E. coli were seen in all phospholipid classes studied, including phosphatidyl ethanolamine (Fig. 3C). In the control BB26-36 cells, the major phosphatidyl ethanolamine was 33:1 (Fig. 3C), as has previously been reported (Oursel et al., 2007). Rescue of BB26-36 cells by P. yoelii apiG3PAT resulted in a change of the major phosphatidyl ethanolamine lipid species from C33:1 to C34:1 (Fig. 3C). This result demonstrates that although the apiG3PAT can rescue the BB26-36 cells, the heterologous expression of the apiG3PAT in E. coli dramatically alters the phospholipid properties of the rescued cells.
The predicted LPAAT localizes to the endoplasmic reticulum and not the apicoplast and is expressed throughout the parasite life cycle
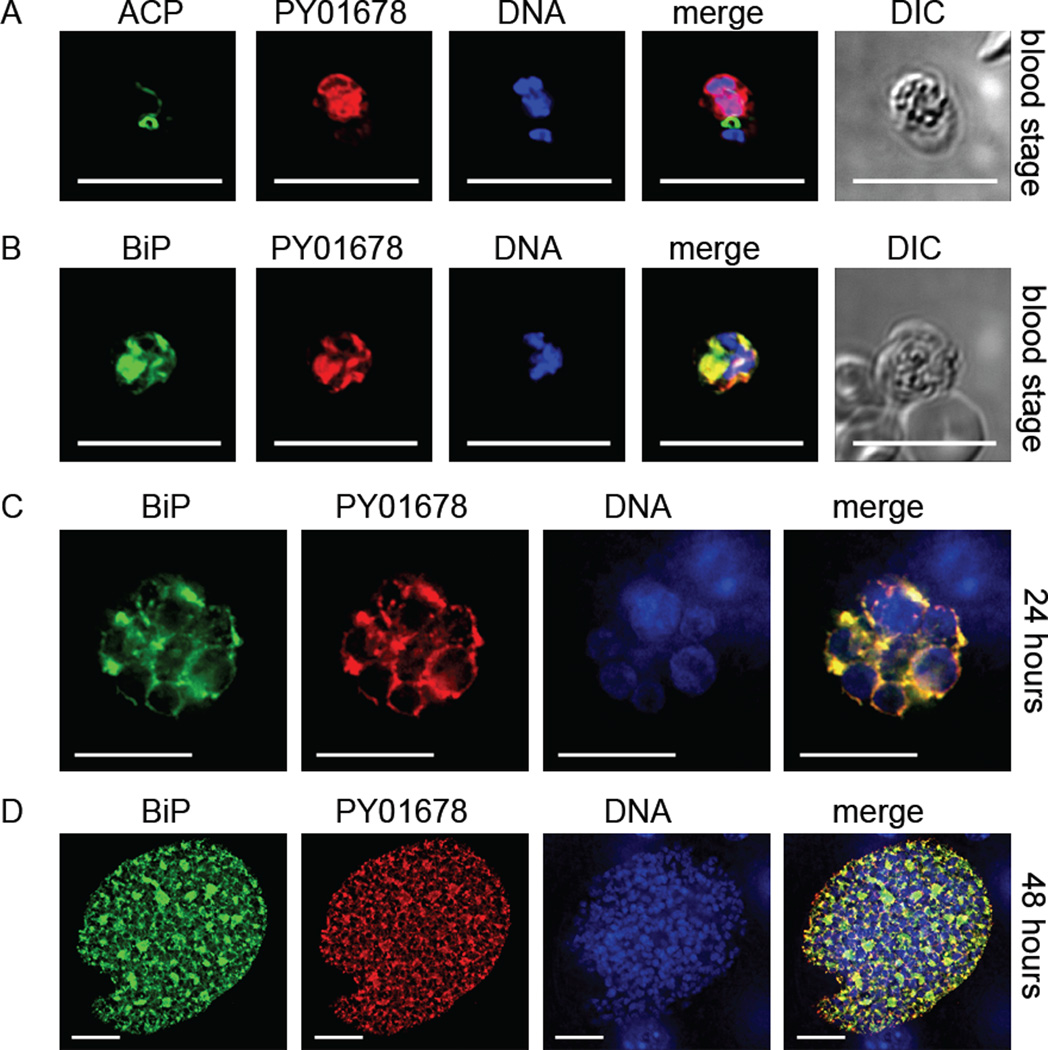
PY01678 and its Plasmodium orthologs are predicted by PlasmoDB.org to be the apicoplast-targeted LPAAT. However, no non-apicoplast-targeted LPAAT is predicted to be present in the Plasmodium genome, which is surprising as there are predicted genes coding for non-apicoplast-targeted G3PDH and G3PAT. We assumed that PY01678, like apiG3PDH and apiG3PAT would be essential only for liver stage development. However, unexpectedly, we were unable to knockout PY01678 (Fig. S3C), suggesting the gene was essential for blood stage replication. We thus decided to confirm the apicoplast-targeted localization of PY01678, and created a 4× myc epitope-tagged transgenic parasite Py 01678myc (Fig. S1C). In agreement with our gene knockout failure, PY01678 was in fact strongly expressed in blood stage parasites but surprisingly was found in the ER, not the apicoplast (Fig. 4A), as demonstrated by its co-localization with the ER chaperone binding immunoglobulin protein (BiP) (Fig. 4B). Western blotting of Py 01678myc mixed blood stage lysates confirmed the expression of a myc-tagged protein of approximately 39 kDa (Fig. S4), which would be predicted based on the amino acid length of the tagged protein. Thus, we concluded that PY01678 was not the apicoplast-targeted LPAAT, was essential for parasite blood stage replication and expressed in the ER. In liver stages, PY01678 ER expression was strong at 24 hours and 48 hours (Fig. 4C, D). The results suggest that PY01678 is an ER-targeted enzyme that is essential for blood stage replication and also highly expressed by the liver stage parasite and thus likely necessary for this life cycle stage also.
Figure 4.
A predicted lysophosphatidic acid acyltransferase (LPAAT) is expressed in the blood stage and liver stage endoplasmic reticulum (ER) but not the apicoplast. The expression of the apicoplast-predicted LPAAT was assessed using the epitope-tagged PY01678myc parasite. Blood stage parasites expressed PY01678 that did not co-localize with the apicoplast marker acyl carrier protein (ACP) (A) but did co-localize with the ER marker binding immunoglobulin protein (BiP) (B). At 24 hours (C) and 48 hours (D) of liver stage development, strong PY01678 expression was seen and as for blood stage expression, expression co-localized with BiP. DNA was stained with 4',6-diamidino-2-phenylindole and differential interference contrast (DIC) images were captured. Scale bar: 10 µm.
Because we were unable to find evidence of an apiLPAAT, we postulated that apiG3PAT might also have LPAAT activity. To determine if this was the case, we attempted to rescue an LPAAT-deficient E. coli strain, JC201 (Coleman, 1990, Cullinane et al., 2005), with apiG3PAT. Similar to our rescue strategy utilizing BB26-36, recombinant P. yoelii apiG3PAT, E. coli PlsB (native E. coli G3PAT as a negative control), E. coli PlsC (native E. coli LPAAT, a positive control) or an empty vector product (a further negative control) were expressed from a plasmid utilizing a strong, constitutive Lac promoter. Bacteria harboring one of each of these plasmids were cloned, and two clones of each type were grown in triplicate in permissive (37°C) or non-permissive conditions (42°C). Bacteria expressing the native E. coli LPAAT (EcPlsC) grew well in both permissive and non-permissive conditions, whereas those expressing the empty vector product, the native E. coli G3PAT (EcPlsB) or the extraplastidial P. yoelii G3PAT grew at 37°C but did not grow at all at 42°C. Taken together, this suggests that P. yoelii apiG3PAT specifically provides a G3PAT activity to the parasite apicoplast and does not have a dual function as an LPAAT. To further analyze the function of PY01678, we set out to determine if this gene product was able to rescue JC201. Indeed, JC201 harboring recombinant expression of PY01678 grew in in non-permissive conditions, suggesting that PY01678 has LPAAT activity, suggesting that PY01678 codes for a functional LPAAT.
A second predicted parasite LPAAT shows partial localization to the ER of blood stage and liver stage parasites
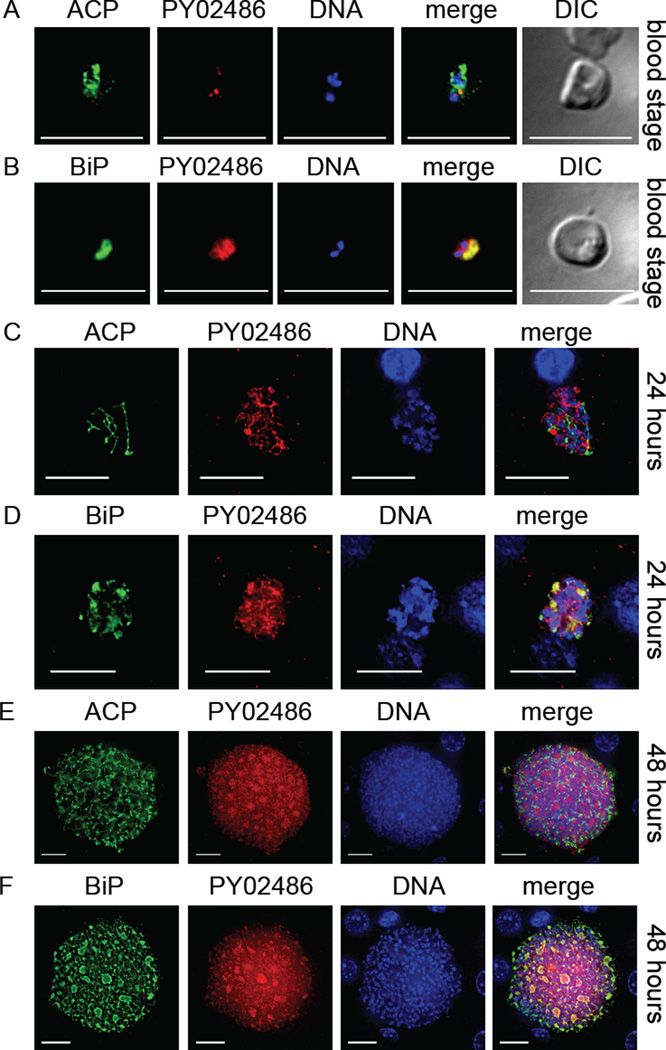
We hypothesized that since PY01678 did not localize to the apicoplast that the Plasmodium genome would code for an undiscovered apiLPAAT. We thus conducted in silico analysis of the Plasmodium genome and uncovered a further hypothetical LPAAT, PY02486, based on the presence of G3PAT-like signature motif. We successfully created a transgenic parasite (Py 02486myc) expressing a 4× myc epitope tag at the C-terminus of PY02486 as before (Fig. S1D). To determine the expression pattern of PY02486, we analyzed blood stage and liver stage development of Py 02486myc. In blood stages, Py 02486myc expression was weak and did not appear to co-localize with the apicoplast marker ACP (Fig. 5A) but partially co-localized with the ER marker BiP (Fig. 5B). Western blotting of blood stage lysate failed to detect the tagged PY02486 (Fig. S4) and we assume this was due to the low level expression. To examine liver stage expression, BALB/cAnN mice were injected with Py 02486myc sporozoites and their livers analyzed by IFA at 24 (Fig. 5C, D) and 48 hours (Fig. 5E, F) after sporozoite injection. As for blood stage expression, Py 02486myc expression was weak and did not appear to co-localize with the apicoplast marker ACP (Fig. 5C, E) but did partially co-localize with the ER marker BiP (Fig. 5D, F). Additionally, the recombinant expression of PY02486 in the LPAAT-deficient E. coli strain JC201 did not rescue the bacteria when grown at the non-permissive 42°C. These combined results suggest that PY02486 does not possess LPAAT activity and does not exclusively localize to the apicoplast.
Figure 5.
A predicted lysophosphatidic acid acyltransferase (LPAAT) (PY02486) is expressed in the blood stage and liver stage and partially co-localizes to the endoplasmic reticulum (ER) but does not predominantly localize to the apicoplast. The expression of the predicted LPAAT was assessed with the transgenic epitope-tagged PY02486myc parasite by IFA. In blood stages, weak PY02486 expression was seen that did not predominantly co-localize to the apicoplast lumen-expressed acyl carrier protein (ACP) (A) but did partially co-localize with the ER marker binding immunoglobulin protein (BiP) (B). A similar localization and weak staining pattern in relation to blood stage expression was also seen during liver stage development at both 24 hours (C, D) and 48 hours (E, F). DNA was stained with 4',6-diamidino-2-phenylindole and differential interference contrast (DIC) images were captured. Scale bar: 10 µm.
DISCUSSION
The parasitic life style of Plasmodium necessitates scavenging the majority of the nutrients it requires for growth and replication from its host. However, at certain times during life cycle progression, the parasite relies on its own biosynthetic machinery to provide essential macromolecules for cellular growth and proliferation. The apicoplast, a secondary plastid, houses the parasite’s only pyruvate dehydrogenase complex (Foth et al., 2005), necessary for the formation of acetyl CoA, as well as the enzymes for FAS II and isoprenoid biosynthesis. The pyruvate dehydrogenase complex appears to be necessary only for the production of FAS II precursors in rodent malaria parasites because P. yoelii and P. berghei parasites lacking genes coding for pyruvate dehydrogenase complex subunits as well as FAS II enzymes all arrest late in liver stage development during the life cycle (Pei et al., 2010, Vaughan et al., 2009, Yu et al., 2008, Nagel et al., 2013). In this study we provide evidence that the PDH/FAS II production of fatty acids is likely necessary for the production of phosphatidic acid and ultimately phospholipids, because both Py apig3pdh(−) and Py apig3pat(−) parasites also arrest late in liver stage development. A further apicoplast-targeted enzymatic pathway downstream of FAS II is lipoic acid synthesis and recently it was shown that the P. berghei lipoic acid protein ligase LipB was also necessary for liver stage maturation, and interestingly P. berghei lipb(−) liver stages showed aberrant apicoplast formation (Falkard et al., 2013), similar to what we observed for Py apig3pdh(−) and Py apig3pat(−) liver stages. It is tempting to hypothesize that the lack of FAS II leads to the attenuated apicoplast development due to the lack of phospholipids necessary for its membrane expansion.
As well as being unable to complete liver stage development, the attenuated Py apig3pdh(−) and Py apig3pat(−) liver stages were also powerful immunogens, able to elicit sterile protection from wildtype challenge after only two immunizations. Our results suggest that immunization with Py apig3pdh(−) and Py apig3pat(−) parasites, which remain resident in the liver for up to 65 hours, would result in the same degree of protection as that seen with the Py fabb/f(−) parasite but this remains to be formally demonstrated.
Excitingly, recent research has shown that the P. falciparum apicoplast can be isolated from asexual blood stage trophozoites and its lipid composition analyzed (Botte et al., 2013). Of interest, placing blood stage parasites in a lipid poor environment actually caused FAS II to be turned on and evidence of de novo fatty acid synthesis was detected in the apicoplast (Botte et al., 2013). It would be interesting to see if these fatty acids are then being incorporated into phosphatidic acid to meet phospholipid requirements of the lipid-deprived parasite. Based on our own results, we would indeed hypothesize that both FAS II and apicoplast-targeted phosphatidic acid biosynthesis would be turned on together in the lipid-deprived blood stage parasites.
When considering the need for FAS II in the liver stage P. yoelii parasite, the question arises, why does the liver stage parasite have to biosynthesize its own fatty acids? Biosynthesis of phospholipids utilizes phosphatidic acid as a precursor and enzymes for phosphatidic acid biosynthesis are predicted to be present in the apicoplast. Indeed, we have shown that apiG3PDH and apiG3PAT are essential and are expressed solely in the P. yoelii liver stage apicoplast. Thus, we conclude that apiG3PAT is incorporating FAS II-derived fatty acids into lysophosphatidic acid, presumably for downstream phosphatidic acid synthesis. It is tempting to hypothesize that the rapidly expanding liver stage apicoplast is relying on FAS II, downstream phosphatidic acid biosynthesis and ultimately phospholipid synthesis to expand its own four membranes. This could explain why at 30 hours post sporozoite inoculation, the Py apig3pdh(−) and Py apig3pat(−) liver stages are a similar size to their wildtype counterpart but are already showing signs of defective apicoplast replication.
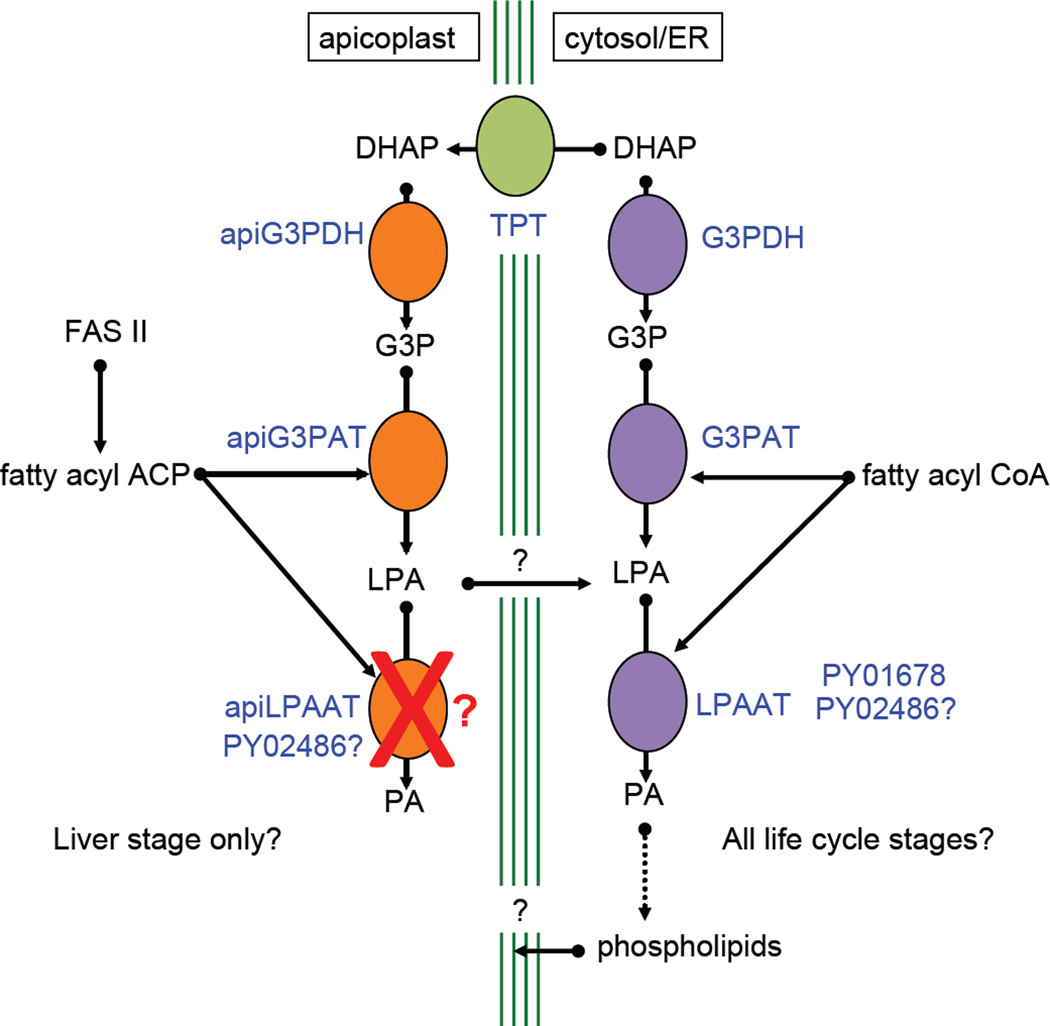
Due to the fact we were able to rescue a G3PAT-deficient E. coli with apiG3PAT, suggests that the apiG3PAT has a preference for acyl-ACP and not acyl-CoA, providing further evidence that the apiG3PAT is of prokaryotic origin. Surprisingly neither of the P. yoelii LPAAT-like genes we studied targeted soley to the apicoplast. PY01678 appeared to be solely expressed in the ER was able to rescue LPAAT-deficient E. coli, suggesting that PY01678 is a true ER-resident LPAAT. Therefore, in order for apicoplast-derived lysophosphatidic acid to be converted to phosphatidic acid, it perhaps needs to leave the organelle to be then acted upon by ER-resident LPAAT. It is possible that PY02486 could traffic between organelles, based on the fact that it did not exclusively co-localize with either the ER or the apicoplast. There is no evidence to date that such a mechanism for the shuttling of lysophosphatidic acid exists in other plastid-harboring organisms, but attrition of pathways within parasites is common. As a model (Fig. 6), we propose that apiG3PDH and apiG3PAT are active during liver stage development and apiG3PAT utilizes FAS II-derived fatty acids to form lysophosphatidic acid. This lysophosphatidic acid then leaves the plastid, ultimately being converted to phosphatidic acid by ER-localized LPAAT that is then metabolized into phospholipids that could ultimately become part of the apicoplast membrane. At this point, it is not clear how the phospholipid would transport back to the apicoplast but recent research on T. gondii has shown the existence of membrane contact sites between the apicoplast’s outermost membrane and the ER (Tomova et al., 2009). These sites could thus allow for the subcellular distribution of lipids in the parasite. Assuming the same sites are present in Plasmodium, they could facilitate the movement of apicoplast-synthesized lipids to and from the ER.
Figure 6.
Model for phosphatidic acid (PA) synthesis in Plasmodium. In the cytoplasm/endoplasmic reticulum (ER), extraplastidial PA synthesis likely takes place in all life cycle stages. Dihydroxyacetone phosphate (DHAP) is converted to glycerol 3-phosphate (G3P) by G3P dehydrogenase (G3PDH), then to lysophosphatidic acid (LPA) by G3P acyltransferase (G3PAT) and finally to PA by LPAAT. In the apicoplast, enzymes involved in plastidial PA synthesis (apiG3PDH and apiG3PAT) are active only during liver stage development. DHAP is transported across the four apicoplast membranes by the triose phosphate transporters (TPT). No exclusive apiLPAAT appears to be present in the Plasmodium genome and we hypothesize that apicoplast-generated LPA could leave the plastid, is acted on by extraplastidial LPAAT in the cytosol/ER (possibly PY016768 or PY02486) to form PA in the cytosol/ER, which is then converted to phospholipids. These phospholipids could then become incorporated into the four membranes of the replicating liver stage apicoplast. Liver stage-expressed enzymes of type II fatty acid synthesis (FAS II) produce the fatty acyl acyl carrier protein (ACP) moiety, which is used by apiG3PAT to produce LPA. Extraplastidial G3PAT and LPAAT use fatty acyl coenzyme A (CoA) moieties. TPT is shown in green, the apicoplast-targeted enzymes in orange and the cytosol/ER enzymes in purple. Enzymes/transporters are in blue text.
It is currently not known if FAS II and downstream phosphatidic acid biosynthesis are required for P. falciparum liver stage development. An argument could be made that the longer period of human malaria liver stage maturation compared to rodent malaria liver stage maturation (seven-to-ten days versus two days) may not require FAS II. Alternatively however, the massive expansion of the liver stage parasite within the human hepatocyte, which is significantly smaller than the mouse hepatocyte, may indeed require the parasite’s own de novo fatty acid synthetic machinery. A significant expansion of parasite biomass also takes place during sporozoite development yet the rodent malarias do not rely on FAS II for this replication. Interestingly, recent research has determined that the P. falciparum parasite requires apicoplast-targeted pyruvate dehydrogenase complex activity for midgut sporozoite development (Cobbold et al., 2013). Because the pyruvate dehydrogenase complex is required to produce the acetyl CoA that feeds into FAS II and thus it is likely that unlike the rodent malarias, FAS II is also required for P. falciparum sporozoite production. Assuming lipid biosynthesis is essential for P. falciparum liver stage development, the pathways involved will be attractive targets for anti-malarial prophylaxis and parasites lacking genes involved in this pathway could be potent immunogens for use as experimental vaccines.
EXPERMENTAL PROCEDURES
Experimental animals and parasite production
Six- to eight-week-old female Swiss Webster (SW) mice from Harlan (Indianapolis, IN) were used for parasite life cycle maintenance and production of transgenic parasites. Six- to eight- week-old female BALB/cAnN mice from Harlan were used for assessments of parasite infectivity and indirect immunofluorescence assays. BALB/cJ mice from the Jackson laboratory (Bar Harbor, ME) were used to assess the ability of gene knockout parasites to act as experimental vaccines. P. yoelii 17XNL (Py17XNL, a non-lethal strain) wildtype and transgenic parasites were cycled between SW mice and Anopheles stephensi mosquitoes. Infected mosquitoes were maintained on sugar water at 24°C and 70% humidity. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Seattle Biomedical Research Institute has an OLAW Animal Welfare Assurance (A3640-01). The protocol was approved by the Seattle Biomedical Research Institute Institutional Animal Care and Use Committee (Protocol #: SK-07).
Reverse genetics of Py17XNL parasite
Gene targeting constructs for transgenic parasite production were designed as previously described (Lindner et al., 2013, Vaughan et al., 2009). Briefly, two regions of the targeted locus were PCR amplified with the Phusion polymerase (NEB) supplemented with 5 mM MgCl2 with locus-specific primers (oligonucleotides used in this study are listed in Supplemental Table S2). The two PCR products were gel purified (Gel Extraction Kit, Qiagen), precipitated by ethanol, and fused by Sequence Overlap Extension PCR (‘SOE PCR’) without supplemented MgCl2. This PCR product was digested at the 5′ and 3′ ends at the exogenous restriction sites designed in the primers, gel purified, precipitated by ethanol, and inserted into a modified pDEF vector designed for genetic disruptions or for gene modifications via double-cross-over recombination to replace the locus.
Wildtype Py17XNL parasites were genetically modified using standard methods as previously described (Jongco et al., 2006, Lindner et al., 2011). The presence of transgenic parasites was assessed by genotyping PCR. Limiting dilution infection of female SW mice was used to isolate at least two independent clones of each transgenic knockout parasite. Transfer pyrimethamine-selected populations of epitope-tagged transgenic parasites used for analysis and were not cloned.
Verification of Genomic and Coding Sequences of P. yoelii apiG3PAT and LPAAT
Targeted sequencing of the P. yoelii apiG3PAT and LPAAT genomic loci and cDNA was conducted to identify the missing sequences in the current build of the Py17XNL reference sequence (PlasmoDB.org). Primer pairs that are specific for these regions-of-interest (listed in Supplemental Table S2) were used to amplify them by PCR. The resulting products were sequenced and used to generate new gene models.
Western blotting
Equal amounts (100 µg) of mixed blood stage protein lysates of wildtype and myc-tagged parasite lines were boiled in SDS-PAGE buffer containing 5% v/v beta-mercaptoethanol for 10 minutes and subsequently separated on an SDS-polyacrylamide (4–20% gradient) gel and transferred to a PVDF membrane. The membrane was probed with a rabbit polyclonal antibody to c-myc and a secondary IRDye 680 antibody (LI-COR Biotechnology). Protein bands were detected by scanning the membrane on an Odyssey imaging system (LI-COR Biotechnology).
Immunofluorescence analysis
Liver stage
BALB/cAnN mice were injected intravenously with approximately 106 sporozoites and livers were harvested from euthanized mice at several time points post infection. Livers were perfused with 1xPBS, fixed in 4% v/v paraformaldehyde (PFA) in 1xPBS and lobes were cut into 50 µm sections using a Vibratome apparatus (Ted Pella Inc., Redding, CA). For IFA, sections were permeabilized in 1xTBS containing 3% v/v H2O2 and 0.25% v/v Triton X-100 for 30 min at room temperature. Sections were then blocked in 1xTBS containing 5% v/v dried milk (TBS-M) for at least 1 h and incubated with primary antibody in TBS-M at 4°C overnight. After washing in 1xTBS, fluorescent secondary antibodies were added in TBS-M for 2 h at room temperature in a similar manner as above. After further washing, the section was incubated in 0.06% w/v KMnO4 for 2 minutes to quench background fluorescence. The section was then washed with 1xTBS and stained with 1µg/ml 4′, 6-diamidino-2-phenylindole (DAPI) in 1xTBS for 5–10 minutes at room temperature to visualize DNA and mounted with FluoroGuard anti-fade reagent (Bio-Rad, Hercules, CA). Preparations were analyzed for fluorescence using a fluorescence inverted microscope (Eclipse TE2000-E; Nikon), and images were acquired using Olympus 1 × 70 Delta Vision deconvolution microscopy.
Blood stage
Infected red blood cells were processed for IFA using a previously described method (Tonkin et al., 2004). Red blood cells were pelleted initially (and between all steps) at 2000g in a microcentrifuge at room temperature for 1 minute. Cells were washed twice in 1xPBS, fixed in 1xPBS + 4% v/v PFA + 0.0075% v/v glutaraldehyde for 30 minutes at room temperature, and permeabilized in 1xPBS + 0.2% v/v Triton X-100 for 10 minutes at room temperature. A 1xPBS + 3% w/v bovine serum albumin (BSA) (blocking solution) was applied at 4°C overnight. Primary antibodies were diluted in blocking solution and incubated for 1 hour with end-over-end rotation at room temperature. Following two washes with 1xPBS, fluorescent secondary antibodies were diluted in blocking solution and incubated with cells for 30 minutes with end-over-end rotation at room temperature and shielding from light. Nucleic acid was then stained with DAPI in 1xPBS for 5–10 minutes at room temperature. Cells were washed three times with 1xPBS, and mixed 1:1 with VectaShield (Vector Laboratories) and applied to a glass slide and coverglass slip. Images were acquired as before.
Sporozoites
Sporozoites were isolated and fixed with 4% v/v PFA in 1xPBS for 20 minutes at room temperature. Fixed sporozoites were applied to a polylysine-treated microscope slide, which was housed in a wet chamber at 4°C overnight. Sporozoites were washed twice with 1xPBS and then permeabilized and blocked with 2% w/v BSA plus 0.2% v/v Triton X-100 in 1xPBS for 1 h at 37°C. IFA was performed as for blood stages but the solutions were applied directly to the slide. Images were acquired as before.
Sporozoite Immunization and Challenge
P. yoelii sporozoites were isolated from the salivary glands of infected A. stephensi mosquitoes. BALB/cJ mice were immunized with sporozoites arising from two individual clones of both the Py g3pdh- and Py g3pat- parasite. Control (immunized with salivary gland isolate from non-infectious mosquitoes) and immunized mice were challenged intravenously with 10,000 wildtype P. yoelii sporozoites. Parasitized red blood cells were identified by Giemsa-stained thin blood smears from 3 days after challenge until 14 days after challenge. Protection was defined as the absence of blood stage parasites. At least 20 fields of >200 red blood cells were examined for each mouse to be designated as protected.
Rescue of G3PAT- and LPAAT-Deficient E. coli Strains
E. coli strains that are deficient in G3PAT activity (BB26-36 strain, Coli Genetic Stock Center #5348, Yale University, New Haven, CT (McIntyre et al., 1977)) or LPAAT activity (JC201 strain, provided by Dr. Fergal O’Gara, National University of Ireland, Cork, (Coleman, 1990, Cullinane et al., 2005)) were used to test if these activities exist in Plasmodium yoelii apiG3PAT (P. yoelii apiG3PAT) and extraplastidial G3PAT (P. yoelii G3PAT), the presumptive apiLPAAT (PY01678) and the possible LPAAT PY02486. Codon optimized sequences (sequences shown in Supplemental Table 3) for P. yoelii apiG3PAT (the bipartite leader was removed, amino acids 1 thru 74 of the full length protein), P. yoelii extraplastidial G3PAT (the signal sequence was removed, amino acids 1 thru 20 of the full length protein), PY01678 and PY02486 were synthesized (GenScript) and ligated into a modified pET-28b(+) vector (sequence available in Supplemental Table S3) with the inducible T7 promoter replaced by the strong constitutive Lac promoter. Additionally, genes coding for E. coli G3PAT (EcPlsB) and LPAAT (EcPlsC) were amplified from E. coli DH5alpha genomic DNA (sequences available in Supplemental Table S3 and oligonucleotide sequences used for amplification available in Supplemental Table S2) and ligated into the modified pET-28b(+) plasmid for use as positive and negative controls. Clones of the deficient bacteria (experimentally verified to exhibit their respective defects) were made chemically competent by the calcium chloride method (Sambrook et al., 2006). These bacteria were then transformed with the expression plasmids in order to express C-terminally 6xHis-tagged variants of the protein of interest or a plasmid that harbors an empty multiple cloning site (“Empty Vector”). Transformed bacterial populations were grown in permissive conditions (LB agar/LB media, 37°C) in order to permit all isolates to be grown, and two independent clones of each bacterial strain that harbors each plasmid-of-interest to be isolated. Rescue of G3PAT activity was measured by the ability of BB26-36 to grow in minimal M9 liquid media in the absence of glycerol as previously described (Feng et al., 2011, McIntyre et al., 1977). Rescue of LPAAT activity was measured by the ability of JC201 to grow at elevated temperatures (42°C) in LB broth as previously described (Coleman, 1990, Cullinane et al., 2005). In all experiments, bacteria were grown to saturation in permissive conditions (LB media, 37°C), pelleted, washed three times in sterile 1xPBS, and resuspended in growth media at the same OD (BB26-36: M9 minimal media; JC201: LB media). Cultures were inoculated at an approximate 1:200 dilution with the washed bacteria (OD was the same for each at the beginning of the culture set up) and growth was determined at 24 hours by measurement of the OD600 (Nanodrop, Thermo Scientific). All experiments were conducted in triplicate with both independent clones.
Lipidomics of the E. coli BB26-36 strain expressing P. yoelii apiG3PAT
Bacterial growth and lipid extraction
BB26-36 rescued with plasmid coding for P. yoelii apiG3PAT or cells containing empty plasmid vector were grown for 24 hrs at 37°C in minimal media with kanamycin (to retain the plasmids). Cells containing empty vector bacteria also received supplemented glycerol to permit growth. Bacterial pellets were resuspended in water and lysed by zirconia bead disruption. Cell lysate was assayed for total protein content using the BCA method (Pierce). A volume containing 127 pmoles of 17:0-14:1 phosphatidyl ethanolamine (LIPID MAPS ID LMGP02010005, Avanti Polar Lipids, Alabaster, AL), 100 pmoles of 17:0-14:1 phosphatidyl glycerol (LMGP04010007, Avanti), and 100 pmoles of 17:0-14:1 phosphatidic acid (LMGP10010006, Avanti) was added to a volume of lysate corresponding to a fixed amount of total protein (50 µg). Total lipid was chloroform-methanol extracted using the Folch method (Folch et al., 1957). Prior to LC/MS total lipid extracts were evaporated under N2 gas and dissolved in 100 µL initial LC solvent (see below) for phosphatidic acid and phosphatidyl glycerol analysis and further diluted 10-fold for phosphatidyl ethanolamine analysis.
LC/ESI-MS/MS analysis
Normal-phase chromatography was carried out on an Agilent 1100 Binary HPLC with a 2.0 inner diameter × 250 mm, 5 µm Luna silica column (Phenomenex, Torrance, CA) kept at 25°C and a flow rate of 300 µL/min. The system was equilibrated with 100% solvent A (isopropanol/hexane/water (58:40:2 v/v/v) containing 5 mM ammonium acetate) and an Agilent 1200 autosampler was used to inject 20 µL of the lipid extracts for phosphatidic acid and phosphatidyl glycerol analysis and 5 µL for phosphatidyl ethanolamine analysis. Solvent A was maintained at 100% for 5.0 min, followed by a 5.0 min linear gradient to 50% solvent B (isopropanol/hexane/water (50:40:10 v/v/v/) containing 5 mM ammonium acetate), then a 15.0 min linear gradient to 100% solvent B and held at 100% solvent B for 15.0 min. An API 4000 QTRAP hybrid quadropole/linear ion trap mass spectrometer (Applied Biosystems) equipped with a Turbo V electrospray ionization source and operated in multiple-reaction monitoring (MRM) mode at unit resolution was used for analysis of the LC eluent. Phosphatidyl ethanolamine lipid transitions were monitored in positive ion mode with the following instrument settings: IS = 5500 V, CUR = 30 psi, GS1 = 50 psi, GS2 = 50 psi, CAD = MED, TEM = 450°C, ihe = ON, EP = 10 V, DP = 70 V, CXP = 10 V. phosphatidic acid and phosphatidyl glycerol lipid transitions were monitored in negative ion mode with the following instrument settings: IS = −4500 V, CUR = 30 psi, GS1 = 50 psi, GS2 = 50 psi, CAD = MED, TEM = 450°C, ihe = ON, EP = −10 V, DP = −120 V, CXP = −5 V. MRM transition lists were developed based on the typical phospholipids and fatty acyl combinations observed from E. coli (Oursel et al., 2007). Molecular composition of the lipid species were determined, where lipid species were annotated as: <lipid class> <sum of carbons in the esterified fatty acids>:<sum of unsaturated bonds in the fatty acids> (e.g. phosphatidyl ethanolamine 33:1). Precursor and fragment ion m/z values and further instrument parameters are provided in Supplemental Table S1. Product ion (MS2) scan data were also obtained for select phosphatidyl glycerol lipid precursors for fatty acid identification in negative ion mode with collision energies of −48 V. The Multiquant™ 2.1 software package (AB Sciex) was used to integrate phosphatidic acid, phosphatidyl ethanolamine, and phosphatidyl glycerol MRM peak areas.
Supplementary Material
Acknowledgements
We would like to thank the Seattle BioMed insectary and vivarium for their help in maintaining the malaria life cycle. NIH funding (R56 AI080685) to SHIK and AMV supported this work.
Abbreviations
- ACP
acyl carrier protein
- apiG3PAT
apicoplast-targeted glycerol 3-phosphate acyl transferase
- apiLPAAT
apicoplast-targeted lysophosphatidic acid acyl transferase
- BiP
binding immunoglobulin protein
- CoA
coenzyme A
- CSP
circumsporozoite protein
- DHAP
dihydroxyacetone phosphate
- ER
endoplasmic reticulum
- FAS II
type II fatty acid synthesis
- G3PAT
glycerol 3-phosphate acyl transferase
- IFA
immunofluorescence assay
- LPAAT
lysophosphatidic acid acyl transferase
- MSP1
merozoite surface protein 1
- PPM
parasite plasma membrane
- PVM
parasitophorous vacuole membrane
- TPT
triose phosphate transporter
REFERENCES
- Botte CY, Yamaryo-Botte Y, Rupasinghe TW, Mullin KA, Macrae JI, Spurck TP, et al. Atypical lipid composition in the purified relict plastid (apicoplast) of malaria parasites. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1301251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold SA, Vaughan AM, Lewis IA, Painter HJ, Camargo N, Perlman DH, et al. Kinetic flux profiling elucidates two independent acetyl-CoA biosynthetic pathways in Plasmodium falciparum. J Biol Chem. 2013 doi: 10.1074/jbc.M113.503557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J. Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3- phosphate acyltransferase activity. J Biol Chem. 1990;265:17215–17221. [PubMed] [Google Scholar]
- Cullinane M, Baysse C, Morrissey JP, O'Gara F. Identification of two lysophosphatidic acid acyltransferase genes with overlapping function in Pseudomonas fluorescens. Microbiology. 2005;151:3071–3080. doi: 10.1099/mic.0.27958-0. [DOI] [PubMed] [Google Scholar]
- Falkard B, Kumar TR, Hecht LS, Matthews KA, Henrich PP, Gulati S, et al. A key role for lipoic acid synthesis during Plasmodium liver stage development. Cell Microbiol. 2013;15:1585–1604. doi: 10.1111/cmi.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Cronan JE. The Vibrio cholerae fatty acid regulatory protein, FadR, represses transcription of plsB, the gene encoding the first enzyme of membrane phospholipid biosynthesis. Mol Microbiol. 2011;81:1020–1033. doi: 10.1111/j.1365-2958.2011.07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI. The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast. Mol Microbiol. 2005;55:39–53. doi: 10.1111/j.1365-2958.2004.04407.x. [DOI] [PubMed] [Google Scholar]
- Funes S, Davidson E, Reyes-Prieto A, Magallon S, Herion P, King MP, Gonzalez-Halphen D. A green algal apicoplast ancestor. Science. 2002;298:2155. doi: 10.1126/science.1076003. [DOI] [PubMed] [Google Scholar]
- Gornicki P. Apicoplast fatty acid biosynthesis as a target for medical intervention in apicomplexan parasites. Int J Parasitol. 2003;33:885–896. doi: 10.1016/s0020-7519(03)00133-4. [DOI] [PubMed] [Google Scholar]
- Jongco AM, Ting LM, Thathy V, Mota MM, Kim K. Improved transfection and new selectable markers for the rodent malaria parasite Plasmodium yoelii. Mol Biochem Parasitol. 2006;146:242–250. doi: 10.1016/j.molbiopara.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Khan SM, Janse CJ, Kappe SH, Mikolajczak SA. Genetic engineering of attenuated malaria parasites for vaccination. Curr Opin Biotechnol. 2012;23:908–916. doi: 10.1016/j.copbio.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Kohler S, Delwiche CF, Denny PW, Tilney LG, Webster P, Wilson RJ, et al. A plastid of probable green algal origin in Apicomplexan parasites. Science. 1997;275:1485–1489. doi: 10.1126/science.275.5305.1485. [DOI] [PubMed] [Google Scholar]
- Lindner SE, Llinas M, Keck JL, Kappe SH. The primase domain of PfPrex is a proteolytically matured, essential enzyme of the apicoplast. Mol Biochem Parasitol. 2011;180:69–75. doi: 10.1016/j.molbiopara.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Mikolajczak SA, Vaughan AM, Moon W, Joyce BR, Sullivan WJ, Jr, Kappe SH. Perturbations of Plasmodium Puf2 expression and RNA-seq of Puf2-deficient sporozoites reveal a critical role in maintaining RNA homeostasis and parasite transmissibility. Cell Microbiol. 2013;15:1266–1283. doi: 10.1111/cmi.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre TM, Chamberlain BK, Webster RE, Bell RM. Mutants of Escherichia coli defective in membrane phospholipid synthesis. Effects of cessation and reinitiation of phospholipid synthesis on macromolecular synthesis and phospholipid turnover. J Biol Chem. 1977;252:4487–4493. [PubMed] [Google Scholar]
- Murata N, Tasaka Y. Glycerol-3-phosphate acyltransferase in plants. Biochim Biophys Acta. 1997;1348:10–16. doi: 10.1016/s0005-2760(97)00115-x. [DOI] [PubMed] [Google Scholar]
- Murray CJ, Rosenfeld LC, Lim SS, Andrews KG, Foreman KJ, Haring D, et al. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet. 2012;379:413–431. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- Nagel A, Prado M, Heitmann A, Tartz S, Jacobs T, Deschermeier C, et al. A new approach to generate a safe double-attenuated Plasmodium liver stage vaccine. Int J Parasitol. 2013;43:503–514. doi: 10.1016/j.ijpara.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oursel D, Loutelier-Bourhis C, Orange N, Chevalier S, Norris V, Lange CM. Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization. Rapid communications in mass spectrometry: RCM. 2007;21:1721–1728. doi: 10.1002/rcm.3013. [DOI] [PubMed] [Google Scholar]
- Pei Y, Tarun AS, Vaughan AM, Herman RW, Soliman JM, Erickson-Wayman A, Kappe SH. Plasmodium pyruvate dehydrogenase activity is only essential for the parasite's progression from liver infection to blood infection. Mol Microbiol. 2010;75:957–971. doi: 10.1111/j.1365-2958.2009.07034.x. [DOI] [PubMed] [Google Scholar]
- Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, et al. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW, Sambrook J. The condensed protocols from Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. p. v. 800 p. [Google Scholar]
- Stanway RR, Witt T, Zobiak B, Aepfelbacher M, Heussler VT. GFP-targeting allows visualization of the apicoplast throughout the life cycle of live malaria parasites. Biol Cell. 2009;101:415–430. doi: 10.1042/BC20080202. [DOI] [PubMed] [Google Scholar]
- Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, Dondorp AM, et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci U S A. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun AS, Vaughan AM, Kappe SH. Redefining the role of de novo fatty acid synthesis in Plasmodium parasites. Trends Parasitol. 2009;25:545–550. doi: 10.1016/j.pt.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Tomova C, Humbel BM, Geerts WJ, Entzeroth R, Holthuis JC, Verkleij AJ. Membrane contact sites between apicoplast and ER in Toxoplasma gondii revealed by electron tomography. Traffic. 2009;10:1471–1480. doi: 10.1111/j.1600-0854.2009.00954.x. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, et al. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol. 2004;137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Vaughan AM, O'Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, et al. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Ralph SA, Reed MB, Su V, Douglas JD, Minnikin DE, et al. A type II pathway for fatty acid biosynthesis presents drug targets in Plasmodium falciparum. Antimicrob Agents Chemother. 2003;47:297–301. doi: 10.1128/AAC.47.1.297-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Health Organization; 2012. World Malaria Report 2012. [Google Scholar]
- Yao J, Rock CO. Phosphatidic acid synthesis in bacteria. Biochim Biophys Acta. 2013;1831:495–502. doi: 10.1016/j.bbalip.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Kumar TR, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, et al. The Fatty Acid Biosynthesis Enzyme FabI Plays a Key Role in the Development of Liver-Stage Malarial Parasites. Cell Host Microbe. 2008;4:567–578. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.