Abstract
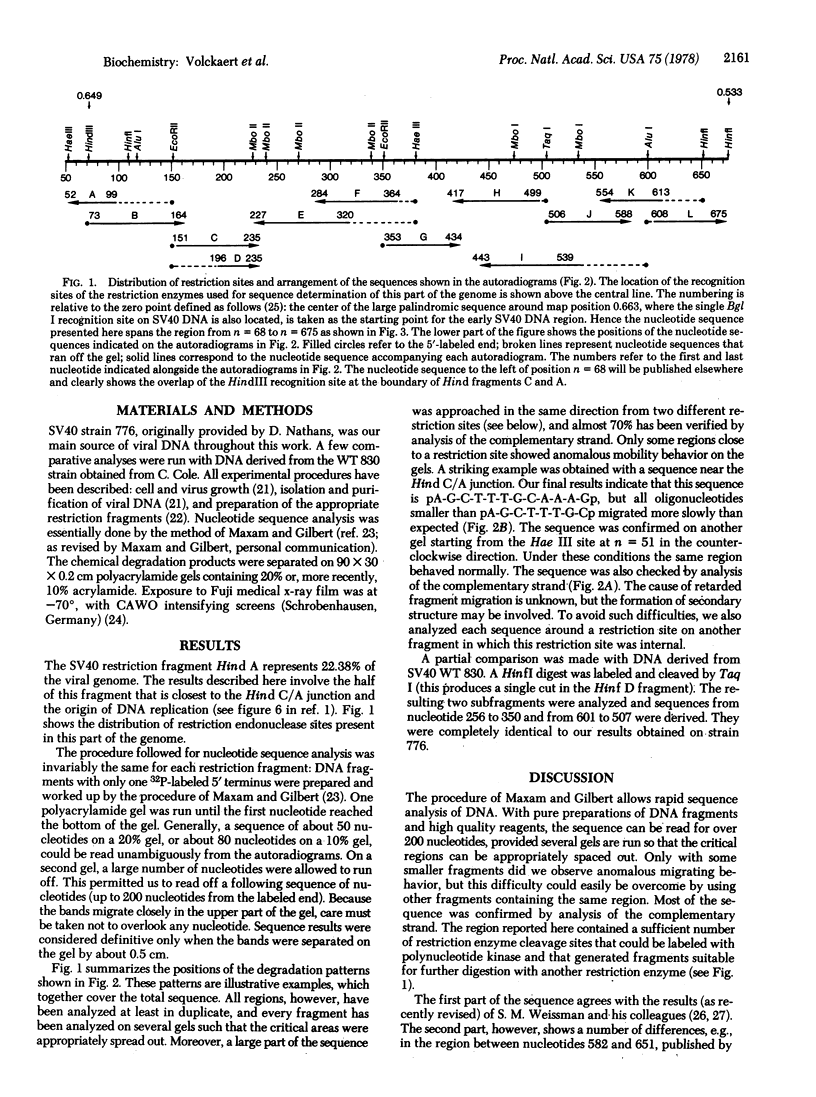
The nucleotide sequence of the segment of simian virus 40 DNA between standard map positions 0.53 and 0.65, i.e., approximately half of the restriction fragment Hind A, is reported. This segment is located near the beginning of the early region and is transcribed counterclockwise. There is a potential initiating ATG signal at 13 nucleotides from the Hind C-Hind A junction in the strand with the same polarity as the early mRNA. From this signal on, an open reading frame is present which would allow the synthesis of a polypeptide of 174 amino acids until a TAA termination codon is reached at nucleotide 602 (map position 0.547). This polypeptide, revealed by the DNA sequence, corresponds almost certainly to small-t antigen. Correlation of the deduced amino acid sequence with the NH2-terminal sequences of small-t and large-T (tumor) antigens of simian virus 40, as established by Paucha et al. [Paucha, E., Mellor, A., Harvey, R., Smith, A. E., Hewick, R. M. & Waterfield, M. D. (1978) [Proc. Natl. Acad. Sci. USA 75, 2165-2169], strongly argues that both proteins are indeed initiated at the ATG triplet. Because the DNA region between 0.547 and 0.534 is blocked for translation in all three reading frames by multiple termination condons, we conclude that the large-T antigen must be coded for by two noncontiguous DNA segments: the segment from 0.65 to around 0.60, which small-t and large-T antigens share, and another segment starting at some point after position 0.534 and continuing counterclockwise until it terminates at map position 0.174. Small-t antigen is methionine-rich and has a remarkably high number of cysteine residues clustered mainly in its COOH-terminal half. It is rich in both basic and acidic residues, the former being slightly in excess.
Keywords: oncogenes, early functions, tumor (T) antigen
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Dhar R., Laub O., Horowitz M., Khoury G. Novel mechanism for RNA maturation: the leader sequences of simian virus 40 mRNA are not transcribed adjacent to the coding sequences. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3686–3690. doi: 10.1073/pnas.74.9.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. L., Martin R. G., Chang C., Mora P. T., Livingston D. M. Nuclear preparations of SV40-transformed cells contain tumor-specific transplantation antigen activity. Virology. 1977 Jan;76(1):420–425. doi: 10.1016/0042-6822(77)90314-2. [DOI] [PubMed] [Google Scholar]
- Berget S. M., Moore C., Sharp P. A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll R. B., Smith A. E. Monomer molecular weight of T antigen from simian virus 40-infected and transformed cells. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2254–2258. doi: 10.1073/pnas.73.7.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celma M. L., Dhar R., Pan J., Weissman S. M. Comparison of the nucleotide sequence of the messenger RNA for the major structural protein of SV40 with the DNA sequence encoding the amino acids of the protein. Nucleic Acids Res. 1977 Aug;4(8):2549–2550. [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Gelinas R. E., Broker T. R., Roberts R. J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977 Sep;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Landers T., Goff S. P., Manteuil-Brutlag S., Berg P. Physical and genetic characterization of deletion mutants of simian virus 40 constructed in vitro. J Virol. 1977 Oct;24(1):277–294. doi: 10.1128/jvi.24.1.277-294.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford L. V., Cole C. N., Smith A. E., Paucha E., Tegtmeyer P., Rundell K., Berg P. Organization and expression of early genes of simian virus 40. Proc Natl Acad Sci U S A. 1978 Jan;75(1):117–121. doi: 10.1073/pnas.75.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R., Subramanian K. N., Pan J., Weissman S. M. Structure of a large segment of the genome of simian virus 40 that does not encode known proteins. Proc Natl Acad Sci U S A. 1977 Mar;74(3):827–831. doi: 10.1073/pnas.74.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn A. R., Hassell J. A. A novel method to map transcripts: evidence for homology between an adenovirus mRNA and discrete multiple regions of the viral genome. Cell. 1977 Sep;12(1):23–36. doi: 10.1016/0092-8674(77)90182-9. [DOI] [PubMed] [Google Scholar]
- Fareed G. C., Davoli D. Molecular biology of papovaviruses. Annu Rev Biochem. 1977;46:471–522. doi: 10.1146/annurev.bi.46.070177.002351. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck M. M., Staneloni R. J., Benjamin T. L. Hr-t and ts-a: two early gene functions of polyoma virus. Virology. 1977 Apr;77(2):610–624. doi: 10.1016/0042-6822(77)90486-x. [DOI] [PubMed] [Google Scholar]
- Fried M., Griffin B. E. Organization of the genomes of polyoma virus and SV40. Adv Cancer Res. 1977;24:67–113. doi: 10.1016/s0065-230x(08)61013-1. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Ford J. Sequence arrangement of the 5' ends of simian virus 40 16S and 19S mRNAs. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4982–4985. doi: 10.1073/pnas.74.11.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Nathans D. The genome of simian virus 40. Adv Virus Res. 1977;21:85–173. doi: 10.1016/s0065-3527(08)60762-9. [DOI] [PubMed] [Google Scholar]
- Kitchingman G. R., Lai S. P., Westphal H. Loop structures in hybrids of early RNA and the separated strands of adenovirus DNA. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4392–4395. doi: 10.1073/pnas.74.10.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig D. F. Two adenovirus mRNAs have a common 5' terminal leader sequence encoded at least 10 kb upstream from their main coding regions. Cell. 1977 Sep;12(1):9–21. doi: 10.1016/0092-8674(77)90181-7. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. A map of temperature-sensitive mutants of simian virus 40. Virology. 1975 Jul;66(1):70–81. doi: 10.1016/0042-6822(75)90179-8. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C., Gilboa E., Revel M., Winocour E. Cell-free translation of simian virus 40 early messenger RNA coding for viral T-antigen. Proc Natl Acad Sci U S A. 1977 Feb;74(2):457–461. doi: 10.1073/pnas.74.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb J. A. Identification of simian virus 40 tumor and U antigens. Proc Natl Acad Sci U S A. 1977 Feb;74(2):447–451. doi: 10.1073/pnas.74.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell K., Collins J. K., Tegtmeyer P., Ozer H. L., Lai C. J., Nathans D. Identification of simian virus 40 protein A. J Virol. 1977 Feb;21(2):636–646. doi: 10.1128/jvi.21.2.636-646.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Deficiency in histone acetylation in nontransforming host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1092–1096. doi: 10.1073/pnas.73.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staneloni R. J., Fluck M. M., Benjamin T. L. Host range selection of transformation-defective hr-t mutants of polyoma virus. Virology. 1977 Apr;77(2):598–609. doi: 10.1016/0042-6822(77)90485-8. [DOI] [PubMed] [Google Scholar]
- Thimmappaya B., Weissman S. M. The early region of SV40 DNA may have more than one gene. Cell. 1977 Aug;11(4):837–843. doi: 10.1016/0092-8674(77)90295-1. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Contreras R., Soeda E., Van de Voorde A., Fiers W. Nucleotide sequence of simian virus 40 Hind H restriction fragment. J Mol Biol. 1977 Mar 5;110(3):467–510. doi: 10.1016/s0022-2836(77)80109-5. [DOI] [PubMed] [Google Scholar]
- Yang R. C., Van de Voorde A., Fiers W. Cleavage map of the simian-virus-40 genome by the restriction endonuclease III of Haemopholus aegyptius. Eur J Biochem. 1976 Jan 2;61(1):101–117. doi: 10.1111/j.1432-1033.1976.tb10002.x. [DOI] [PubMed] [Google Scholar]