Abstract
The role of material surface properties in the direct interaction with bacteria and the indirect route via host defense cells is not fully understood. Recently, it was suggested that nanostructured implant surfaces possess antimicrobial properties. In the current study, the adhesion and biofilm formation of Staphylococcus epidermidis and human monocyte adhesion and activation were studied separately and in coculture in different in vitro models using smooth gold and well-defined nanostructured gold surfaces. Two polystyrene surfaces were used as controls in the monocyte experiments. Fluorescent viability staining demonstrated a reduction in the viability of S. epidermidis close to the nanostructured gold surface, whereas the smooth gold correlated with more live biofilm. The results were supported by scanning electron microscopy observations, showing higher biofilm tower formations and more mature biofilms on smooth gold compared with nanostructured gold. Unstimulated monocytes on the different substrates demonstrated low activation, reduced gene expression of pro- and anti-inflammatory cytokines, and low cytokine secretion. In contrast, stimulation with opsonized zymosan or opsonized live S. epidermidis for 1 hour significantly increased the production of reactive oxygen species, the gene expression of tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), IL-6, and IL-10, as well as the secretion of TNF-α, demonstrating the ability of the cells to elicit a response and actively phagocytose prey. In addition, cells cultured on the smooth gold and the nanostructured gold displayed a different adhesion pattern and a more rapid oxidative burst than those cultured on polystyrene upon stimulation. We conclude that S. epidermidis decreased its viability initially when adhering to nanostructured surfaces compared with smooth gold surfaces, especially in the bacterial cell layers closest to the surface. In contrast, material surface properties neither strongly promoted nor attenuated the activity of monocytes when exposed to zymosan particles or S. epidermidis.
Keywords: nanotopography, staphylococci, host defense, bacteria, zymosan, macrophage
Introduction
Biomaterial-associated infections are highly problematic for the patient, health care system, and society. The consequences of biomaterial-associated infections can be devastating and include potentially life-threatening systemic infections, tissue injury, device malfunction, and ultimately, a need to remove the implant.1,2 Under normal conditions, microorganisms that enter the body are kept under control by the immune system. The first lines of defense include a variety of factors, such as epithelial barriers, complement proteins, acute phase proteins, and cytokines, together with phagocytic cells such as monocytes, macrophages, and neutrophils. However, when a foreign material is present, such as an implant, the host defense system becomes hampered. It has been suggested that an “immuno-incompetent” zone is formed around the implant.3 In addition, adherent bacteria may form a biofilm that protects them from phagocytic uptake, bactericidal and opsonizing antibodies, as well as antibiotic treatment.3 In such cases, removal of the implant may be the only option to eradicate the infection.
Staphylococcus species, especially Staphylococcus epidermidis and Staphylococcus aureus, are the predominant species found in biomaterial-associated infections, accounting for about 66% of the microorganisms.1 Unfortunately, more and more bacteria are acquiring resistance to antibiotics,4 further necessitating preventive measures with a view to reducing infection rates.
Materials with nanotopographic features have been explored with respect to the adhesion and function of various cell types, such as fibroblasts, osteoblasts, mesenchymal stem cells, and keratinocytes.5–10 Less literature is available on the activities of inflammatory cells on defined, nanostructured surfaces,11–13 even though these cells are among the first to encounter an implanted device and play a decisive role in the acceptance of the implant. It has also been suggested that nanostructured materials play a role in bacterial adhesion. In contrast to eukaryotic cells, bacteria have a rigid cell wall with a limited capability to deform upon attachment, implying that they do not react to structures smaller than themselves.14 However, some studies in fact show a decrease in bacterial adhesion when exposed to nanotextured surfaces,15–17 but the results are contradictory and the contribution from surface chemistry cannot always be excluded.14
In the present study, we have investigated whether nanostructures have an effect on bacterial adhesion and biofilm formation and/or have an influence on the behavior of immune cells in response to microbial stimuli. Selected for this purpose were smooth gold-sputtered silica wafers that can be modified with well-defined nanoparticles (35–40 nm) in a systematic manner, using thiol chemistry. These model surfaces possess the same chemical characteristics18 and therefore enable a direct comparison of the role played by surface nanotexture in the behavior of inflammatory cells and microorganisms. Both a strong biofilm-producing strain of S. epidermidis and primary monocytes isolated from human blood donors were used in the study. Zymosan, a cell wall product from Saccharomyces cerevisiae, was used as a nonliving microbial control stimulus.
Materials and methods
The material surfaces used in the study were a smooth gold surface and different nanostructured gold surfaces. Tissue culture plastic (TCP) and tissue culture-treated plastic cover slips (Thx), (Thermanox®; Thermo Fisher Scientific, Waltham, MA, USA), which, similar to the gold surfaces can be transferred between wells, were used as control surfaces in the monocyte cell culture experiments.
Surface preparation
Synthesis of gold nanoparticles
Gold nanoparticles were prepared using a reduction of HAuCl4 (Sigma-Aldrich, St Louis, MO, USA) by sodium citrate (Sigma-Aldrich), using a modification of a previously described protocol.19 In short, particles with an average size of 38 nm were synthesized by heating the HAuCl4 (2.4 mM) solution to 60°C and adding 60°C tribasic sodium citrate solution (3.9 mM) (1:1). The synthesis was allowed to continue for 1 hour under stirring conditions. The particle stock solution was stored at 8°C until use.
Preparation of nanostructured gold surfaces
Gold-sputtered silicon wafers (200 nm gold on a supporting layer of 10 nm titanium) were purchased from Litcon AB (Gothenburg, Sweden). The substrates were washed for 15 minutes at 80°C in basic piranha solution containing 3:1:1 Milli-Q water, NH3 (24%), and H2O2 (30%) and washed in an excess of Milli-Q water before incubation in 20 mM aqueous solution of cysteamine (Sigma-Aldrich) for 2 hours. The surfaces were then rinsed with excess Milli-Q water and incubated in the gold nanoparticle stock solution at room temperature overnight, resulting in immobilization of the nanoparticles, and thereby a nanostructured surface. After incubation, surfaces were washed in an excess of Milli-Q water. Immediately before use, all the surfaces (smooth and nanostructured) were cleaned in an ultraviolet/ozone chamber for 15 minutes, washed in basic piranha for 10 minutes at 70°C, rinsed with an excess of water, and finally blown dry in a gentle stream of N2 (g). As shown earlier,20 the cleaning steps lead to removal of the organic layers on the nanoparticles and the gold substrate, resulting in a partial sintering of the particles to the substrate.
In an initial subset of experiments, exclusively with S. epidermidis (live and dead fluorescence microplate readings), surfaces with high and low surface coverage of nanoparticles (nanodense gold [AuND] and nanolight gold [AuNL] respectively), were prepared by controlling the electrostatic repulsion between the particles. The distance between colloids in an electrolyte depends primarily on the size of the electric double layer of counter ions surrounding the colloids. The interparticle distance between gold nanoparticles suspended in an electrolyte can thus be controlled by changing the ionic strength of the electrolyte, as described earlier.20,21 Briefly, the gold nanoparticle stock solution was centrifuged at 1,000× g for 90 minutes, and the pellet was resuspended in Milli-Q water or 10 mM sodium citrate (tri-basic) buffer at pH 4. Cysteamine-functionalized gold substrates were then incubated in the nanoparticle solutions for 3 hours and washed as described earlier before use.
Surface analysis
Surfaces were viewed in a Zeiss 982 Gemini digital scanning electron microscope (SEM; Carl Zeiss SMT GmbH, Oberkochen, Germany) in the secondary electron mode, using the in-lens detector mode. Nanoparticle size and surface coverage (projected area) were calculated from SEM images through image analysis in ImageJ software (National Institutes of Health, Bethesda, MD, USA); the images were thresholded to remove the background surface, and by assuming spherical particles, the average particle size and surface area coverage were calculated from the pixel count. In addition, surface roughness was evaluated using a Bruker Dimension 3100 atomic force microscope with an nsc 15 tip (MicroMash, NanoAndMore GmbH, Wetzlar, Germany) in the tapping mode in ambient air. Water contact angles were measured on the experimental substrates to assess surface wettability and to confirm the efficacy of the washing protocol. A 5-μL ultrapure water droplet (Milli-Q, 18.2 MΩ) was applied to the surface, and a side view image of the droplet was captured with high-magnification macrophotography. Contact angles were then measured using the angle tool in ImageJ software.
Bacterial adhesion and biofilm formation on nanotopographic versus smooth surfaces
Bacterial strains and culture
The biofilm producer strain S. epidermidis ATCC 35984, obtained from the Culture Collection University of Gothenburg (CCUG 31568), was used in this study. Single colonies from overnight cultures on Columbia horse blood agar plates (Media Department, Clinical Microbiology Lab, Sahlgrenska University Hospital, Gothenburg, Sweden) were suspended in 4 mL Roswell Park Memorial Institute (RPMI) 1640 medium containing GlutaMAX™ (Gibco, Life Technologies, Carlsbad, CA, USA) until an optical density (OD; 546 nm) of 0.25 (=108 colony-forming units [CFU]/mL). An inoculum suspension was prepared by diluting the OD suspension to 105 CFU/mL in pre-warmed RPMI medium. The RPMI medium was chosen because it was the most suitable medium to culture human monocytes and because it also supported the growth of S. epidermidis.
Live and dead fluorescence microplate readings
To examine the relative amount of adherent live and dead S. epidermidis after 24 hours, an inoculum of 105 CFU/mL in RPMI medium was added to smooth gold, AuND, and AuNL surfaces (n=3). After 24 hours of static incubation at 37°C, the surfaces were carefully washed with 0.9% sterile saline (3 × 1 mL) and incubated with 250 μL premixed staining solution from the FilmTracer™ LIVE/DEAD® Biofilm Viability kit (Invitrogen, Life Technologies, Carlsbad, CA, USA) for 30 minutes in the dark. The kit provides a two-color fluorescence assay (SYTO® 9 and propidium iodide) of bacterial viability in which all the cells are stained fluorescent green, and cells with damaged membranes are counterstained to fluoresce red. The surfaces were then washed, transferred to black 24-well plates (lumox® multiwell; Sarstedt AG and Co, Nümbrecht, Germany), and 500 μL saline was added to each surface. The microtiter plate was read by a FLUOstar Omega microplate reader (BMG Labtech, Ortenberg, Germany) for fluorescence, multichromatic, and top optic reading, using excitation filter 485 nm and emission filter 520 nm. A scan matrix of 20 × 20 was used, together with gains set at 1,500 for SYTO9 and 2,000 for propidium iodide fluorophores. Nonstained surfaces served as blanks. The experiment was repeated four times.
Viable counts
To test the initial adhesion and biofilm formation capacity of S. epidermidis on smooth and nanostructured gold surfaces, the following static adhesion experiment was performed twice: an inoculum of 105 CFU/mL S. epidermidis was prepared in RPMI medium, and a total of 1 mL was added to smooth and nanostructured gold surfaces (n=3). The surfaces and the bacterial suspension were incubated for 2, 24, or 48 hours at 37°C under static conditions. After each time was reached, the surfaces were washed to remove nonadherent bacterial cells. The surfaces were transferred to new tubes containing 1 mL 0.9% saline + 0.1% triton-X, sonicated for 30 seconds at 40 kHz, and hard vortexed for 1 minute to dislodge the adherent bacteria and break aggregates. Subsequent SEM analysis of the surfaces showed a good detachment procedure, with most of the surface area cleaned from bacteria. The sonicated suspension was assessed by quantitative cultures on blood agar plates. The CFU per surface was quantified by adding 0.1 mL of the sonicated suspension to serial dilutions until 10−6 in 0.9% saline + 0.1% triton-X. From the undiluted solution and the six dilutions, CFU counting was performed (double measurements).
SEM
The initial adherence and biofilm formation of S. epidermidis on smooth and nanostructured gold were assessed morphologically, using SEM. Samples were washed with Hank’s Balanced Salt Solution (HBSS) and fixated in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.15 M sodium cacodylate buffer (pH 7.2) overnight at 4°C. The samples were washed with 0.15 M sodium cacodylate buffer and postfixated with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer for 2 hours at 4°C. Contrast enhancement was performed with 1% thiocarbohydrazide for 10 minutes at room temperature, followed by incubation in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer for 1 hour at 4°C. Dehydration was performed in a graded series of ethanol (70%–99.5%) and critical point drying by hexamethyldisilazane evaporation. The samples were mounted on stubs and sputtered with palladium before viewing in a Zeiss 982 Gemini SEM operated at 3 kV.
Focused ion beam SEM
Samples prepared for morphological observations of S. epidermidis on smooth and nanostructured gold surfaces were analyzed in a dual-beam focused ion beam (FIB) system (Versa 3D; FEI, Eindhoven, The Netherlands) operated at 16.0 kV (Ga+ ions) and 10 kV (electrons). When single bacterial cells were localized, cross-sections of the cells were exposed by FIB milling and imaged by SEM. A total of six single bacterial cells per surface were evaluated.
Confocal laser scanning microscopy (CLSM)
To determine the capacity of S. epidermidis to form biofilm when growing on smooth and nanostructured gold surfaces, 103–104 CFU/mL S. epidermidis was incubated statically on the surfaces for 2, 24, and 48 hours. After each adhesion time, the surfaces (n=1) were carefully washed to remove nonadherent cells. A total of 200 μL FilmTracer™ LIVE/DEAD® staining solution was added to the biofilms for 30 minutes at room temperature under dark conditions. The surfaces were then carefully washed and placed in 60 mm Petri plates covered in saline for in situ visualization under a LSM 710 microscopy system (Carl Zeiss) fitted with a Plan-Apochromat 20x/1.0 water objective. For SYTO 9 a 488 nm argon ion laser was used and emitted fluorescence was filtered through a 505 to 530 nm band-pass. Propidium iodide was excited with a 561 nm diode-pumped solid-state laser and emitted fluorescence was filtered through a 585 to 690 nm band-pass. Five different equidistant random spots (four in the corners and one in the center of the surface) were chosen to perform z-stacks every 3 μm. Three independent experiments were performed. Confocal biofilm images were analyzed using the COMSTAT2 (http://www.comstat.dk/; Lyngby, Denmark) software, which converts biofilm image stacks into three-dimensional outputs for quantitative analysis. The parameters that were analyzed were biomass, maximum thickness, and area occupied at the surface (z=0 μm) by live and dead cells.22 Biomass is the amount of the image stack that is covered by bacteria (live and dead); the maximum thickness represents the compacted thickness of the biofilm image stack, and the area occupied by bacteria in the layer closest to the surface is the area occupied by biomass (cells) in the first stack image (the substratum). The images were thresholded with a threshold of 30 for 2 hours, 35 for 24 hours, and 40 for 48 hours.
Monocyte coculture experiments
Monocyte isolation, characterization, and culture
Human monocytes were isolated from buffy coats obtained from six blood donors, using Ficoll separation followed by negative selection on a magnetic column (MACS; Miltenyi Biotec, Bergisch Gladbach, Germany; viability, 98.7%±1.9%). For characterization, samples of 500,000 cells were stained with CD14-PE (CD14-phycoerythrin) and CD45-FITC (CD45-fluorescein isothiocyanate) antibodies and analyzed using flow cytometry (BD FACSCalibur™; BD Biosciences, San Jose, CA, USA), and the monocyte purity was estimated as 85.4%±4.7% (Figure S1). The cells were seeded on nanostructured or smooth gold squares (8 × 8 mm), on cell culture-treated Thx plastic cover slips (diameter, 13 mm; Nunc™, Thermo Fisher Scientific) and on tissue culture-treated polystyrene (TCP) in 24- or 48-well plates (Falcon™ [BD Biosciences] or Nunc™ [Thermo Fisher Scientific]) to best fit the size of the different materials. The experimental procedure is outlined in Figure 1. One million cells in 1 mL RPMI medium supplemented with 5% fetal bovine serum were seeded in all wells and cultured in humidified air with 5% CO2 at 37°C. Monocytes were allowed to adhere to the surfaces for 18 hours, after which the medium was removed, the surfaces were transferred to new wells, and 1 mL fresh medium was added. After a further 24 hours of culture, the cells were divided into three groups: unstimulated cells, zymosan-stimulated cells, or S. epidermidis-stimulated cells. Monocytes were stimulated by adding serum-opsonized zymosan A particles (final concentration, 2 × 107 particles/mL; Sigma-Aldrich) or serum-opsonized S. epidermidis (final concentration, 108 CFU/mL) for 1 hour, corresponding to a particle-to-cell ratio of 20 particles and 102 bacterial cells, respectively, per seeded monocyte. Zymosan was opsonized in active human serum in phosphate-buffered saline (1:1) for 1 hour at 37°C. From an overnight culture of S. epidermidis on blood agar, a colony was inoculated in tryptic soy broth and incubated at 37°C for 4 hours under shaking (150 rpm) until exponential growth (in duplicate), and opsonized in 10% active human serum in HBSS for 5 minutes at 37°C. All analyses were performed on cell culture replicates (n=2–3) and cells from four (S. epidermidis-stimulated), five (zymosan-stimulated) or six (unstimulated) blood donors were used.
Figure 1.
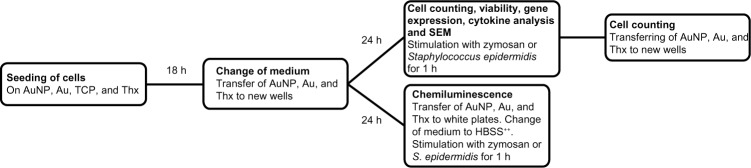
Schedule for coculture experiments.
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip; SEM, scanning electron microscopy; HBSS++, Hanks Balanced Salt Solution with Ca2+ and Mg2+; h, hours.
Monocyte viability
Cell viability was assessed using the centrifuged cell medium to evaluate lactate dehydrogenase (LDH) content (C-laboratory; Sahlgrenska University Hospital, Gothenburg, Sweden). LDH is a marker of cell membrane injury that can be measured using a spectrophotometric evaluation of the LDH-mediated conversion of lactate to pyruvate. The detection limit of the instrument was 0.17 μKatal/L. Values below the detection limit were set at 0.16 μKatal/L.
Monocyte quantification
The adhesion of monocytes on smooth gold, nanostructured gold, TCP, and Thx was assessed after 18 hours and after the 1-hour stimulation period to compare the cell adhesion of unstimulated, zymosan-stimulated, and S. epidermidis-stimulated samples. Cells in the supernatant and on the plastic (TCP) below the materials in the well were also counted separately. The quantification of cells was performed using the Nucleocounter® system (ChemoMetec A/S, Allerød, Denmark). The samples were treated with lysis buffer and stabilization buffer. Lysed samples were loaded in a Nucleocassette™ precoated with fluorescent propidium iodide that stains the cell nuclei and were then quantified in the NucleoCounter®.
SEM
Adherent monocytes with and without the stimulation of zymosan or S. epidermidis on smooth gold, nanostructured gold, and Thx were fixated and prepared as described above. The surfaces were visualized using a mixture of secondary and backscattered electrons in a Zeiss 982 Gemini SEM operated at 3 kV.
Real-time reverse transcription polymerase chain reaction (qRT-PCR)
RNA was extracted from a total of 137 samples obtained from 4–6 different blood donors, resulting in 8–15 surface samples from each group. Cells on smooth and nanostructured gold, TCP, and Thx were lysed in 200 μL RLT lysis buffer (Qiagen, Hilden, Germany) and frozen at −80°C before total RNA extraction, using the NucleoSpin® RNA XS kit (Macherey-Nagel, Düren, Germany), as described in the manufacturer’s instructions. The RNA quality and concentration were determined for selected samples, using an Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA, USA) for pico profile and a nanospectrophotometer (Pearl; NanoPhotometer™, Implen GmbH, Munich, Germany).
Total RNA was converted to cDNA using a TATAA GrandScript cDNA Synthesis Kit (TATAA Biocenter AB, Gothenburg, Sweden) in 10 μL reactions. The samples were diluted 15×, and qRT-PCR analysis was performed in duplicate 10 μL reactions on the QuantStudio 12K Flex platform (Life Technologies), using a TATAA SYBR® GrandMaster Mix (TATAA Biocenter AB) and primers (final concentration, 400 nM) for 10 target genes and two reference genes. The genes of interest were IL-1β, IL-6, IL-10, TNF-α, integrin-β1 (CD29), integrin-β2 (CD18), integrin-αv (CD51), integrin-αm (CD11b), superoxide dismutase 2 (SOD2), and Nox2 (cytochrome b-245). Peptidylprolyl isomerase A and ribosomal protein large P0 were identified as the best reference genes in the TATAA reference gene panel on the basis of analysis in GenEx software (MultiD Analyses AB, Gothenburg, Sweden), using the NormFinder and the geNorm algorithms. Raw data were analyzed in QuantStudio 12K Flex Software 1.1.2 (Life Technologies) and processed in GenEx, using the relative comparative Cq method.
Cytokine determination
After the centrifugation of medium from cells cultured on smooth and nanostructured gold surfaces, TCP, and Thx surfaces, with or without zymosan or S. epidermidis, the supernatant was analyzed with respect to IL-1β, IL-6, IL-10, and TNF-α, using commercial enzyme-linked immunosorbent assay kits (Quantikine®, R&D Systems, Minneapolis, MN, USA), according to the manufacturer’s instructions. The detection limits are less than 1 pg/mL, 0.7 pg/mL, 3.9 pg/mL, and 5.5 pg/mL, respectively. The optical density was measured with a microplate reader and translated to cytokine levels, using the accompanying software.
Reactive oxygen species measured by luminol-mediated chemiluminescence
Monocytes/macrophages cultured on smooth and nanostructured gold surfaces, TCP, and Thx (n=2–3) were evaluated for their ability to produce reactive oxygen species (ROS) on stimulation with zymosan or S. epidermidis. The surfaces were transferred to white 24-well plates (Visiplate™ TC, PerkinElmer, Waltham, MA, USA) and HBSS++ (with Ca2+ and Mg2+) with zymosan or S. epidermidis stimulus added to a final volume of 1.96 mL before insertion into a microplate reader (37°C) equipped with luminescence optics and a 3 mm light guide for increased sensitivity (gain: 4,000). A kinetic program was run for 80–90 cycles (1 minute per cycle) in which light emission from each well was measured every minute. In the fifth cycle, 40 μL luminol was automatically injected (final concentration, 5 × 10−5 M; catalogue number 09253, Fluka/Sigma-Aldrich), followed by 5 seconds of shaking, for the detection of ROS formed in the samples. Nonstimulated samples were used as controls (n=2). On termination of the experiment, the cells were quantified. One well without seeded cells but with HBSS and luminol was used as a blank. The obtained data points were divided by the total cell number in each well, and the values were analyzed in MATLAB®, software version R2011a (MathWorks, Inc, Natick, MA, USA), where graphs were created using the Smoothing Spline method (smoothing parameter 0.001). The curve for each sample was then analyzed for total chemiluminescence (CL) produced (integral under the curve), peak CL, and time to peak CL for x-values between 3 and 80 min.
Focused ion beam SEM
Samples prepared for morphological observations of interactions between monocytes/macrophages and zymosan and S. epidermidis, respectively, were analyzed in a dual-beam FIB system (Strata DB 235; FEI) operated at 30.0 kV (Ga+ ions) and 5 kV (electrons). When cells were localized, a protective layer of platinum was sputtered over the cell surface, and cross-sections of the cells were exposed by FIB milling and then imaged by SEM.
Statistics
All results are presented as the mean ± standard deviation. The data were statistically evaluated by one-way analysis of variance, followed by Tukey’s post hoc test or an independent sample t-test (two-tailed), using 95% confidence intervals in PASW/Statistics 18.0 (IBM Corporation, Armonk, NY, USA) or SPSS version 21 (IBM Corporation). The evaluation of gene expression data was based on the logarithmic Cq values. Statistical differences in the graphs and tables are denoted by letters, where values/bars in the same table/graph that share the same letters are significantly different (P<0.05).
Results
Materials
Material characterization
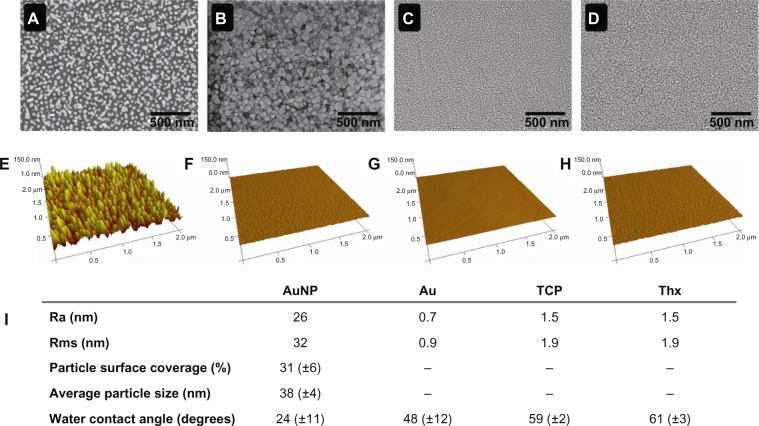
The physicochemical properties of the experimental surfaces are summarized in Figure 2. Surface roughness was increased on the smooth gold substrates from 0.9 to 32 nm (rms) by the immobilized gold nanoparticles. On TCP and Thx, the roughness was low, as expected (1.9 nm rms). Smooth gold, TCP, and Thx had water-contact angles in the same range, whereas nanostructured gold demonstrated a considerably more hydrophilic surface. The lower contact angle on nanostructured surfaces is in accordance with Wenzel’s theory of increased wetting on nanostructured hydrophilic surfaces with a given surface chemistry.23 Average particle size and surface coverage of the nanostructured gold surface were 38±4 nm and 31%±6%, respectively. On the AuND and AuNL surfaces, the average particle size was 39±2 nm, and the particle surface coverage was 28% and 17%, respectively.
Figure 2.
Material characterization of AuNP (A), Au (B), TCP (C), and Thx (D) with SEM. AFM characterization of AuNP (E), Au (F), TCP (G), and Thx (H). Physicochemical properties are summarized (I).
Abbreviations: AFM, atomic force microscopy; Au, smooth gold surface; AuNP, nanostructured gold surface; SEM, scanning electron microscopy; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip.
Bacterial adhesion and biofilm formation on nanotopographic versus smooth surfaces
Live and dead fluorescence microplate readings
After 24 hours of incubation, significantly more live bacteria (Figure 3) were detected on smooth gold surfaces compared with AuNL and AuND. No differences were seen in the amount of dead bacteria.
Figure 3.
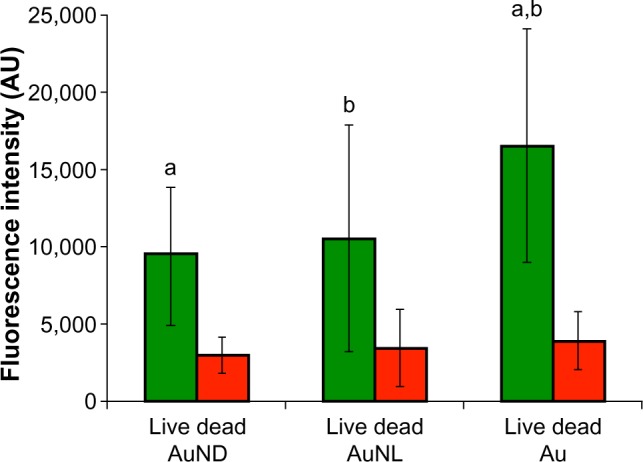
Live and dead fluorescence staining of adherent Staphylococcus epidermidis on AuNP and Au after 24 hours of static incubation in Roswell Park Memorial Institute medium.
Notes: Data represent the mean ± standard deviation (n=4). Significant differences between surfaces are indicated by letters a and b, where bars that share the same letters are significantly different (P<0.05).
Abbreviations: Au, smooth gold surface; AU, arbitrary unit; AuND, nanostructured gold surface (Nano Dense); AuNL, nanostructured gold surface (Nano Light).
Viable counts
Similar amounts of viable S. epidermidis adhered to both smooth and nanostructured gold surfaces when recovered after 2, 24, and 48 hours (Figure 4). The main increase in viable numbers occurred during the period from 2 to 24 hours, when the formation of the biofilm and accumulation phase commonly take place. From 24 to 48 hours, there was a small increase in viable numbers on both surfaces; this may be due to a lack of nutrients (no change of media) and/or dispersal events from the biofilm.
Figure 4.
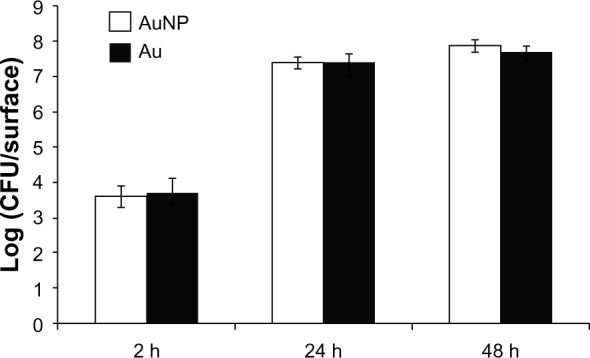
Viable counts of adherent Staphylococcus epidermidis on AuNP and Au after 2, 24, and 48 hours of static incubation in Roswell Park Memorial Institute medium.
Note: Data represent the mean ± standard deviation (n=2).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; CFU, colony-forming units; h, hours.
S. epidermidis morphology
SEM analysis of S. epidermidis on smooth and nanostructured gold surfaces revealed very few attached cells on both surfaces after 2 hours. After 24 and 48 hours, the amount of bacteria had increased considerably, and there was a general impression that bacterial cells on smooth gold had an earlier onset of biofilm formation compared with nanostructured gold. After 24 hours, the bacterial cells on smooth gold displayed intercellular slime connecting biofilm; this was observed on nanostructured gold only after 48 hours (Figure 5). In addition, more mature biofilms with higher tower formations were observed on smooth gold compared with nanostructured gold after 48 hours (Figure 5). Composite SEM images of the surfaces can be found in the supplementary material (Figure S1).
Figure 5.
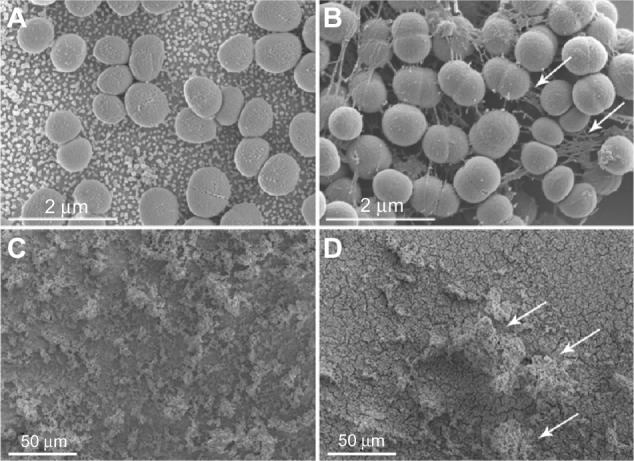
Staphylococcus epidermidis adhering to AuNP and Au after 24 and 48 hours, as visualized with SEM. After 24 hours, S. epidermidis on smooth gold surfaces (B) displayed more cell-connecting slime (some indicated by arrows) compared with nanostructured gold (A). A mature biofilm with high tower formations (arrows) was seen on smooth surfaces (D) after 48 hours, whereas the bacteria on nanostructured gold (C) were more horizontally scattered.
Abbreviations: AuNP, nanostructured gold surface; Au, smooth gold surface; SEM, scanning electron microscopy.
To further investigate the bacteria–surface interface, cross-sections of S. epidermidis on nanostructured gold and smooth gold were made by using the FIB slice and view (Figure 6). An animation of sequential cutting through the bacterial cell can be found in the supplementary material (Figure S3). A distinct difference in the area of contact between the bacterial cell wall and the surface could be seen. S. epidermidis displayed very few discrete attachment points on the nanostructured gold surface, whereas continuous attachment, and thus a higher contact area to the surface, was seen on the smooth gold surface.
Figure 6.
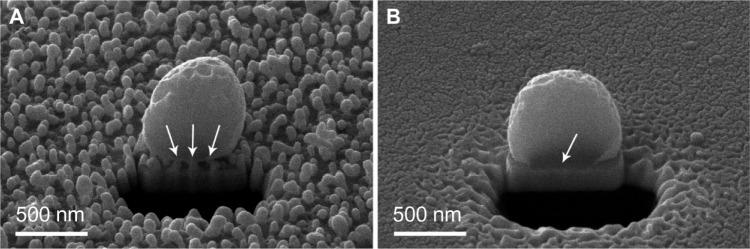
Bacteria–surface interface on AuNP and Au as visualized after ion milling in a FIB-SEM.
Notes: The attachment of Staphylococcus epidermidis to AuNP relies on a few discrete attachment points between the bacterial cell wall and the immobilized nanoparticles (arrows in A), whereas the cell wall is in continuous contact with the smooth surface (arrow in B).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; FIB-SEM, focused ion beam-scanning electron microscopy.
CLSM
Figure 7 shows the results of the analysis of the CLSM images taken from biofilms. After 2 hours of static adhesion of S. epidermidis to smooth and nanostructured gold surfaces, significantly more dead biomass was found on nanostructured surfaces compared with smooth ones. Furthermore, the area occupied by dead cells in close contact with the surface was also larger on nanostructured surfaces than on smooth surfaces. Moreover, after 24 hours of biofilm formation, the area occupied by dead cells on nanostructured surfaces was also significantly larger (Figure 8).
Figure 7.
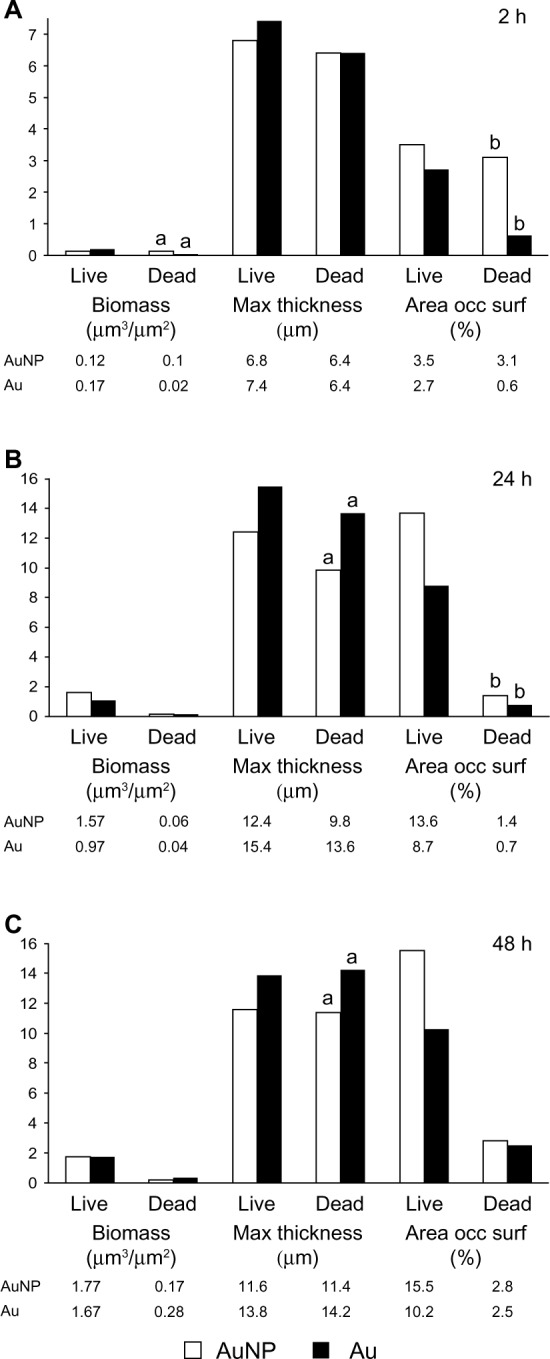
CLSM analysis of Staphylococcus epidermidis biofilms grown on Au and AuNP surfaces for 2 (A), 24 (B), and 48 (C) hours under static incubation in Roswell Park Memorial Institute medium. The total biomass, the maximum thickness of the biofilm, and the area occupied at the surface by live and dead cells were analyzed using COMSTAT2 software.
Notes: Data represent the mean (n=3). Significant differences between surfaces are indicated by letters a and b, where bars that share the same letters are significantly different (P<0.05).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; CLSM, confocal laser scanning microscopy; h, hours; occ surf, occupied surface.
Figure 8.
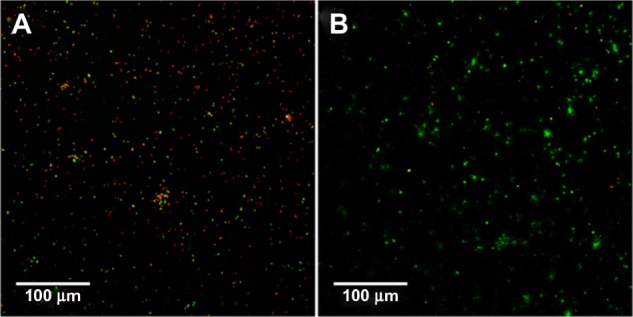
Live and dead fluorescence staining of Staphylococcus epidermidis at the surface interface (z=0–3 μm) on AuNP (A) and Au (B) surfaces after 24 hours of static incubation in Roswell Park Memorial Institute medium, as analyzed by CLSM.
Note: Green indicates live cells; red indicates dead cells.
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; CLSM, confocal laser scanning microscopy.
In overall terms, thicker biofilms of both live and dead cells formed on smooth surfaces compared with nanostructured surfaces after 24 and 48 hours.
Monocyte coculture experiments
Monocyte viability
Cell viability, as measured by LDH, was high on all materials, irrespective of zymosan or S. epidermidis stimulation, with average values between 0.2 and 0.4 μKatal/L (Table S1).
Monocyte adhesion
Significantly more cells attached to TCP compared with all other materials, whereas Thx had significantly fewer adherent cells compared with all other materials (Figure 9). Interestingly, on stimulation, the adhesion pattern differed between the materials: cell attachment increased on TCP and decreased on both smooth and nanostructured gold when compared with nonchallenged cells (Figure 9). Despite these differences in adherent cells, the number of supernatant cells increased with zymosan stimulation, and even more with S. epidermidis stimulation for all materials.
Figure 9.
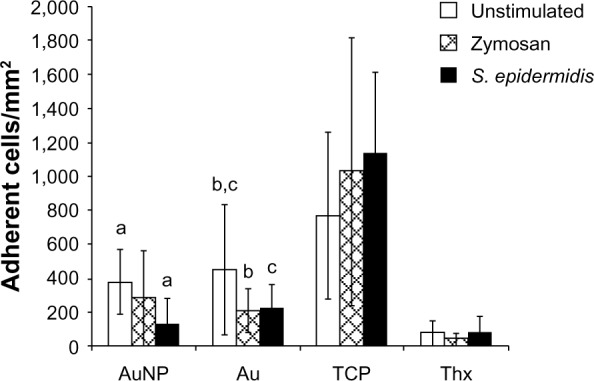
Number of adherent monocytes per mm2 on AuNP, Au, TCP, and Thx after 18 hours of primary adhesion followed by 24 hours of culture and a 1 hour challenge with either opsonized zymosan particles or opsonized Staphylococcus epidermidis. Unstimulated cells were used as controls.
Notes: Data represent the mean ± standard deviation (n=4–6). Significant differences between treatments are indicated by letters a–c, where bars that share the same letters are significantly different (P<0.05).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip.
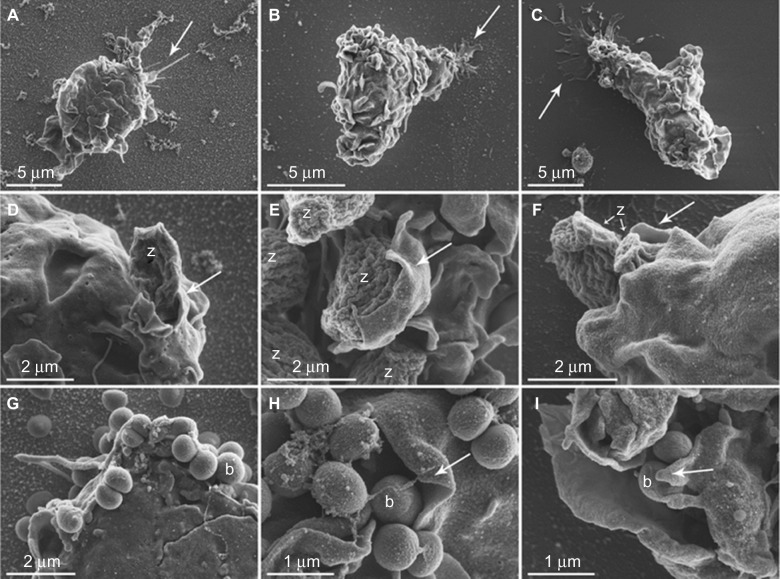
Adherent cells on smooth and nanostructured gold surfaces and Thx were viewed in SEM with and without the addition of stimuli (Figure 10). The cells had a large number of contacts with the different prey, sometimes with extending filopodia reaching out for their target. Many prey were also attached directly to the cell body membrane, and some of them were halfway toward being phagocytosed into the monocyte/macrophage. A general impression was that the cell surface attachment tended to be more filopodia-dependent on the nanostructured gold surfaces, whereas cells on the smooth gold and Thx surfaces had a much larger part of their plasma membrane in close contact with the surface. In addition, presumably dead cells (apoptotic bodies) were seen free on the surface and in the process of being phagocytosed by cells on all surfaces. The induction of apoptosis in monocytes/macrophages is normal in in vitro environments,24 and the process per se is crucial for the correct maintenance of the immune system.
Figure 10.
Monocyte morphology and interaction with opsonized zymosan particles or Staphylococcus epidermidis on AuNP, Au and Thx as visualized with SEM.
Notes: Monocytes on AuNP (A), Au (B), and Thx (C) surfaces in general demonstrated a rounded morphology with a moderate number of membrane ruffles/ridges with nonpreferential directions. The cell–material surface interaction was morphologically evident in areas with cytoplasmic extensions, in the form of either distinct, thin spikes of variable length (arrow in A) or thin, spread extensions (arrows in B and C). Cells with a polarized morphology were also observed (examples in B and C), suggestive of being in the process of attachment, detachment, or migration. Details of monocyte–zymosan particle interactions are observed morphologically in (D–F). Typically, oval-shaped zymosan particles with fine, undulating ridges were clearly distinguishable. Several zymosan (z) particles were enveloped by pseudopods (arrows). A large number of Staphylococcus epidermidis (0.5–1-μm round-shaped cocci) were detected in close association with the monocyte membrane (G–I). Multiple bacteria (some of which are denoted b) were clustered and enveloped by large, thin-walled pseudopods (arrows) in an apparent phagocytic process.
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; SEM, scanning electron microscopy; Thx, tissue culture-treated plastic cover slip.
Monocyte gene expression
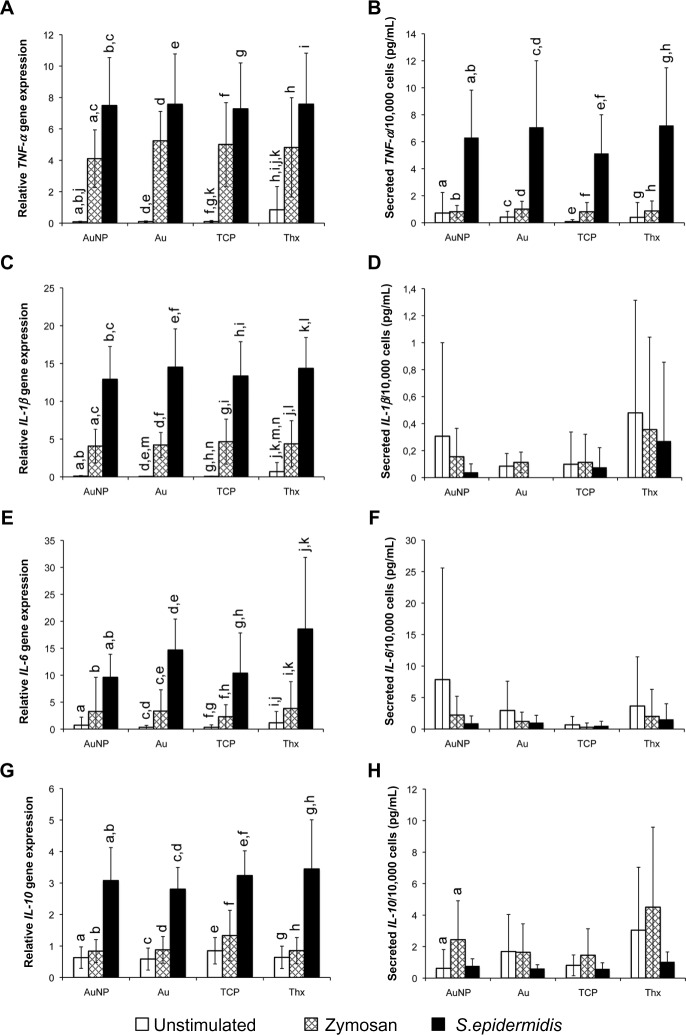
To explore whether the different material surfaces induced different gene expression, a panel of four cytokines was used (Figure 11). When comparing the different materials in unstimulated conditions, Thx showed a significantly higher expression of TNF-α and IL-1β genes. However, nanostructured gold did not induce a different response. After a 1 hour stimulation period with either zymosan or S. epidermidis, no differences in the gene expression were noted between any of the surfaces. Nevertheless, the stimulation provoked a significant increase in the proinflammatory cytokine genes TNF-α, IL-1β, and IL-6 on stimulation with both zymosan and S. epidermidis, with higher levels for cells stimulated with S. epidermidis on all materials. The anti-inflammatory IL-10 was significantly higher for S. epidermidis-stimulated samples on all materials, but not for zymosan.
Figure 11.
Gene expression (left) and cytokine secretion (right) of pro- and anti-inflammatory cytokines from monocytes on AuNP, Au, TCP, and Thx. The gene expression of TNF-α (A), IL-1β (C), IL-6 (E), and IL-10 (G) was analyzed from adherent cells after 1 hour of stimulation with either opsonized zymosan particles or opsonized Staphylococcus epidermidis. The secretion of TNF-α (B), IL-1β (D), IL-6 (F), and IL-10 (H) proteins is presented as the amount of cytokines per 10,000 total cells. Unstimulated cells were used as controls.
Notes: Data represent the mean ± standard deviation (n=4–6). Significant differences between stimulus or material surfaces are indicated by letters a–n, where bars that share the same letters are significantly different (P<0.05).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; IL, interleukin; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip; TNF-α; tumor necrosis factor-alpha.
The data relating to the gene expression of SOD2 and Nox2, which is relevant for cell oxidative metabolism, can be found in Figure 12. Unstimulated cells showed a significantly higher SOD2-expression on Thx. This difference did, however, level out on stimulation, when the expression was significantly induced, both for zymosan and even more for S. epidermidis. Nox2 gene expression, in contrast, was relatively stable for both smooth and nanostructured gold surfaces, whereas a significant decrease in expression was noted for TCP and Thx when stimulated with S. epidermidis.
Figure 12.
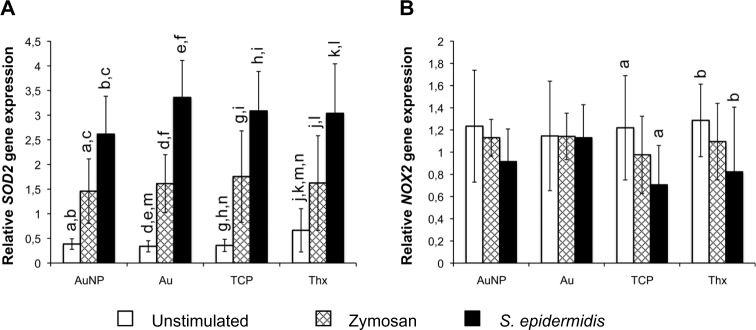
Gene expression of oxidative burst-related enzymes SOD2 (A) and Nox2 (B) in adherent monocytes on AuNP, Au, TCP, and Thx after 1 hour of stimulation with either opsonized zymosan particles or opsonized Staphylococcus epidermidis. Unstimulated cells were used as controls.
Notes: Data represent the mean ± standard deviation (n=4–6). Significant differences between stimulus or material surfaces are indicated by letters a–l, where bars that share the same letters are significantly different (P<0.05).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; SOD2, superoxide dismutase 2; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip.
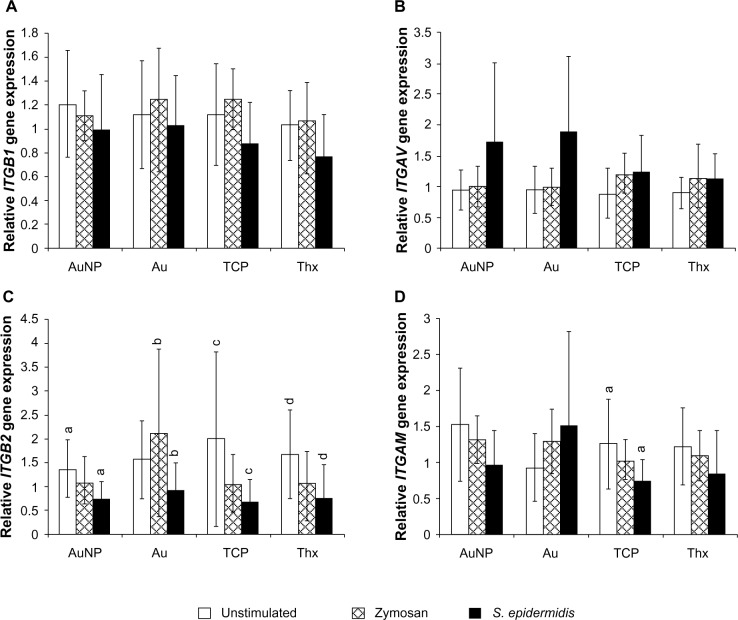
Four different cell adhesion markers were analyzed: integrin-β1, integrin-β2, integrin-αv, and integrin-αM. Heterodimers of integrin-β2 and integrin-αM constitute the complement receptor 3 (CR3), which is implicated in the phagocytosis of serum-opsonized microbes, for example. However, the gene expression data revealed a decrease in the expression of the CR3 subunits on stimulation, with significant differences between unstimulated and S. epidermidis-stimulated samples for integrin-β2 on all surfaces (Figure 13). No significant differences were found for integrin-β1 and integrin-αv.
Figure 13.
Gene expression of integrin-β1 (A), integrin-αv (B), integrin-β2 (C), and integrin-αM (D) in adherent monocytes on AuNP, Au, TCP, and Thx after 1 hour of stimulation with either opsonized zymosan particles or opsonized Staphylococcus epidermidis. Unstimulated cells were used as controls.
Notes: Data represent the mean ± standard deviation (n=4–6). Significant differences between stimulus or material surfaces are indicated by letters a–d, where bars that share the same letters are significantly different (P<0.05).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip; ITGB1, integrin-β1; ITGB2, integrin-β2; ITGAV, integrin-αv; ITGAM, integrin-αM.
Monocyte cytokine secretion
The cytokines detected in the supernatants of stimulated or unstimulated cells on different material surfaces are shown in Figure 11. Cells on all surfaces were induced to secrete significantly more TNF-α protein on S. epidermidis stimulation. Zymosan also induced a small TNF-α increase, albeit not one significantly different to unstimulated samples. A more pronounced production of IL-10 was noted for zymosan-stimulated samples on nanostructured gold surfaces. However, the amounts of IL-1β and IL-6 were similar for all surfaces and stimuli.
Monocyte oxidative response and phagocytosis
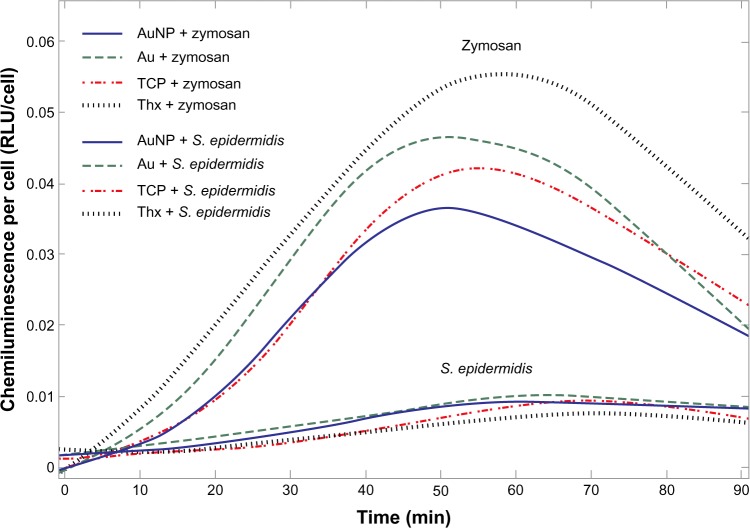
The stimulation of the cells with opsonized zymosan particles or opsonized S. epidermidis resulted in a significant increase in ROS production (Figure 14), whereas the CL production of the unstimulated cells was on the same level as the HBSS blanks. The amount of CL produced was shown to be linearly dependent on the number of cells, allowing for the use of CL per cell when comparing activity on different materials. Moreover, the opsonization of zymosan and S. epidermidis caused a higher oxidative response than the use of nonopsonized prey (data not shown). Interestingly, cells challenged with zymosan always produced a significantly higher response than cells challenged with S. epidermidis (both total ROS response and peak ROS; Figure 14 and Table 1). The amount of total ROS response per cell (integral under the curve) differed between materials and prey (Table 1). Cells on Thx had the highest activity when stimulated with zymosan, whereas cells on TCP had the highest activity when stimulated with S. epidermidis. The time to peak was not consistent between the different stimuli but was always faster for the two gold surfaces compared with the two plastic surfaces.
Figure 14.
Representative graph of monocyte ROS production on AuNP, Au, TCP, and Thx in response to opsonized zymosan particles and opsonized Staphylococcus epidermidis, as measured by CL.
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; CL, chemiluminescence; ROS, reactive oxygen species; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip; RLU, relative luminescence units.
Table 1.
Production of reactive oxygen species (as measured by chemiluminescence) in monocytes cultured on AuNP, Au, TCP, and Thx surfaces in response to opsonized zymosan particles or opsonized Staphylococcus epidermidis
| Zymosan
|
Staphylococcus epidermidis
|
|||||
|---|---|---|---|---|---|---|
| Total ROS (RLU/cell, 10−2) | Peak ROS (RLU/cell, 10−3) | Time to peak (min) | Total ROS (RLU/cell, 10−2) | Peak ROS (RLU/cell, 10−3) | Time to peak (min) | |
| AuNP | 205±59a | 44±14b | 56.7±6.2c | 49±24e | 10±4h | 60.2±7.7k |
| Au | 226±62 | 48±14 | 55.7±6.0d | 45±18f | 9±3i | 54.0±11.5l,m |
| TCP | 224±90 | 47±18 | 59.5±6.6 | 74±46e–g | 16±7h–j | 69.5±11.1k,l |
| Thx | 266±84a | 57±20b | 62.5±8.7c,d | 47±29g | 10±5j | 67.1±9.6m |
Notes: Data represent the mean ± standard deviation (n=4–5). Significant differences between material surfaces are indicated by letter superscripts a–m, where values that share the same letters are significantly different (P<0.05).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; ROS, reactive oxygen species; RLU, relative luminescence units; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip; min, minutes.
Increased ROS levels indicate interaction between the cells and zymosan/S. epidermidis, but to demonstrate the internalization of the prey, the FIB slice and view was employed. Although the cells did not always have visible contact with the prey on the cell surface, a cut through the cell revealed internalized particles and bacteria in the cell interior, irrespective of the surface that was used (Figure 15). S. epidermidis was clearly seen as small, compact, round cocci, whereas the zymosan was more structured and more difficult to discern. Unstimulated cells did not reveal similar structures in the cell interior.
Figure 15.

Phagocytosis of opsonized zymosan particles and opsonized Staphylococcus epidermidis by monocytes. The internalization of zymosan particles (B) and S. epidermidis (C) was evident on all surfaces, exemplified here by monocytes on AuNP, after 1 hour of stimulation. Unstimulated monocytes lacked the presence of internal prey (A). The cell surface was protected by a thin platinum film before ion milling the cell in a FIB-SEM.
Notes: Zymosan (B) and S. epidermidis (C), some of which are marked by arrows, are internalized by monocytes.
Abbreviations: AuNP, nanostructured gold surface; FIB-SEM, focused ion beam-scanning electron microscopy.
Discussion
There are at least two possible mechanisms by which material surfaces can reduce the incidence of medical device–related infections. First, the material surface may have direct effects on microorganisms. Second, the surface may have indirect effects on microorganisms by promoting antimicrobial host defense mechanisms. In an in vivo situation, both these components are likely to be present, as well as proteins and other cell types. In this study, we attempted to model these events in vitro by exploring the behavior of bacteria and monocytes/macrophages separately, as well as in combination. We have chosen both to evaluate one specific type of nanofeature, consisting of 35–40 nm sized gold particles immobilized on smooth gold surfaces, and to compare it with its smooth counterpart. Previous studies have shown that these surfaces have identical surface chemistry and that the nanostructured surface consists of solidly attached (partially sintered) structures,18,20 ensuring that any influence on the bacteria or monocytes derives solely from the changes in surface nanotopography.
Surface nanotopography influences bacterial adhesion and biofilm formation
The results of live/dead fluorescence plate readings demonstrated that the total number of live cells was significantly lower on the nanostructured gold surfaces (AuND and AuNL) compared with on the smooth gold surfaces after 24 hours of adhesion. This also correlated with the results obtained from CLSM, in which the live/dead cell biomass ratio was 9× higher at the substrate interface (2 hours) on smooth surfaces compared with nanostructured ones. This is in accordance with previous observations of reduced biofilm formation and bacterial adhesion on nanostructured substrates.17,25,26
For a given surface chemistry, the degree of hydrophilicity will be influenced by nanostructures. On a hydrophilic material, the presence of nanostructures will lead to increased hydrophilicity.23 This is exemplified by the lower water-contact angle for the nanostructured gold surfaces compared with the angle for the smooth ones. Hydrophobic bacteria, such as S. epidermidis, are known to detach more easily and form less biofilm on a hydrophilic surface.27–29 We therefore suggest that increased hydrophilicity, induced by nanotopography, is a factor contributing to the lower amounts of live bacteria seen on the nanostructured surfaces in our experiments. Moreover, a small decrease in live bacteria was found when increasing the density of nanoparticles from 17% to 28%. Nanotopography effects on bacterial adhesion have previously been shown for titanium substrates, where increased surface nanoroughness was shown to reduce bacterial adhesion.17
We do not exclude the possibility that effects apart from nanotopography may have influenced the differences in bacterial behavior on the smooth and nanostructured gold surfaces. In the in vivo environment, protein adsorption is a crucial step in both eukaryotic and prokaryotic cell attachment/adhesion.30,31 The effect of surface chemical properties such as hydrophilicity and surface charge, and of nanotopography on protein adsorption, is commonly acknowledged, and they are known determinants of the proteins that adsorb to a surface and in which manner.32–34 However, in this study, S. epidermidis was cultured in protein-free RPMI medium on the different experimental surfaces to minimize effects resulting from protein–surface interactions.
In overall terms, higher tower formations and thicker biofilms were found on the smooth surfaces than on nanostructured ones, as revealed by SEM and CLSM. In addition, the fact that a larger surface area was covered by both live and dead bacterial cells on nanostructured gold surfaces indicates that S. epidermidis spread more in the horizontal plane on nanostructured gold surfaces and in the vertical plane on smooth surfaces (thicker biofilms).
The initial adhesion events are important, as this is when the direct effects of the surface are more evident and restrictions from nutrients and oxygen are usually not an issue. The CLSM observation that a significantly larger area of the nanostructured gold surface interface was covered by dead cells suggests some type of bactericidal effect on direct contact with the gold nanotopography. This is supported by the fact that the initial adhesion (2 hours) of S. epidermidis also resulted in a significantly larger dead biomass (429% higher) at the nanosurface when compared with dead biomass on smooth surface. In contrast, on the smooth surface, there was a tendency toward more live biomass. To the best of our knowledge, this is the first report of a bactericidal effect induced solely by nanoparticles immobilized on a surface, as gold is not bactericidal per se.35 We consider one important factor to be the smaller contact area between the rigid cell wall of the bacteria and the surface, as verified by FIB slice and view, resulting in fewer sites available for adhesion receptor–ligand connections on nanostructured surfaces. This may in turn lead to a different microenvironment that is formed in the confined volume under the bacterial cells and in between the nanoparticles, resulting in, for example, accumulation of waste products, change of pH, and limited flow of nutrients because of the formation of a stagnant layer at the interface. Release of nanoparticles from the surface is highly unlikely to be the explanation for the lost bacterial viability because of the firm attachment to the substrate; in fact, the nanoparticles are partly sintered to the underlying gold substrate, as seen in Hulander et al.20 In addition, no visible signs of nanoparticle loss were seen in SEM when comparing the surfaces before and after culture. On the basis of these observations, one important question is whether nanoparticles immobilized on a surface directly interact with the bacterial cell wall and induce cell death. At present, no such assumption has been verified. The vertical dimension and aspect ratio of the present, immobilized particles may be too small to produce a mechanism of this kind.
Differential cell adhesion on gold versus plastic on microbial stimulation
Different adhesion patterns of the monocytes/macrophages were seen on smooth and nanostructured gold surfaces compared with TCP: stimulation with zymosan and S. epidermidis induced a loss of adherent cells on smooth and nanostructured gold surfaces, despite the fact that nanostructured gold presented a more hydrophilic substrate for cells. One hypothesis is that this is related to the gold chemistry and its effect on the subsequent protein adsorption that occurs on all surfaces before cell attachment. Albeit speculative, one explanation could be the use of different adhesion receptors on gold versus polystyrene (TCP). For example, some receptors are used for both adhesion and phagocytosis,36 and if these events coincide, this may result in detachment from the surface. Previous in vitro studies have demonstrated that integrin-β2 receptors play an important role in the adhesion to material surfaces, NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) transcription factor activation and IL-1β, IL-8, macrophage inflammatory protein (MIP)-1α, and MIP-1β expression.37–40 Moreover, in vivo studies have revealed a significantly higher expression of integrin-β2 in cells adherent to an implant surface with a combined micro- and nanoscale surface texture, rather than a relatively smooth surface.41 The present results show that the integrin-β2 gene expression in adherent cells decreased markedly after stimulation with zymosan and S. epidermidis, suggesting a relationship between reduced integrin expression and loss of adherence. In contrast, reduced integrin-β2 expression after stimulation with the different prey was also observed for the polymer substrates, suggesting that the interaction between gold and monocytes involves other integrins and attachment factors than for the polymer substrates. No firm evidence for this assumption was found after analyzing the gene expression of integrin-β1, integrin-αv, and integrin-αm in adherent cells. Further studies, using markers for additional integrins, along with blocking experiments with antibodies, would therefore be of interest.
Monocyte activation in response to different substrates and microbial stimulation
Next, we analyzed the activity of the cells by measuring the luminol-mediated chemiluminescence (detecting both intra- and extracellular ROS)42 and the gene and protein expression of selected pro- and anti-inflammatory cytokines. Polymorphonuclear cells and monocytes are among the first cell types that adhere to the surface of implants after insertion in vivo. The oxidative response and secretion of molecules are influenced by both the properties of the material and the component of their microenvironment.43–45 The early involvement of these cells plays an important role in the inflammatory events and the repair and/or regeneration of soft tissues and bone in association with the implant.46,47 In addition, when these implant–tissue interface cells encounter and interact with live bacteria, components of their cell walls, and other molecules, it is of paramount importance that an efficient immune response is elicited. One main observation in this study was that the amount of CL was dependent on both the type of material surface and the stimulus presented to the monocytes. After stimulation with zymosan and S. epidermidis, the total ROS production was higher in cells on polystyrene surfaces than on smooth and nanostructured gold surfaces. In contrast, both smooth and nanostructured gold surfaces caused a more rapid response in cells challenged with zymosan and S. epidermidis, indicating that the material surface properties influence both the speed and magnitude of the mounted oxidative response. As noted for cell adhesion, chemistry appears to play a more important role than nanotopography. Similarly, chemistry also played the most decisive role when examining TiO surfaces with 110/160 nm hemispherical protrusions with different surface densities/coverage, where most differences in monocyte/macrophage adhesion and cytokine production and secretion were noted between TiO and TCP, and structured versus smooth TiO did not differ.48
In the present study, zymosan promoted higher ROS production than S. epidermidis. This is in agreement with Wagner and Bryers, who showed that both opsonized zymosan and exogenous LPS were more potent CL inducers in human monocytes than live S. epidermidis.49 Serum-opsonized zymosan elicited a higher total CL in macrophages than opsonized S. epidermidis and Pseudomonas aeruginosa. Further, the opsonization of S. epidermidis caused a higher oxidative response than nonopsonized S. epidermidis, indicating that the activation of the cells via complement and Fc receptors plays a role. Moreover, the gene expression of both pro- and anti-inflammatory cytokines, as well as SOD, was significantly higher in S. epidermidis-stimulated samples, indicating that cells are more active when challenged with live opsonized bacteria than with opsonized zymosan.
Possible S. epidermidis molecular candidates for eliciting ROS production in monocytes stem from the work of Martínez-Martínez et al50 showing that S. epidermidis cell wall peptidoglycan is the crucial CL-inducing component in human polymorphonuclear cells. The mechanism of CL response induced by S. epidermidis may involve the toll-like receptor 2 in the activation of the NF-κB, as demonstrated for peptidoglycan and lipoteichoic acid from gram-positive bacteria.51
The live, opsonized S. epidermidis efficiently induced the gene expression of proinflammatory TNF-α, IL-1β, and IL-6 and anti-inflammatory IL-10. This effect, in contrast to that observed for ROS production, was higher than with opsonized zymosan. Because this effect was observed for cells on all types of material substrate, no specific role for chemistry and/or nanotopography was suggested. One exception was the Thx surface, which promoted a higher expression of proinflammatory cytokines in unstimulated cells than the other surfaces.
At protein level, our results demonstrate a significant increase in the secretion of TNF-α by cells stimulated by live, opsonized S. epidermidis. Again, no specific modulatory effect of substrate properties was noted. In contrast, the secretion of IL-1β and IL-6 did not increase after stimulation. However, the 1-hour stimulation period could be too short to detect the secretion of IL-1 and IL-6. Previously, the gram-negative LPS stimulation of human whole blood induced different time patterns for secreted TNF-α and IL-6 proteins, with a somewhat delayed gene expression and protein secretion for IL-6 in comparison with TNF-α.52 Likewise, the storage of precursor IL-1 intracellularly in human monocytes is followed by the continuous release of IL-1β, starting 2 hours after synthesis.53 Zymosan stimulation promoted significantly higher secreted levels of IL-10 in monocytes on nanostructured gold surfaces, suggesting that the nanofeatures had anti-inflammatory potential. Gold ions released from the surfaces could result in an anti-inflammatory reaction,54 but measurements of gold in the media (data not shown) indicated low yet equal levels in smooth and nanostructured gold samples, whereas elevated levels of IL-10 were only seen for nanostructured gold samples, indicating that the effect is caused by the nanofeatures, rather than gold ions leaking from the surface. Other studies have found a lower gene expression of IL-1β and TNF-α on nanomodified TiO surfaces (rms 4.8) compared with smooth surfaces, using a murine macrophage cell line,55 and less TNF-α protein as well as more rounded human monocytes on 20 nm pores compared with 200 nm pores,11 indicating that small surface nanofeatures may have a down-regulatory effect on monocytes/macrophages.
Taken together, the present data show that in unstimulated conditions, the different chemistries and topographies of the surfaces did not result in major differences in monocyte viability, surface-induced CL, gene expression, and the secretion of pro- and anti-inflammatory cytokines. In contrast, a role for both smooth and nanostructured gold substrates was detected in specific inflammatory events when exposed to phagocytic prey; that is, adhesion, rapidity of CL induction, and anti-inflammatory IL-10 secretion.
Conclusion
The difference in surface chemistry (gold vs polystyrene) appears to have a greater effect on the monocytes/macrophages than nanotopography when it comes to both adhesion and activity, when comparing unstimulated cells with zymosan-and S. epidermidis-stimulated cells. The response levels of the cells were more determined by the origin of the stimuli. However, the amount of live S. epidermidis was shown to decrease on nanostructured surfaces compared with smooth ones. In addition, direct contact by S. epidermidis with the nanostructured surfaces correlated with a loss in bacterial viability. We therefore suggest that the smaller contact area of the bacterial cell wall with the surface as well as the increase in surface hydrophilicity caused by the nanostructures are important factors for the reduced adhesion and biofilm formation on the nanostructured gold surfaces.
Supplementary materials
Monocyte characterization and purity
Flow cytometry was used to determine monocyte purity after isolation from buffy coats. Samples of 500,000 cells were centrifuged, and the cell pellet was dissolved in 50 μL human serum immunoglobulin G (IgG; Sigma-Aldrich, St Louis, MO, USA) for 15 minutes at 4°C to block unspecific binding. Antibodies or isotypic controls (phycoerythrin mouse anti-human CD14, fluorescein isothiocyanate mouse anti-human CD45, phycoerythrin mouse IgG1 κ isotype control and fluorescein isothiocyanate mouse IgG1 κ isotype control, all from BD Biosciences, San Jose, CA, USA; 10 μL) and buffer (1% serum in phosphate-buffered saline) were then added in a total volume of 50 μL to label the cells. After 30 minutes of incubation at 4°C, the cells were washed twice in buffer and then fixed in 2% paraformaldehyde for 30 minutes. After centrifugation, the cell pellet was dissolved in 1 mL buffer, and the samples were stored in the dark at 4°C until analyzed in a flow cytometer (BD FACSCalibur™; BD Biosciences). Unlabeled cells and isotypic controls were used as controls.
The instrument settings were made on all events to optimize cell separation and by gating on the monocyte population in a forward scatter-side scatter plot to adjust for autofluorescence and nonspecific binding (background fluorescence). On analysis, acquisition was stopped at 10,000 events in a gate surrounding the monocytic population in the forward scatter-side scatter plot. The number of CD14+CD45+ cells (monocyte population, red) and CD45+ cells (all blood cells, green) was quantified and the proportion of monocytes was calculated. Isotypic controls were run to ensure that no nonspecific binding of the antibodies occurred (Figure S1). The monocyte purity varied between 73.6% and 89.5% for different donors, averaging 85.4% (±4.7%).
Staphylococcus epidermidis adhesion and biofilm formation on nanostructured versus smooth gold surfaces analyzed by scanning electron microscopy
Scanning electron microscopy images were captured at 500 μm intervals in three lines running across the surface, generating a total of 45 images per surface, taken at 5,000× magnification. The images were stitched together to make a collage (Figure S2).
S. epidermidis–surface interaction visualized by slice and view in focused ion beam-scanning electron microscopy
A stack of slice and view images of the sequential cutting into the bacterial cell to visualize the adhesion mechanism on nanostructured versus smooth gold surface were merged into an animation (Figure S3), using Image J (ImageJ 1.45, National Institutes of Health, Bethesda, MD, USA).
Cell viability
Cell viability was high on all materials, regardless of stimuli (Table S1), as evaluated by lactate dehydrogenase measurements (low values indicate a small degree of cell injury).
Dot plot graphs showing the size and granularity (A) and fluorescence (B) of CD14-PE-labeled and CD45-FITC-labeled newly isolated blood cells. The histogram plots (C and D) illustrate the number of counts at different fluorescence intensities, confirming that the nonspecific labeling of the cells (isotypic controls shown in gray line) was avoided.
Abbreviations: PE, phycoerythrin; FITC, fluorescein isothiocyanate.
Multiple SEM images taken with 5000x magnification stitched together for an overview of Staphylococcus epidermidis growth and biofilm formation on nanostructured and smooth Au surfaces after 2, 24, and 48 hours of static incubation.
Note: For a better view of this figure please see: http://bildbank.gu.se/pincollection.jspx?collectionName={a8788011-1997-405f-ae6c-32916424851b}#1390903824525_0.
Abbreviations: Nano, nanostructured gold surface; SEM, scanning electron microscopy; smooth, smooth gold surface.
Table S1.
Cell viability measured by lactate dehydrogenase activity (μKatal/L)
| Unstimulated | Zymosan | S. epidermidis | |
|---|---|---|---|
| AuNP | 0.31±0.07 | 0.29±0.09 | 0.26±0.06 |
| Au | 0.27±0.06 | 0.34±0.17 | 0.26±0.07 |
| TCP | 0.40±0.10 | 0.44±0.15 | 0.34±0.05 |
| Thx | 0.26±0.08 | 0.32±0.15 | 0.22±0.05 |
Notes: Media controls on the different materials averaged between 0.18 and 0.20 μKatal/L. Data represent the mean ± standard deviation (n=4–6).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip; S. epidermidis, Staphylococcus epidermidis.
Acknowledgments
The authors would like to acknowledge the Center for Cellular Imaging at the Sahlgrenska Academy, University of Gothenburg, for the use of CLSM equipment and for the support of the staff, and the Electron Microscopy Unit, University of Gothenburg, where the SEM was performed. Financial support was obtained from the BIOMATCELL Vinn Excellence Center of Biomaterials and Cell Therapy, supported by VINNOVA and the Västra Götaland Region, the Swedish Research Council (grant K2012-52X-09495-25-3), the Hjalmar Svensson Foundation, IngaBritt and Arne Lundberg Foundation, and the Adlerbertska Foundation.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Darouiche RO. Treatment of infections associated with surgical implants. N Engl J Med. 2004;350(14):1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- 2.Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004;17(2):255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gristina AG. Implant failure and the immuno-incompetent fibro-inflammatory zone. Clin Orthop Relat Res. 1994;(298):106–118. [PubMed] [Google Scholar]
- 4.Campoccia D, Montanaro L, Arciola CR. The significance of infection related to orthopedic devices and issues of antibiotic resistance. Biomaterials. 2006;27(11):2331–2339. doi: 10.1016/j.biomaterials.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 5.Colon G, Ward BC, Webster TJ. Increased osteoblast and decreased Staphylococcus epidermidis functions on nanophase ZnO and TiO2. J Biomed Mater Res A. 2006;78(3):595–604. doi: 10.1002/jbm.a.30789. [DOI] [PubMed] [Google Scholar]
- 6.Dalby MJ, Giannaras D, Riehle MO, Gadegaard N, Affrossman S, Curtis AS. Rapid fibroblast adhesion to 27 nm high polymer demixed nano-topography. Biomaterials. 2004;25(1):77–83. doi: 10.1016/s0142-9612(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 7.Dalby MJ, McCloy D, Robertson M, et al. Osteoprogenitor response to semi-ordered and random nanotopographies. Biomaterials. 2006;27(15):2980–2987. doi: 10.1016/j.biomaterials.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Kubo K, Tsukimura N, Iwasa F, et al. Cellular behavior on TiO2 nanonodular structures in a micro-to-nanoscale hierarchy model. Biomaterials. 2009;30(29):5319–5329. doi: 10.1016/j.biomaterials.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 9.Puckett SD, Lee PP, Ciombor DM, Aaron RK, Webster TJ. Nanotextured titanium surfaces for enhancing skin growth on transcutaneous osseointegrated devices. Acta Biomater. 2010;6(6):2352–2362. doi: 10.1016/j.actbio.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Smith LL, Niziolek PJ, Haberstroh KM, Nauman EA, Webster TJ. Decreased fibroblast and increased osteoblast adhesion on nanostructured NaOH-etched PLGA scaffolds. Int J Nanomedicine. 2007;2(3):383–388. [PMC free article] [PubMed] [Google Scholar]
- 11.Ferraz N, Hong J, Santin M, Karlsson Ott M. Nanoporosity of alumina surfaces induces different patterns of activation in adhering monocytes/macrophages. Int J Biomater. 2010;2010:402715. doi: 10.1155/2010/402715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohiuddin M, Pan HA, Hung YC, Huang GS. Control of growth and inflammatory response of macrophages and foam cells with nanotopography. Nanoscale Res Lett. 2012;7(1):394. doi: 10.1186/1556-276X-7-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wójciak-Stothard B, Curtis A, Monaghan W, MacDonald K, Wilkinson C. Guidance and activation of murine macrophages by nanometric scale topography. Exp Cell Res. 1996;223(2):426–435. doi: 10.1006/excr.1996.0098. [DOI] [PubMed] [Google Scholar]
- 14.Anselme K, Davidson P, Popa AM, Giazzon M, Liley M, Ploux L. The interaction of cells and bacteria with surfaces structured at the nanometre scale. Acta Biomater. 2010;6(10):3824–3846. doi: 10.1016/j.actbio.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Mitik-Dineva N, Wang J, Truong VK, et al. Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus attachment patterns on glass surfaces with nanoscale roughness. Curr Microbiol. 2009;58(3):268–273. doi: 10.1007/s00284-008-9320-8. [DOI] [PubMed] [Google Scholar]
- 16.Mitik-Dineva N, Wang J, Truong VK, et al. Differences in colonisation of five marine bacteria on two types of glass surfaces. Biofouling. 2009;25(7):621–631. doi: 10.1080/08927010903012773. [DOI] [PubMed] [Google Scholar]
- 17.Puckett SD, Taylor E, Raimondo T, Webster TJ. The relationship between the nanostructure of titanium surfaces and bacterial attachment. Biomaterials. 2010;31(4):706–713. doi: 10.1016/j.biomaterials.2009.09.081. [DOI] [PubMed] [Google Scholar]
- 18.Hulander M, Lundgren A, Berglin M, Ohrlander M, Lausmaa J, Elwing H. Immune complement activation is attenuated by surface nanotopography. Int J Nanomedicine. 2011;6:2653–2666. doi: 10.2147/IJN.S24578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimling J, Maier M, Okenve B, Kotaidis V, Ballot H, Plech A. Turkevich method for gold nanoparticle synthesis revisited. J Phys Chem B. 2006;110(32):15700–15707. doi: 10.1021/jp061667w. [DOI] [PubMed] [Google Scholar]
- 20.Hulander M, Lundgren A, Faxälv L, et al. Gradients in surface nanotopography used to study platelet adhesion and activation. Colloids Surf B Biointerfaces. 2013;110:261–269. doi: 10.1016/j.colsurfb.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Lundgren AO, Björefors F, Olofsson LG, Elwing H. Self-arrangement among charge-stabilized gold nanoparticles on a dithiothreitol reactivated octanedithiol monolayer. Nano Lett. 2008;8(11):3989–3992. doi: 10.1021/nl802543g. [DOI] [PubMed] [Google Scholar]
- 22.Heydorn A, Nielsen AT, Hentzer M, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146(Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 23.Sheng YJ, Jiang S, Tsao HK. Effects of geometrical characteristics of surface roughness on droplet wetting. J Chem Phys. 2007;127(23):234704. doi: 10.1063/1.2804425. [DOI] [PubMed] [Google Scholar]
- 24.Kiener PA, Davis PM, Starling GC, et al. Differential induction of apoptosis by Fas-Fas ligand interactions in human monocytes and macrophages. J Exp Med. 1997;185(8):1511–1516. doi: 10.1084/jem.185.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ercan B, Taylor E, Alpaslan E, Webster TJ. Diameter of titanium nanotubes influences anti-bacterial efficacy. Nanotechnology. 2011;22(29):295102. doi: 10.1088/0957-4484/22/29/295102. [DOI] [PubMed] [Google Scholar]
- 26.Machado MC, Tarquinio KM, Webster TJ. Decreased Staphylococcus aureus biofilm formation on nanomodified endotracheal tubes: a dynamic airway model. Int J Nanomedicine. 2012;7:3741–3750. doi: 10.2147/IJN.S28191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An YH, Friedman RJ. Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J Biomed Mater Res. 1998;43(3):338–348. doi: 10.1002/(sici)1097-4636(199823)43:3<338::aid-jbm16>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 28.Boks NP, Norde W, van der Mei HC, Busscher HJ. Forces involved in bacterial adhesion to hydrophilic and hydrophobic surfaces. Microbiology. 2008;154(Pt 10):3122–3133. doi: 10.1099/mic.0.2008/018622-0. [DOI] [PubMed] [Google Scholar]
- 29.Braem A, Van Mellaert L, Mattheys T, et al. Staphylococcal biofilm growth on smooth and porous titanium coatings for biomedical applications. J Biomed Mater Res A. 2013 May 10; doi: 10.1002/jbm.a.34688. Epub. [DOI] [PubMed] [Google Scholar]
- 30.Nilsdotter-Augustinsson A, Claesson C, Lindgren PE, Lundqvist-Gustafsson H, Ohman L. Adherence of Staphylococcus epidermidis to extracellular matrix proteins and effects of fibrinogen-bound bacteria on oxidase activity and apoptosis in neutrophils. APMIS. 2005;113(5):361–373. doi: 10.1111/j.1600-0463.2005.apm_113508.x. [DOI] [PubMed] [Google Scholar]
- 31.Wilson CJ, Clegg RE, Leavesley DI, Pearcy MJ. Mediation of biomaterial-cell interactions by adsorbed proteins: a review. Tissue Eng. 2005;11(1–2):1–18. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 32.Roach P, Farrar D, Perry CC. Surface tailoring for controlled protein adsorption: effect of topography at the nanometer scale and chemistry. J Am Chem Soc. 2006;128(12):3939–3945. doi: 10.1021/ja056278e. [DOI] [PubMed] [Google Scholar]
- 33.Steiner G, Tunc S, Maitz M, Salzer R. Conformational changes during protein adsorption. FT-IR spectroscopic imaging of adsorbed fibrinogen layers. Anal Chem. 2007;79(4):1311–1316. doi: 10.1021/ac061341j. [DOI] [PubMed] [Google Scholar]
- 34.Xu LC, Siedlecki CA. Effects of surface wettability and contact time on protein adhesion to biomaterial surfaces. Biomaterials. 2007;28(22):3273–3283. doi: 10.1016/j.biomaterials.2007.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran N, Tran PA. Nanomaterial-based treatments for medical device-associated infections. Chemphyschem. 2012;13(10):2481–2494. doi: 10.1002/cphc.201200091. [DOI] [PubMed] [Google Scholar]
- 36.Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 37.Collie AM, Bota PC, Johns RE, Maier RV, Stayton PS. Differential monocyte/macrophage interleukin-1β production due to biomaterial topography requires the β2 integrin signaling pathway. J Biomed Mater Res A. 2011;96(1):162–169. doi: 10.1002/jbm.a.32963. [DOI] [PubMed] [Google Scholar]
- 38.Perez RL, Roman J. Fibrin enhances the expression of IL-1 beta by human peripheral blood mononuclear cells. Implications in pulmonary inflammation. J Immunol. 1995;154(4):1879–1887. [PubMed] [Google Scholar]
- 39.Rezzonico R, Chicheportiche R, Imbert V, Dayer JM. Engagement of CD11b and CD11c beta2 integrin by antibodies or soluble CD23 induces IL-1beta production on primary human monocytes through mitogen-activated protein kinase-dependent pathways. Blood. 2000;95(12):3868–3877. [PubMed] [Google Scholar]
- 40.Rezzonico R, Imbert V, Chicheportiche R, Dayer JM. Ligation of CD11b and CD11c beta(2) integrins by antibodies or soluble CD23 induces macrophage inflammatory protein 1alpha (MIP-1alpha) and MIP-1beta production in primary human monocytes through a pathway dependent on nuclear factor-kappaB. Blood. 2001;97(10):2932–2940. doi: 10.1182/blood.v97.10.2932. [DOI] [PubMed] [Google Scholar]
- 41.Omar O, Lennerås M, Svensson S, et al. Integrin and chemokine receptor gene expression in implant-adherent cells during early osseointegration. J Mater Sci Mater Med. 2010;21(3):969–980. doi: 10.1007/s10856-009-3915-x. [DOI] [PubMed] [Google Scholar]
- 42.Jancinová V, Drábiková K, Nosál R, Racková L, Májeková M, Holománová D. The combined luminol/isoluminol chemiluminescence method for differentiating between extracellular and intracellular oxidant production by neutrophils. Redox Rep. 2006;11(3):110–116. doi: 10.1179/135100006X116592. [DOI] [PubMed] [Google Scholar]
- 43.Lindblad M, Lestelius M, Johansson A, Tengvall P, Thomsen P. Cell and soft tissue interactions with methyl- and hydroxyl-terminated alkane thiols on gold surfaces. Biomaterials. 1997;18(15):1059–1068. doi: 10.1016/s0142-9612(97)00029-x. [DOI] [PubMed] [Google Scholar]
- 44.Suska F, Gretzer C, Esposito M, et al. In vivo cytokine secretion and NF-kappaB activation around titanium and copper implants. Biomaterials. 2005;26(5):519–527. doi: 10.1016/j.biomaterials.2004.02.066. [DOI] [PubMed] [Google Scholar]
- 45.Suska F, Svensson S, Johansson A, et al. In vivo evaluation of noble metal coatings. J Biomed Mater Res B Appl Biomater. 2010;92(1):86–94. doi: 10.1002/jbm.b.31492. [DOI] [PubMed] [Google Scholar]
- 46.Omar OM, Lennerås ME, Suska F, et al. The correlation between gene expression of proinflammatory markers and bone formation during osseointegration with titanium implants. Biomaterials. 2011;32(2):374–386. doi: 10.1016/j.biomaterials.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 47.Suska F, Emanuelsson L, Johansson A, Tengvall P, Thomsen P. Fibrous capsule formation around titanium and copper. J Biomed Mater Res A. 2008;85(4):888–896. doi: 10.1002/jbm.a.31575. [DOI] [PubMed] [Google Scholar]
- 48.Rice JM, Hunt JA, Gallagher JA, Hanarp P, Sutherland DS, Gold J. Quantitative assessment of the response of primary derived human osteoblasts and macrophages to a range of nanotopography surfaces in a single culture model in vitro. Biomaterials. 2003;24(26):4799–4818. doi: 10.1016/s0142-9612(03)00381-8. [DOI] [PubMed] [Google Scholar]
- 49.Wagner VE, Bryers JD. Poly(ethylene glycol)-polyacrylate copolymers modified to control adherent monocyte-macrophage physiology: interactions with attaching Staphylococcus epidermidis or Pseudomonas aeruginosa bacteria. J Biomed Mater Res A. 2004;69(1):79–90. doi: 10.1002/jbm.a.20115. [DOI] [PubMed] [Google Scholar]
- 50.Martínez-Martínez L, Timmerman CP, Fleer A, Verhoef J. Chemiluminescence of human polymorphonuclear leucocytes after stimulation with whole cells and cell-wall components of Staphylococcus epidermidis. J Med Microbiol. 1993;39(3):196–203. doi: 10.1099/00222615-39-3-196. [DOI] [PubMed] [Google Scholar]
- 51.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem. 1999;274(25):17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 52.DeForge LE, Remick DG. Kinetics of TNF, IL-6, and IL-8 gene expression in LPS-stimulated human whole blood. Biochem Biophys Res Commun. 1991;174(1):18–24. doi: 10.1016/0006-291x(91)90478-p. [DOI] [PubMed] [Google Scholar]
- 53.Hazuda DJ, Lee JC, Young PR. The kinetics of interleukin 1 secretion from activated monocytes. Differences between interleukin 1 alpha and interleukin 1 beta. J Biol Chem. 1988;263(17):8473–8479. [PubMed] [Google Scholar]
- 54.Eisler R. Chrysotherapy: a synoptic review. Inflamm Res. 2003;52(12):487–501. doi: 10.1007/s00011-003-1208-2. [DOI] [PubMed] [Google Scholar]
- 55.Lee S, Choi J, Shin S, et al. Analysis on migration and activation of live macrophages on transparent flat and nanostructured titanium. Acta Biomater. 2011;7(5):2337–2344. doi: 10.1016/j.actbio.2011.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dot plot graphs showing the size and granularity (A) and fluorescence (B) of CD14-PE-labeled and CD45-FITC-labeled newly isolated blood cells. The histogram plots (C and D) illustrate the number of counts at different fluorescence intensities, confirming that the nonspecific labeling of the cells (isotypic controls shown in gray line) was avoided.
Abbreviations: PE, phycoerythrin; FITC, fluorescein isothiocyanate.
Multiple SEM images taken with 5000x magnification stitched together for an overview of Staphylococcus epidermidis growth and biofilm formation on nanostructured and smooth Au surfaces after 2, 24, and 48 hours of static incubation.
Note: For a better view of this figure please see: http://bildbank.gu.se/pincollection.jspx?collectionName={a8788011-1997-405f-ae6c-32916424851b}#1390903824525_0.
Abbreviations: Nano, nanostructured gold surface; SEM, scanning electron microscopy; smooth, smooth gold surface.
Table S1.
Cell viability measured by lactate dehydrogenase activity (μKatal/L)
| Unstimulated | Zymosan | S. epidermidis | |
|---|---|---|---|
| AuNP | 0.31±0.07 | 0.29±0.09 | 0.26±0.06 |
| Au | 0.27±0.06 | 0.34±0.17 | 0.26±0.07 |
| TCP | 0.40±0.10 | 0.44±0.15 | 0.34±0.05 |
| Thx | 0.26±0.08 | 0.32±0.15 | 0.22±0.05 |
Notes: Media controls on the different materials averaged between 0.18 and 0.20 μKatal/L. Data represent the mean ± standard deviation (n=4–6).
Abbreviations: Au, smooth gold surface; AuNP, nanostructured gold surface; TCP, tissue culture-treated polystyrene; Thx, tissue culture-treated plastic cover slip; S. epidermidis, Staphylococcus epidermidis.