Abstract
Accurate measurements of the abundances, synthesis rates and degradation rates of cellular proteins are critical for understanding how cells and organisms respond to changes in their environments. Over the past two decades, there has been increasing interest in the use of mass spectrometry for proteomic analysis. In many systems, however, protein diversity as well as cell and tissue heterogeneity limit the usefulness of mass spectrometry-based proteomics. As a result, researchers have had difficulty in systematically identifying proteins expressed within specified time intervals, or low abundance proteins expressed in specific tissues or in a few cells in complex microbial systems. In this review, we present recently-developed tools and strategies that probe these two subsets of the proteome: proteins synthesized during well-defined time intervals – temporally resolved proteomics – and proteins expressed in predetermined cell types, cells or cellular compartments – spatially resolved proteomics – with a focus on chemical and biological mass spectrometry-based methodologies.
Key Terms: Protein Synthesis, BONCAT, SILAC, APEX
Introduction
Messenger RNA (mRNA) profiling at the systems level (transcriptomics) with microarray or deep-sequencing technologies offers a high-throughput route to the analysis of gene expression.26,91 However, transcriptomic methods are blind to post-transcriptional phenomena such as translational regulation, protein modification, protein-protein interactions or protein interactions with other molecular components. Furthermore, because mRNA abundance correlates poorly with protein abundance,15,82,89 reliable quantitative information about changes in protein abundance cannot be derived from microarray analysis.
Recent advances in genomic sequencing and high-resolution mass spectrometry have enabled rapid progress in the study of proteins and their abundances, modifications, interactions and functions.1,93 Sample analysis workflows that employ high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) use site-specific endopeptidases (e.g., trypsin or Lys-C) to convert mixtures of proteins into peptides.60 Microscale or nanoscale high-performance liquid chromatography (HPLC)54 and/or ion exchange (IEX) chromatography20 separates the peptides and injects them into the mass spectrometer. After an initial ion scan (MS1), peptide ions fragment in the mass spectrometer and yield secondary “MS/MS” (MS2) spectra. Peptide search engines like Mascot or Andromeda match the MS/MS spectra with theoretical MS/MS spectra and recover the corresponding peptide sequences from a database.12,73 Finally, proteins are identified using two or more unique peptides. Mass spectrometry-based proteomic methods have been applied to a wide range of problems including deciphering the protein composition of organelles,9 systematically mapping protein-protein interactions,63 and large-scale decoding of post-translational events in response to stimuli.24
As of this writing, no eukaryotic proteome has been mapped with 100% coverage.63–64 The dynamic range of protein expression can span many orders of magnitude (e.g., up to 12 orders of magnitude in serum11). The resulting variations in protein abundance challenge the dynamic range and sequencing speed of contemporary mass spectrometers. Sequence coverage becomes increasingly important in the analysis of higher eukaryotes where proteins exhibit high levels of sequence homology due to evolution of protein families, alternative splicing and differential processing.3,8,69 Furthermore, while genome-wide transcriptome analyses are now routinely performed on small samples of RNA, even from single cells,68 low abundance proteins cannot be amplified to improve identification rates. To expand the dynamic range of identified proteins, researchers have focused on information-rich subsets of the proteome, such as the glycoproteome,35 phosphoproteome85 and ubiquitome.86 In this review, we highlight recent work that leverages mass spectrometry and chemical biology to examine two additional subsets of the proteome: proteins synthesized during predetermined time intervals – temporally resolved proteomics – and proteins expressed in specific cell types, cells or cellular compartments – spatially resolved proteomics.
Temporally Resolved Proteomic Analysis
Stable-Isotope Labeling with Amino Acids in Cell Culture (SILAC)
Interest in quantitative modeling and analysis of biological processes has motivated the development of tools for quantitative mass spectrometry-based proteomics such as “label-free” and isotopically labeled protein profiling.4–5 Comparison of protein abundances in different samples is most accurately accomplished by using stable isotopes.4,5,78 Stable-isotope labeling of peptides relies on either chemical methods that attach isotopically labeled linkers to peptides (e.g., ICAT27 or iTRAQ92) or metabolic methods that incorporate isotopically labeled amino acids into peptides (e.g., 15N-labeling40 or SILAC61).
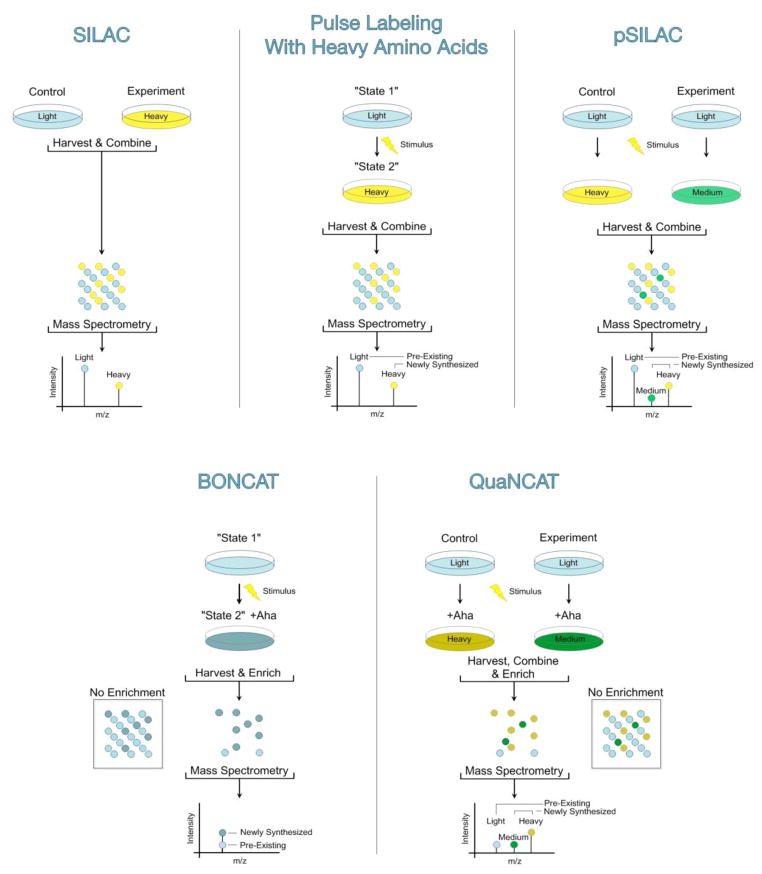
Stable-isotope labeling by amino acids in cell culture (SILAC, Figure 1, Top Left) introduces the label at the amino acid level and overcomes complex isotopic clusters found in alternative metabolic methods like 15N-labeling. Distinct isotopologs of the labeled amino acids – typically “light” arginine (12C614N41H1416O2) and “light” lysine (12C614N21H1416O2) in one sample, and “heavy” arginine (13C615N41H1416O2) and “heavy” lysine (13C615N21H1416O2) in the other – are added to the samples of interest. After several generations of growth, all cellular proteins have uniformly incorporated the labeled amino acids. Cells are lysed and subjected to proteolysis, and the resulting peptide pools are mixed. In theory, both the “heavy” and “light” forms of each peptide should behave identically with respect to processing and LC-MS/MS analysis. However, the origin of each peptide can be determined from the mass of its isotope label, and the relative signal intensities of the “heavy” and “light” peptides indicate the relative abundances of the corresponding proteins in the two samples. Its simplicity and accuracy have made SILAC an increasingly popular method, both for cell culture-based quantitative proteomics,50 and for whole organism quantitative proteomics.15,21,41,43,80
Figure 1.
Cells can be metabolically labeled with a combination of Aha and/or stable isotopic variants of arginine and lysine in 5 workflows: standard SILAC (top left), pulse labeling with heavy amino acids (top middle), pSILAC (top right), BONCAT (bottom left) and QuaNCAT (bottom right).
Repurposing SILAC for Temporally Resolved Proteomic Analysis
Because “old” and “new” copies of cellular proteins are chemically indistinguishable, selective analysis of the subset of the proteome that is expressed within a specified time interval is challenging. A classic method for quantifying specific protein turnover rates involves pulse labeling cells with a radiolabeled amino acid (typically 35S-methionine or cysteine).44,55 Only those proteins synthesized during the pulse incorporate the radioactive element, and their fates can be monitored by standard radioisotope methods. Analogous pulse labeling with stable isotopes can also determine protein turnover or transport (Figure 1, Top Middle).42,53,65
Pulse labeling with amino acids and nucleosides can be combined to yield an unbiased and comprehensive picture of protein and mRNA dynamics. To determine if the cellular abundance of proteins is predominantly controlled at the level of transcription or translation, Schwanhäausser et al. pulse labeled mammalian cells with the nucleoside analog 4-thiouridine (Figure 2) and heavy arginine and lysine to quantify absolute mRNA and protein abundances, half-lives and transcription and translation rates for more than 5000 genes in mouse fibroblast cells.76 The thio-substituted nucleoside 4-thiouridine is not a natural component of nucleic acids but is incorporated into RNA biosynthetically; only newly synthesized RNAs are thio-labeled and can be tagged and purified using commercially available reagents.22,67 Although mRNA and protein levels correlated better than in previous studies,15,16,49,82,89,90 their half-lives showed no overall correlation. Nevertheless, the authors note that genes with similar combinations of mRNA and protein half-lives share common functions, suggesting that half-lives evolved under similar constraints. For example, many genes involved in constitutive processes like translation, respiration and central metabolism have stable mRNAs and proteins, consistent with the requirement for conservation of resources. On the other hand, genes involved in transcription regulation, signaling, chromatin modification and cell cycle-specific processes have unstable mRNAs and proteins, consistent with the requirement for rapid regulation. Correlation of protein abundance with mRNA levels and with translation rates showed that protein copy numbers are determined primarily at the level of translation.
Figure 2.
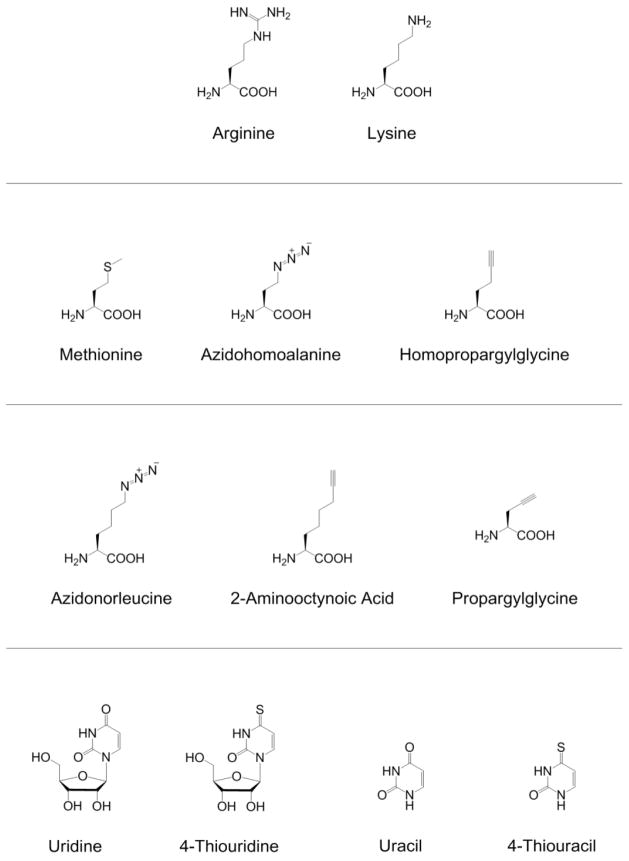
Structures discussed in this review: amino acids for stable isotopic labeling (top row), methionine and analogs that are substrates for wild-type methionyl-tRNA synthetases (second row), methionine analogs that require the expression of mutant methionyl-tRNA synthetases for proteomic incorporation (third row), and uridine and uracil as well as their thio-substituted analogs 4-thiouridine and 4-thiouracil (last row).
Pulsed Stable-Isotope Labeling with Amino Acids in Cell Culture (pSILAC)
Unlike pulse labeling with a single label to determine protein turnover or transport in the same sample, pulsed stable-isotope labeling with amino acids in cell culture (pSILAC, Figure 1, Top Right) quantifies differences in protein synthesis between different samples integrated over the measurement time after the pulse. In pSILAC, cells in two different samples previously cultured in light media are transferred to two different pulse media: one containing heavy amino acids (typically arginine-10: 13C615N41H1416O2 and lysine-8: 13C615N21H1416O2) and the other containing “medium-heavy” amino acids (e.g., arginine-6: 13C614N41H102H416O2 and lysine-4: 12C614N21H102H416O2).75,77 During labeling, only newly synthesized proteins incorporate either the heavy or the medium-heavy amino acids. Peptides derived from heavy or medium-heavy proteins are distinguishable during the initial ion scan from pre-existing, light proteins on the basis of the mass difference introduced by the isotope label. Intensity ratios for heavy and medium-heavy peptides directly indicate the relative abundances of the corresponding newly synthesized proteins in the samples of interest. Light pre-existing peptides are identified but ignored during quantification.
Selbach and coworkers introduced pSILAC in 2008 as a method to measure changes in protein translation involved in cellular iron homeostasis.75 Since then, researchers have used pSILAC to assess protein dynamics associated with microRNA overexpression,34,62,77 monocyte-macrophage differentiation,37 hyperglycemia-induced stress95 and mammalian target of rapamycin inhibition.33 Selbach et al. utilized pSILAC to measure changes in the synthesis of several thousand proteins in HeLa cells in response to either microRNA transfection or endogenous microRNA knockdown.77 HeLa cells cultivated in light media were first transfected with miR-1, miR-155, miR-16, miR-30a, or let-7b, or mock transfected. Eight hours later, cells were transferred to media containing either medium-heavy or heavy amino acids for 24 hours. Unexpectedly, pSILAC in this study and subsequent studies revealed that overexpression of a single microRNA can lead to repression of hundreds of proteins; however the repressive effect was relatively small and rarely exceeded fourfold.34,62 These results cast microRNAs as general orchestrators that tune cellular physiology and metabolism in subtle ways in response to specific cues.
Bio-Orthogonal Non-Canonical Amino Acid Tagging (BONCAT)
Under some circumstances (e.g., for short labeling times), the dynamic range and complexity of the proteome can preclude reliable identification and analysis of low-abundance isotope-labeled proteins in the presence of more abundant pre-existing proteins. Moreover, the use of multiple labels increases the number of distinct peptide forms and the computational complexity associated with protein identification. These limitations can be addressed by selective enrichment and identification of newly synthesized proteins.
In 2006, Dieterich et al. introduced the bio-orthogonal non-canonical amino acid tagging (BONCAT, Figure 1, Bottom Left) strategy to enable selective enrichment and identification of newly synthesized proteins in cells and tissues.17 This strategy relies on bio-orthogonal functional groups – functional groups that react rapidly and selectively with each other but remain inert to the functional groups normally found in biological systems.66 An exemplary bio-orthogonal group, the azide is small, kinetically stable and absent from living systems, yet can be modified easily and selectively through the Staudinger ligation with triarylphosphine reagents,74 the copper(I)-catalyzed “click” cycloaddition with terminal alkynes72,87 or the strain-promoted “click” cycloaddition with strained cyclooctynes.2
BONCAT employs a two-stage procedure. First, pulse labeling of cells or tissues with the azide-bearing methionine (Met, Figure 2) analog azidohomoalanine (Aha, Figure 2) or the alkyne-bearing methionine analog homopropargylglycine (Hpg, Figure 2) enables metabolic incorporation of either azide or alkyne functional groups into the proteome. In bacterial and mammalian systems, wild-type methionyl-tRNA synthetases (MetRSs) are capable of appending the non-canonical amino acid Aha (or Hpg) to cognate transfer RNAs (tRNAMet).17,36 Aha (or Hpg) is thereby incorporated into proteins made during the Aha (or Hpg) pulse. Second, a bio-orthogonal ligation with the complementary reactive group conjugated to a probe enables detection of Aha- or Hpg-tagged proteins. Treatment of azide-tagged proteins with alkyne-functionalized fluorescent dyes permits visualization of newly synthesized proteins.6,7,18 Alternatively, treatment of azide-tagged proteins with alkyne-functionalized affinity reagents allows selective enrichment of proteins made during the Aha pulse. Enriched proteins can then be identified by LC-MS/MS.56,81
BONCAT has been used in mammalian cells,17 in both unicellular38–39 and multicellular organisms,29 and even adapted to measure genome-wide nucleosome turnover dynamics in Drosophila S2 cells.14 Here, we highlight a few recent examples with an emphasis on proteomic discovery. Zhang et al. reported a tandem labeling (Aha and alkyne-functionalized palmitic acid) and detection method to monitor the dynamic acylation of Lck, an N-myristoylated and S-palmitoylated non-receptor tyrosine kinase required for T-cell activation.96 Liu et al. expanded on this concept by enriching proteins made in Jurkat cells during a specified time interval, then progressively monitoring their post-translational modifications over time by quantitative in-gel fluorescence scanning with Aha/Hpg and eight additional azide- and alkyne-functionalized post-translational modification probes.47 The up-regulation of myristoylated protein kinase A was found to be intimately linked to butyric acid-induced apoptosis.
Because local protein synthesis is critical for long-term functional synaptic changes, BONCAT is an attractive tool for neurobiological studies. Tcherkezian et al. examined the colocalization of DCC (Deleted in Colorectal Cancer) transmembrane receptors with newly synthesized proteins in cultured commissural axon growth cones with Aha-labeling.84 Hodas et al. used BONCAT to identify proteins translated in intact hippocampal neuropil sections upon treatment with the selective D1/D5 dopamine receptor agonist SKF81297.30 Yoon et al. developed DIGE-NCAT – a combination of 2D difference gel electrophoresis (2D-DIGE) and BONCAT – to examine changes in the proteome of Xenopus retinal ganglion cell (RGC) axons in response to stimulation with Engrailed-1.94 Engrailed-1 belongs to a family of transcription factors previously shown to cause rapid translation-dependent guidance responses in RGC axons.10 The authors first severed distal portions of the axon bundles from Xenopus eyes and stimulated them with Engrailed-1 for 1 hour along with the addition of Aha. Next, Aha-tagged axonally synthesized proteins were treated with an alkynyl dye and resolved on 2D-DIGE. Compared to control gels, spots with the greatest difference in fluorescence were analyzed by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Surprisingly, this strategy revealed that the intermediate filament protein lamin B2 (LB2) – normally associated with the nuclear membrane – is axonally synthesized in response to stimulation. Coupled with the finding of LB2’s association with mitochondria, these results suggest that LB2-promotion of mitochondrial function is needed for axon maintenance.
Quantitative Non-Canonical Amino Acid Tagging (QuaNCAT)
How can we quantitatively study proteomic changes during short time intervals (e.g., in response to a stimulus) without the complications that arise from the abundance of pre-existing proteins? As labeling times decrease, co-eluting pre-existing peptides increasingly obscure low abundance pSILAC-labeled peptides during the first mass spectrometry scan. To overcome this limitation, pSILAC and BONCAT have been combined in an approach designated Quantitative Non-Canonical Amino Acid Tagging (QuaNCAT, Figure 1, Bottom Right). In a QuaNCAT experiment, two parallel populations of cells in light media are transferred for a limited time to either medium-heavy or heavy media that also contain Aha.19,31 During the labeling period, newly synthesized proteins incorporate either the heavy or medium-heavy amino acid as well as Aha. Through BONCAT enrichment of the combined protein pools, pre-existing light peptides are greatly reduced in abundance. As in pSILAC, ratios of intensities of heavy and medium-heavy peptides directly indicate the relative abundances of newly synthesized proteins in the two samples.
To determine the benefit of selectively enriching and quantifying secreted proteins, Eichelbaum et al. labeled two human cell lines (PC3 and WPMY-1) with Aha and isotopologs of arginine and lysine for 24 hours.19 With enrichment via on-bead azide-alkyne cycloaddition and trypsin digestion, 684 secreted proteins were quantified with high correlation (R = 0.96) between biological duplicates. Without enrichment, only 22 proteins were quantified with low correlation (R = 0.02). The authors identified as many as 500 proteins, even at short labeling times (2 hours), including several known lipopolysaccharide (LPS) effector proteins as well as many other previously unassociated proteins during LPS stimulation of mouse macrophages. Howden et al. tested QuaNCAT by examining changes in expression of more than 600 proteins enriched from freshly isolated human CD4-positive T cells stimulated by activation with phorbol 12-myristate 13-acetate and ionomycin during 2 and 4 hour pulses.31 Many transcription factors and transcriptional regulators, cytokines, activation surface markers, protein chaperones and proteins involved in cytoskeleton dynamics and vesicle transport, were among the proteins found to be substantially increased in expression following stimulation. While less than 1% of the proteome was labeled in a 2 hour pulse, the post-enrichment protein pool consisted of 10–20% heavy and medium-heavy proteins. The ability to resolve protein abundance changes over very short time spans should prove useful when studying systems previously thought to be “unquantifiable” by SILAC such as short-lived progenitor or primary cells.32
O-Propargyl-Puromycin Labeling
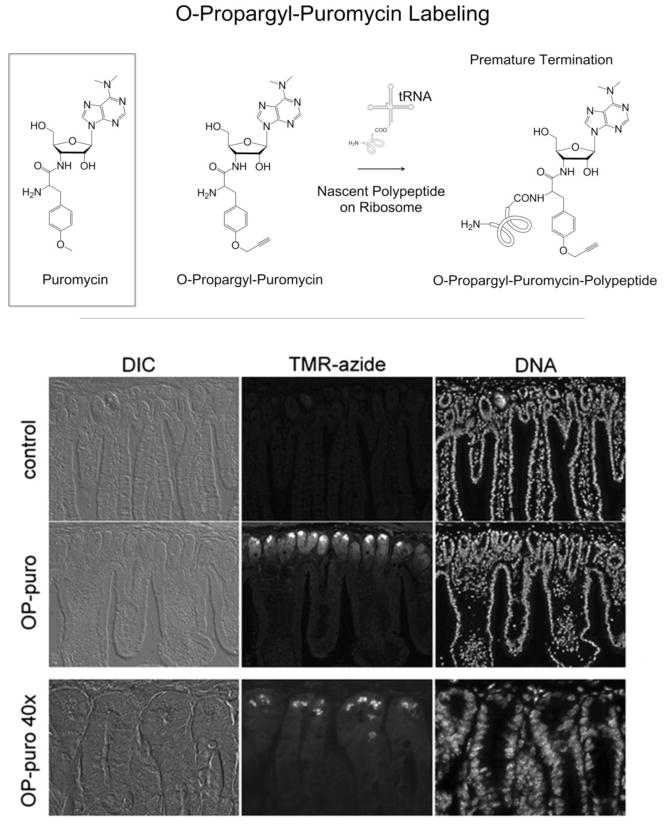
Salic and coworkers have developed an alternative approach to the selective labeling of newly synthesized proteins, in which a puromycin analog (O-propargyl-puromycin, Figure 3, Top) bearing a terminal alkyne is used to label nascent polypeptide chains.46 Puromycin mimics aminoacyl-tRNAs and terminates translational elongation after entering the acceptor site of the ribosome. Salic and coworkers showed that O-propargyl-puromycin not only terminates nascent polypeptide chains but also appends an alkyne label at the C-terminus. Liu and co-workers injected mice intraperitoneally with O-propargyl-puromycin and harvested tissues 1 hour later. Tissues from these mice displayed specific patterns of O-propargyl-puromycin incorporation into nascent proteins. For example, in the small intestine, the most intense labeling occurred in cells in the crypts and at the base of intestinal villi, consistent with the high proliferative and secretory activity of these cells (Figure 3, Bottom).
Figure 3.
(Top) Both puromycin and its alkyne-functionalized analog O-propargyl-puromycin incorporate into nascent polypeptide chains on translating ribosomes, resulting in premature termination of nascent polypeptide chains. (Bottom) Sectioning of mouse small intestine showed that OP-Puro labeling occurred primarily in cells in the crypts and the cells at the base of the villi. (Adapted with permission from Liu et al., Proc. Natl. Acad. Sci. U.S.A., 109, 413–418, 2012. Copyright 2012 Proceedings of the National Academy of Sciences USA.)
Spatially Resolved Proteomic Analysis
Coupling Flow Cytometry and Mass Spectrometry
Proteomic analysis of rare cells in heterogeneous environments presents important, difficult challenges. Combining flow cytometry with quantitative proteomics provides a solution in some systems of this kind. For example, Rechavi et al. combined quantitative proteomics and high-purity cell sorting to discover proteins transferred from human B cells to natural killer cells.70 These authors developed a strategy called “trans-SILAC,” in which one cell type – capable of transferring proteins – is labeled with heavy arginine and lysine and the other cell type – the recipient of transferred proteins – remains unlabeled. To initiate contact- and actin cytoskeleton-dependent protein transfer, the heavy “donor” B cells and freshly isolated light “recipient” natural killer cells were co-incubated for 1.5 hours. Fluorescence-activated cell sorting separated recipient cells from donor cells before the proteomic workflow, and transferred proteins in the recipient cells were identified by their mass shifts. Analysis of the transferred proteins revealed significant enrichment for the annotation term “MHC class II protein complex”, as expected.
Cell-Selective BONCAT
Although some cell types can be isolated for proteomic analysis through cell sorting or laser-capture techniques, others cannot. Subsets of neurons or glia in the central nervous system, for example, are difficult to isolate by dissection or dissociation methods. Or perhaps one would like to analyze proteomic responses of pathogenic bacteria hiding inside host cells, or of specific cells in multicellular animals, without interference from proteins derived from the host or from all of the other cells in the animal. Can we perform these kinds of tasks without prior separation of the cells of interest?
Promising solutions to analogous problems in genomics and transcriptomics rely on spatially restricted enzymatic labeling. In 2010, Henikoff and coworkers developed INTACT (isolation of nuclei tagged in specific cell types) – a method that allows affinity-based isolation of nuclei from individual cell types in a tissue.13 In INTACT, the spatial restriction of the Escherichia coli biotin ligase BirA and a nuclear targeting fusion protein results in biotin-labeled nuclei in the cell type of choice. First described in Arabidopsis, INTACT has been extended to profile gene expression and histone modifications in adult Caenorhabditis muscle79 and Kenyon cells and octopaminergic neurons in the adult Drosophila brain.28
For RNA, Miller et al. developed TU-tagging, a method that allows affinity-based isolation of RNA from individual cell types of a tissue.52 In TU-tagging, the spatial restriction of the Toxoplasma gondii nucleotide salvage enzyme uracil phosphoribosyltransferase enables RNA in the cell type of choice to be labeled with 4-thiouracil (Figure 2). Unlike 4-thiouridine, 4-thiouracil is not recognized by the endogenous biosynthetic machinery. Recently, Gay et al. used TU-tagging to purify transcripts from rare (<5%) cells, such as Tie2:Cre+ brain endothelia/microglia in mice.23
Analogous strategies have been developed for cell-selective proteomic analysis. Because endogenous methionyl-tRNA synthetases charge Aha to cognate tRNAs in all cell types, Aha-based BONCAT is not cell-selective – newly synthesized proteins in all cell types are labeled. To enable cell-selective proteomic analysis, Tirrell and coworkers engineered a family of mutant E. coli MetRSs capable of appending the azide-bearing methionine analog azidonorleucine (Anl, Figure 2) to tRNAMet.45,83 Anl is not a good substrate for any of the wild-type aminoacyl-tRNA synthetases in bacteria to mammals; it is excluded from proteins made in wild-type cells but incorporated readily into proteins made in cells that express an appropriately engineered MetRS. Anl-labeling does not require depletion of methionine if the mutant MetRS activates Anl faster than methionine.
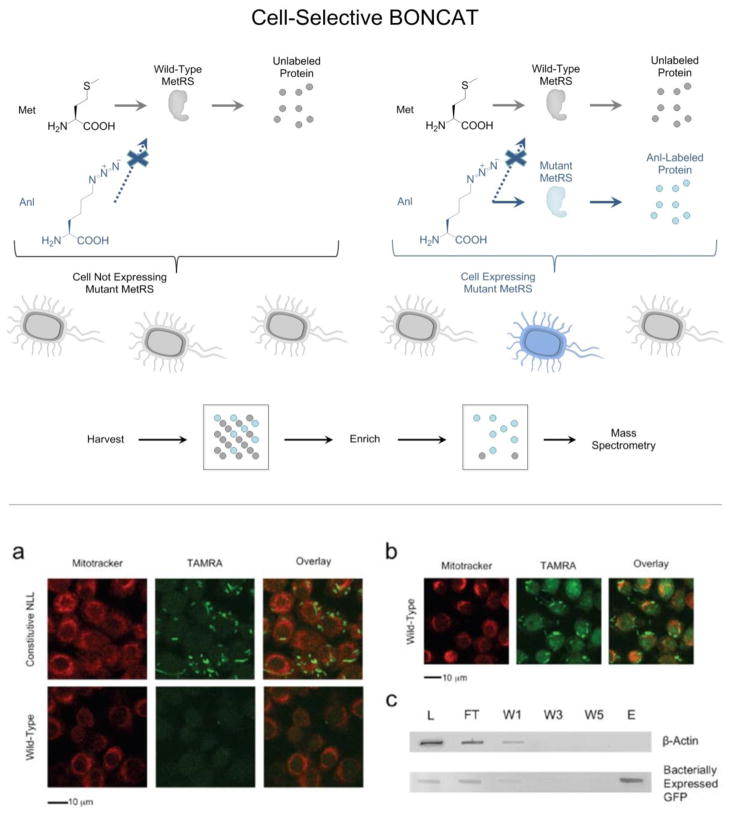
Ngo et al. first achieved cell type-selectivity by outfitting an E. coli strain with the L13N/Y260L/H301L mutant form of the E. coli MetRS (NLL-EcMetRS). Proteins made in this strain could be labeled with Anl in co-culture with murine alveolar macrophages, which were not labeled (Figure 4).59 Bacterial proteins were effectively separated from murine proteins by treatment of mixed lysates with alkyne-functionalized biotin reagents and subsequent affinity chromatography on NeutrAvidin resin.
Figure 4.
(Top) Cell-selective BONCAT performed in a mixture of cells. Restricting expression of a mutant synthetase to a certain cell restricts Anl labeling to that cell (highlighted in blue). Proteins synthesized in cells (highlighted in gray) that do not express the mutant synthetase are neither labeled nor detected following enrichment. (Bottom) Cell-selective labeling in mixtures of bacterial and mammalian cells. (a) In Anl-containing mixed cultures of E. coli and mouse alveolar macrophages, only E. coli cells constitutively expressing the mutant NLL-EcMetRS were labeled by TAMRA-alkyne. Macrophages were labeled with Mitotracker Deep Red and displayed low TAMRA-alkyne background emission. (b) In Aha-containing mixed cultures of E. coli and mouse alveolar macrophages, both wild-type E. coli cells and macrophages exhibited strong TAMRA-alkyne emission; incorporation of Aha occurs in both cell types. (c) Mixed cell lysate was subjected to conjugation with alkyne-functionalized biotin, and labeled proteins were enriched by NeutrAvidin affinity chromatography. Immunoblotting of unbound flow-through (FT), washes (W1, W3, W5) and eluent (E) reveals enrichment of the bacterial marker protein GFP. (Adapted with permission from Ngo et al., Nat. Chem. Biol., 5, 715–717, 2009. Copyright 2009 Nature Publishing Group.)
System-wide identification of bacterial proteins expressed or secreted during infection can provide new insight into mechanisms of bacterial pathogenesis. Grammel et al. studied cultures of Raw264.7 murine macrophages infected with S. typhimurium (a Gram-negative intracellular pathogen) outfitted with the NLL-EcMetRS.25 Cultures were pulse-labeled with 2-aminooctynoic acid (Aoa, Figure 2), an alkyne analog of Anl, for 1 hour. Of the 218 proteins identified, 185 (85 %) were Salmonella proteins and 33 were mouse proteins. Five of the Salmonella proteins (SodM, SsrB, SseA, PipB2, and PhoP) had been previously described as virulence factors.
In more recent work, Ngo and co-workers demonstrated that heterologous expression of the NLL-EcMetRS enables incorporation of Anl into proteins expressed in human (HEK293) cells, permitting enrichment and visualization of proteins made during various stages of the cell cycle.58 Interestingly, Anl replaces methionine selectively at N-terminal positions not at internal sites. Site-selectivity occurs because NLL-EcMetRS catalyzes aminoacylation only of the mammalian initiator tRNAMet, not the mammalian elongator tRNAMet. Through judicious selection of regulatory elements, systems of this kind will enable cell-selective or “cell-state-selective”57 interrogation of protein synthesis in co-cultures of mammalian cells, in virally transfected tissues or even in living animals.
To expand the set of tools available for cell-selective BONCAT, Truong et al. engineered a MetRS variant capable of activating propargylglycine (Pra, Figure 2) but not Anl.88 Pra is an alkynyl amino acid smaller than methionine and is not activated by wild-type aminoacyl-tRNA synthetases or by NLL-EcMetRS. Using directed evolution, Truong et al isolated a MetRS variant (designated propargylglycyl-tRNA synthetase; PraRS, L13P/A256G/P257T/Y260Q/H301F/A331V/Δ548E) capable of near-quantitative replacement of Met by Pra in proteins. In protein mixtures that have been labeled both with Anl and with Pra, treatment with cyclooctyne-functionalized probes selectively tags Anl side chains, and subsequent treatment with azide-functionalized probes selectively tags Pra side chains. By using one promoter to drive expression of NLL-EcMetRS in one cell and a different promoter to drive expression of PraRS in another cell, researchers can perform differential, cell-selective BONCAT in complex multicellular systems without prior separation.
Restricting expression of mutant methionyl-tRNA synthetases by using promoters active only in specific cells or tissues should restrict Anl/Aoa labeling to those cells or tissues. Additional specificity should be possible by exploiting the combinatorial action of multiple regulators. To this end, Mahdavi et al. bisected the NLL-EcMetRS and found several split variants capable of charging Anl to tRNAMet.48 Because labeling requires expression of both the N- and C-terminal fragments of the synthetase, this system allows the investigator to restrict labeling to cells in which two promoters of interest are active.
Ascorbate Peroxidase (APEX) Labeling
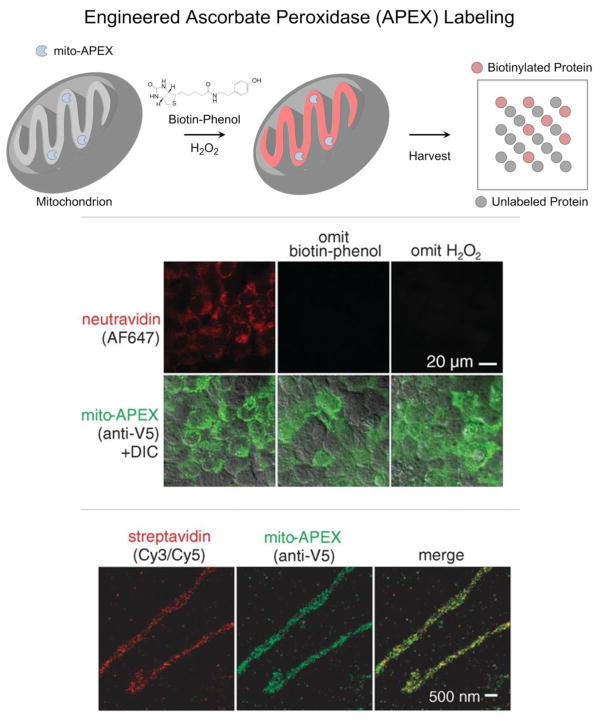
Ting and coworkers have recently used an engineered variant of ascorbate peroxidase (APEX) to selectively tag proteins that localize to the mitochondrial matrix in living cells.71 The Ting laboratory had previously used APEX to catalyze the H2O2-dependent polymerization of diaminobenzidine to provide contrast in electron microscopy.51 They genetically targeted APEX to the mitochondrial matrix of human embryonic kidney (HEK) cells, initiated labeling by adding biotin-phenol and H2O2, and stopped labeling after 1 minute by cell fixation or lysis (Figure 5). APEX processes biotin-phenol in a hydrogen peroxide-dependent manner to yield highly reactive products that covalently link biotin to electron-rich amino acids such as tyrosine, tryptophan, histidine and cysteine. Unlike cell-selective BONCAT, APEX labeling is not time-selective. Any proteins accessible to the phenoxyl radical -- including pre-existing proteins made before the biotin-phenol pulse -- are biotinylated in the 1 minute “snapshot”. Enrichment and mass spectrometry of the biotinylated proteins led to identification of 464 known mitochondrial proteins as well as 31 that were not previously known to localize to mitochondria. Demonstrating APEX-based proteomics as a viable discovery tool, a random subset of these 31 “mitochondrial orphans” were verified by fluorescence imaging to have complete or partial mitochondrial localization.
Figure 5.
(Top) Selective labeling of the mitochondrial matrix proteome in living cells requires 1) genetically targeting APEX to the mitochondrial matrix (mito-APEX), 2) initiating biotinylation by adding biotin-phenol and H2O2 to the medium, and 3) stopping biotinylation by cell fixation or lysis. (Middle) In human embryonic kidney cells, only mitochondria that expressed mito-APEX and were exposed to both biotin-phenol and H2O2 contained biotinylated proteins (stained with NeutrAvidin-Alexa Fluor 647). Both confocal fluorescence imaging (Middle) and stochastic optical reconstruction microscopy (STORM) (Bottom) showed that biotinylated proteins (stained with Streptavidin-Cy3/Cy5 for STORM images) overlapped with mito-APEX only in the mitochondrial matrix. (Adapted with permission from Rhee et al., Science, 339, 1328–1331, 2013. Copyright 2013 The American Association for the Advancement of Science.)
Conclusion
This review highlights chemical biological strategies that allow researchers to identify, isolate and quantitatively analyze proteins within defined windows of time and space. Many of these strategies rely on amino acids with isotopic signatures or functional groups that are not found in biological systems. Incorporation of these “tags” permits the investigator to distinguish proteins made in different time intervals or in different cells or organelles. In combination with these tagging methods, mass spectrometry becomes an even more powerful tool for addressing complex biological questions.
Acknowledgments
Work at Caltech on non-canonical amino acid tagging is supported by National Institutes of Health grant NIH R01 GM062523 and by the Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from U.S. Army Research Office.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Agard N, Prescher J, Bertozzi C. A strain-promoted [3+2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 3.Ahrens CH, Brunner E, Qeli E, Basler K, Aebersold R. Generating and navigating proteome maps using mass spectrometry. Nat Rev Mol Cell Biol. 2010;11:789–801. doi: 10.1038/nrm2973. [DOI] [PubMed] [Google Scholar]
- 4.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 5.Bantscheff M, Lemeer S, Savitski MH, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404:939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- 6.Beatty K, Xie F, Wang Q, Tirrell D. Selective dye-labeling of newly synthesized proteins in bacterial cells. J Am Chem Soc. 2005;127:14150–14151. doi: 10.1021/ja054643w. [DOI] [PubMed] [Google Scholar]
- 7.Beatty K, Liu J, Xie F, Dieterich D, Schuman E, Wang Q, Tirrell D. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed. 2006;45:7364–7367. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- 8.Black DL. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 9.Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics. 2010;9:457–470. doi: 10.1074/mcp.M900429-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunet I, Weinl C, Piper M, Trembleau A, Volovitch M, Harris W, Prochiantz A, Holt C. The transcription factor Engrailed-2 guides retinal axons. Nature. 2005;438:94–98. doi: 10.1038/nature04110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corthals GL, V, Wasinger C, Hochstrasser DF, Sanchez JC. The dynamic range of protein expression: A challenge for proteomic research. Electrophoresis. 2000;21:1104–1115. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1104::AID-ELPS1104>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 12.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 13.Deal RB, Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M. Comprehensive mass spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 16.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dieterich DC, Hodas J, Gouzer G, Shadrin I, Ngo J, Triller A, Tirrell D, Schuman E. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichelbaum K, Winter M, Diaz MB, Herzig S, Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotechnol. 2012;30:984–990. doi: 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- 20.Essader AS, Cargile BJ, Bundy JL, Stephenson JL. A comparison of immobilized pH gradient isoelectric focusing and strong cation-exchange chromatography as a first dimension in shotgun proteomics. Proteomics. 2005;5:24–34. doi: 10.1002/pmic.200400888. [DOI] [PubMed] [Google Scholar]
- 21.Fredens J, Engholm-Keller K, Giessing A, Pultz D, Larsen MR, Højrup P, Møller-Jensen J, Færgeman NJ. Quantitative proteomics by amino acid labeling in C. elegans. Nat Methods. 2011;8:845–847. doi: 10.1038/nmeth.1675. [DOI] [PubMed] [Google Scholar]
- 22.Friedel CC, Dolken L, Ruzsics Z, Koszinowski UH, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gay L, Miller MR, Ventura PB, Devasthali V, Vue Z, Thompson HL, Temple S, Zong H, Cleary MD, Stankunas K, Doe CQ. Mouse TU tagging: a chemical/genetic intersectional method for purifying cell type-specific nascent RNA. Genes Dev. 2013;27:98–115. doi: 10.1101/gad.205278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay TR. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 25.Grammel M, Zhang MM, Hang HC. Orthogonal alkynyl-amino acid reporter for selective labeling of bacterial proteomes during infection. Angew Chem Int Ed. 2010;49:5970–5974. doi: 10.1002/anie.201002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gresham D, Dunham MJ, Botstein D. Comparing whole genomes using DNA microarrays. Nat Rev Genet. 2008;9:291–302. doi: 10.1038/nrg2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 28.Henry GL, Davis FP, Picard S, Eddy SR. Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res. 2012;40:9691–9704. doi: 10.1093/nar/gks671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinz FI, Dieterich DC, Tirrell DA, Schuman EM. Noncanonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem Neurosci. 2012;3:40–49. doi: 10.1021/cn2000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodas JJL, Nehring A, Höche N, Sweredoski MJ, Pieolt R, Hess S, Tirrell DA, Dieterich DC, Schuman EM. Dopaminergic modulation of the hippocampal neuropil proteome identified by bioorthogonal noncanonical amino acid tagging (BONCAT) Proteomics. 2012;12:2464–2476. doi: 10.1002/pmic.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howden AJM, Geoghegan V, Katsch K, Efstathiou G, Bhushan B, Boutureira O, Thomas B, Trudgian DC, Kessler BM, Deiterich DC, Davis BG, Acuto O. QuaNCAT: quantitating proteome dynamics in primary cells. Nat Methods. 2013;10:343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes C, Krijgsveld J. Developments in quantitative mass spectrometry for the analysis of proteome dynamics. Trends Biotechnol. 2012;30:668–676. doi: 10.1016/j.tibtech.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Huo Y, Iadevaia V, Yao Z, Kelly I, Cosulich S, Guichard S, Foster LJ, Proud CG. Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem J. 2012;444:141–151. doi: 10.1042/BJ20112107. [DOI] [PubMed] [Google Scholar]
- 34.Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Röh S, Hoffmann R, Warscheid B, Hermeking H. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics. 2011;10:M111.010462. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: Direct identification of O-GlcNAc-modified proteins. Proc Natl Acad Sci USA. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraft-Terry SD, Gendelman HE. Proteomic biosignatures for monocyte-macrophage differentiation. Cell Immunol. 2011;271:239–255. doi: 10.1016/j.cellimm.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramer G, Sprenger RR, Nessen MA, Roseboom W, Speijer D, de Jong L, de Mattos MJ, Back J, de Koster CG. Proteome-wide alterations in Escherichia coli translation rates upon anaerobiosis. Mol Cell Proteomics. 2010;9:2508–2516. doi: 10.1074/mcp.M110.001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kramer G, Sprenger RR, Back J, Dekker HL, Nessen MA, van Maarseveen JH, de Koning LJ, Hellingwerf KJ, de Jong D, de Koster CG. Identification and quantitation of newly synthesized proteins in Escherichia coli by enrichment of azidohomoalanine-labeled peptides with diagonal chromatography. Mol Cell Proteomics. 2009;8:1599–1611. doi: 10.1074/mcp.M800392-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krijgsveld J, Ketting RF, Mahmoudi T, Johansen J, Artal-Sanz M, Verrijzer CP, Plasterk RHA, Heck AJR. Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat Biotechnol. 2003;21:927–931. doi: 10.1038/nbt848. [DOI] [PubMed] [Google Scholar]
- 41.Krüger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fässler R, Mann M. SILAC mouse for quantitative proteomics uncovers Kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 42.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larance M, Bailly AP, Pourkarimi E, Hay RT, Buchanan G, Coulthurst S, Xirodimas DP, Gartner A, Lamond AI. Stable isotope labeling with amino acids in nematodes. Nat Methods. 2011;8:849–851. doi: 10.1038/nmeth.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larrabee KL, Phillips JO, Williams GJ, Larrabee AR. The relative rates of protein synthesis and degradation in a growing culture of Escherichia coli. Journal of Biological Chemistry. 1980;255:4125–4130. [PubMed] [Google Scholar]
- 45.Link AJ, Vink MKS, Agard NJ, Prescher JA, Bertozzi CR, Tirrell DA. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc Natl Acad Sci USA. 2006;103:10180–10185. doi: 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci USA. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu K, Yang PY, Na Z, Yao SQ. Dynamic monitoring of newly synthesized proteomes: up-regulation of myristoylated protein kinase A during butyric acid induced apoptosis. Angew Chem Int Ed. 2001;50:6776–6781. doi: 10.1002/anie.201102542. [DOI] [PubMed] [Google Scholar]
- 48.Mahdavi A, Segall-Shaprio TH, Kou S, Jindal GA, Hoff KG, Liu S, Chitsaz M, Ismagilov RF, Silberg JJ, Tirrell DA. A genetically encoded AND gate for cell-targeted metabolic labeling of proteins. J Am Chem Soc. 2013;135:2979–2982. doi: 10.1021/ja400448f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 50.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 51.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller MR, Robinson KJ, Cleary MD, Doe CQ. TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat Methods. 2009;6:439–441. doi: 10.1038/nmeth.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milner E, Barnea E, Beer I, Admon A. The turnover kinetics of major histocompatibility complex peptides of human cancer cells. Mol Cell Proteomics. 2006;5:357–365. doi: 10.1074/mcp.M500241-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Mitulovíc G, Mechtler K. HPLC techniques for proteomics analysis-a short overview of latest developments. Brief Funct Genomic Proteomic. 2006;5:249–260. doi: 10.1093/bfgp/ell034. [DOI] [PubMed] [Google Scholar]
- 55.Mosteller R, Goldstein R, Nishimoto K. Metabolism of individual proteins in exponentially growing Escherichia coli. J Biol Chem. 1980;255:2524–2532. [PubMed] [Google Scholar]
- 56.Nessen MA, Kramer G, Back J, Baskin JM, Smeenk LEJ, de Koning LJ, van Maarseveen JH, de Jong L, Bertozzi CR, Hiemstra H, de Koster CG. Selective enrichment of azide-containing peptides from complex mixtures. J Proteome Res. 2009;8:3702–3711. doi: 10.1021/pr900257z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ngo JT, Babin BM, Champion JA, Schuman EM, Tirrell DA. State-selective metabolic labeling of cellular proteins. ACS Chem Biol. 2012;7:1326–1330. doi: 10.1021/cb300238w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ngo JT, Schuman EM, Tirrell DA. Mutant methionyl-tRNA synthetase from bacteria enables site-selective N-terminal labeling of proteins expressed in mammalian cells. Proc Natl Acad Sci USA. 2013;110:4992–4997. doi: 10.1073/pnas.1216375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, Beatty KE, Conner RE, Yoo TH, Dieterich DC, Schuman EM, Tirrell DA. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;5:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olsen JV, Ong S-E, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2007;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 62.Patron JP, Fendler A, Bild M, Jung U, Müller H, ØArntzen M, Piso C, Stephan C, Thiede B, Mollenkopf H-J, Jung K, Kaufmann SHE, Schreiber J. MiR-133b targets antiapoptotic genes and enhances death receptor-induced apoptosis. PLOS ONE. 2012;7:e35, 345. doi: 10.1371/journal.pone.0035345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Picotti P, Clément-Ziza M, Lam H, Campbell DS, Schmidt A, Deutsch EW, Röst H, Sun Z, Rinner O, Reiter L, Shen Q, Michaelson JJ, Frei A, Alberti S, Kusebauch U, Wollscheid B, Moritz RL, Beyer A, Aebersold R. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pratt JM, Petty J, Riba-Garcia I, Robertson DH, Gaskell SJ, Oliver SG, Beynon RJ. Dynamics of protein turnover, a missing dimension in proteomics. Mol Cell Proteomics. 2002;1:579–591. doi: 10.1074/mcp.m200046-mcp200. [DOI] [PubMed] [Google Scholar]
- 66.Prescher J, Bertozzi C. Chemistry in Living Systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 67.Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, Regev A. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rappsilber J, Mann M. What does it mean to identify a protein in proteomics? Trends Biochem Sci. 2002;27:74–78. doi: 10.1016/s0968-0004(01)02021-7. [DOI] [PubMed] [Google Scholar]
- 70.Rechavi O, Kalman M, Fang Y, Vernitsky H, Jacob-Hirsch J, Foster LJ, Kloog Y, Goldstein I. Trans-SILAC: sorting out the non-cell-autonomous proteome. Nat Methods. 2010;7:923–927. doi: 10.1038/nmeth.1513. [DOI] [PubMed] [Google Scholar]
- 71.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rostovtsev V, Green L, Fokin V, Sharpless K. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective ligation of azides and terminal alkynes. Angew Chem Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 73.Sadygov RG, Cociorva D, Yates JR., III Large-scale database searching using tandem mass spectra: Looking up the answer in the back of the book. Nat Methods. 2004;1:195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- 74.Saxon E, Bertozzi C. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 75.Schwanhäusser B, Gossen M, Dittmar G, Selbach M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9:205–209. doi: 10.1002/pmic.200800275. [DOI] [PubMed] [Google Scholar]
- 76.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 77.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 78.Steen H, Mann M. The abc’s (and xyz’s) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 79.Steiner FA, Talbert PB, Kasinathan S, Deal RB, Henikoff S. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 2012;22:766–777. doi: 10.1101/gr.131748.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sury MD, Chen JX, Selbach M. The SILAC fly allows for accurate protein quantification in vivo. Mol Cell Proteomics. 2010;9:2173–2183. doi: 10.1074/mcp.M110.000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szychowski J, Mahdavi A, Hodas JJL, Bagert JD, Ngo JT, Landgraf P, Dieterich DC, Schuman EM, Tirrell DA. Cleavable biotin probes for labeling of biomolecules via azide-alkyne cycloaddition. J Am Chem Soc. 2010;132:18351–18360. doi: 10.1021/ja1083909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanrikulu IC, Schmitt E, Mechulam Y, Goddard WA, III, Tirrell DA. Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc Natl Acad Sci USA. 2009;106:15285–15290. doi: 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thingholm TE, Jensen ON, Larsen MR. Analytical strategies for phosphoproteomics. Proteomics. 2009;9:1451–1468. doi: 10.1002/pmic.200800454. [DOI] [PubMed] [Google Scholar]
- 86.Tomlinson E, Palaniyappan N, Tooth D, Layfield R. Methods for the purification of ubiquitinated proteins. Proteomics. 2007;7:1016–1022. doi: 10.1002/pmic.200601008. [DOI] [PubMed] [Google Scholar]
- 87.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 88.Truong F, Yoo TH, Lampo TJ, Tirrell DA. Two-strain, cell-selective protein labeling in mixed bacterial cultures. J Am Chem Soc. 2012;134:8551–8556. doi: 10.1021/ja3004667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vogel C, Abreu S, de R, Ko D, Le SY, Shaprio BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics. 2006;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 93.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 94.Yoon BC, Jung H, Dwivedy A, O’Hare CM, Zivraj K, Holt CE. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148:752–764. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang L, Zhao H, Blagg BS, Dobrowsky RT. C-terminal heat shock protein 90 inhibitor decreases hyperglycemia-induced oxidative stress and improves mitochondrial bioenergetics in sensory neurons. J Proteome Res. 2012;11:2581–2593. doi: 10.1021/pr300056m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang MM, Tsou LK, Charron G, Raghavan AS, Hang HC. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proc Natl Acad Sci USA. 2010;107:8627–8632. doi: 10.1073/pnas.0912306107. [DOI] [PMC free article] [PubMed] [Google Scholar]