Abstract
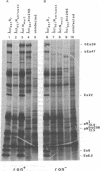
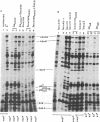
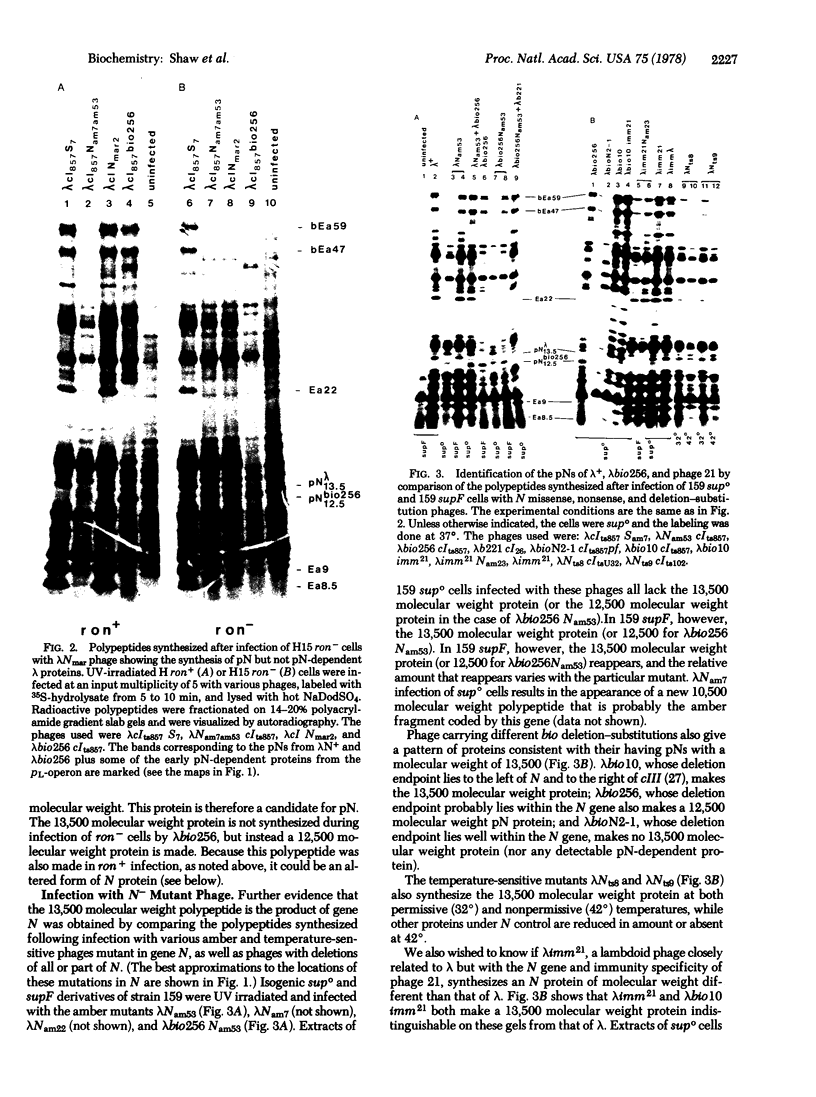
The N gene protein, pN, of bacteriophage λ stimulates early gene transcription by allowing mRNA chain elongation to proceed into genes distal to transcription termination sites normally recognized by the Escherichia coli transcription termination protein ρ. pN has previously eluded detection on sodium dodecyl sulfate/polyacrylamide gels because of its small size, its instability, and the difficulty of distinguishing pN itself both from host proteins and from other early λ proteins whose synthesis depends on pN action. These problems have now been overcome and we find that the major form of pN present in crude cell extracts of infected cells has an apparent molecular weight of 13,500. λbio256, a deletion-substitution mutant terminating in N, codes for a shorter pN of molecular weight 12,500. A nonsense fragment of 10,500 molecular weight coded by λNam7 has also been identified. These conclusions are based on examination of the electrophoretic profiles of the proteins synthesized after infection of UV-irradiated E. coli by various λN- temperature-sensitive, nonsense, and deletion-substitution mutants. It has also been possible to distinguish pN itself from other early λ polypeptides by infecting ron- cells with either λNmar phage allowing pN synthesis but not pN action or λNam phage defective in pN synthesis and pN action.
Our results together with previous data are discussed with respect to the possible existence of multiple molecular weight forms of pN and the location of the coding sequences in the N gene region.
Keywords: transcription antitermination protein, polyacrylamide gel electrophoresis, phage and host mutants
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M., De Crombrugghe B. Release of polarity in Escherichia coli by gene N of phage lambda: termination and antitermination of transcription. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2534–2538. doi: 10.1073/pnas.71.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROWN A., ARBER W. TEMPERATURE-SENSITIVE MUTANTS OF COLIPHAGE LAMBDA. Virology. 1964 Oct;24:237–239. doi: 10.1016/0042-6822(64)90114-x. [DOI] [PubMed] [Google Scholar]
- Brunel F., Davison J. Bacterial mutants able to partly suppress the effect of N mutations in bacteriophage lambda. Mol Gen Genet. 1975;136(2):167–180. doi: 10.1007/BF00272037. [DOI] [PubMed] [Google Scholar]
- CAMPBELL A. Sensitive mutants of bacteriophage lambda. Virology. 1961 May;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- Crawford L. V., Gesteland R. F. Synthesis of polyoma proteins in vitro. J Mol Biol. 1973 Mar 15;74(4):627–634. doi: 10.1016/0022-2836(73)90053-3. [DOI] [PubMed] [Google Scholar]
- Dahlberg J. E., Blattner F. R. Sequence of the promoter-operator proximal region of the major leftward RNA of bacteriophage lambda. Nucleic Acids Res. 1975 Sep;2(9):1441–1458. doi: 10.1093/nar/2.9.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen H. A., Fuerst C. R., Siminovitch L., Thomas R., Lambert L., Pereira da Silva L., Jacob F. Genetics and physiology of defective lysogeny in K12 (lambda): studies of early mutants. Virology. 1966 Oct;30(2):224–241. doi: 10.1016/0042-6822(66)90098-5. [DOI] [PubMed] [Google Scholar]
- Franklin N. C. Altered reading of genetic signals fused to the N operon of bacteriophage lambda: genetic evidence for modification of polymerase by the protein product of the N gene. J Mol Biol. 1974 Oct 15;89(1):33–48. doi: 10.1016/0022-2836(74)90161-2. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Baron L. S. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974 Mar;58(1):141–148. doi: 10.1016/0042-6822(74)90149-4. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Baumann M., Baron L. S. Cooperative effects of bacterial mutations affecting lambda N gene expression. I. Isolation and characterization of a nusB mutant. Virology. 1976 Aug;73(1):119–127. doi: 10.1016/0042-6822(76)90066-0. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Wilgus G. S., Mural R. J. Gene N regulator function of phage lambda immun21: evidence that a site of N action differs from a site of N recognition. J Mol Biol. 1973 Dec 25;81(4):505–516. doi: 10.1016/0022-2836(73)90519-6. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2977–2981. doi: 10.1073/pnas.68.12.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A., Pironio M. Relationship between the N function of bacteriophage lambda and host RNA polymerase. J Mol Biol. 1972 Mar 28;65(2):259–272. doi: 10.1016/0022-2836(72)90281-1. [DOI] [PubMed] [Google Scholar]
- Greenblatt J. Positive control of endolysin synthesis in vitro by the gene N protein of phage lambda. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3606–3610. doi: 10.1073/pnas.69.12.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J. Regulation of the expression of the N gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1973 Feb;70(2):421–424. doi: 10.1073/pnas.70.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix R. W., Casjens S. R. Protein fusion: a novel reaction in bacteriophage lambda head assembly. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1451–1455. doi: 10.1073/pnas.71.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Control of gene expression in bacteriophage lambda. Annu Rev Genet. 1973;7:289–324. doi: 10.1146/annurev.ge.07.120173.001445. [DOI] [PubMed] [Google Scholar]
- Inoko H., Imai M. Isolation and genetic characterization of the nitA mutants of Escherichia coli affecting the termination factor rho. Mol Gen Genet. 1976 Jan 16;143(2):211–221. doi: 10.1007/BF00266924. [DOI] [PubMed] [Google Scholar]
- Kayajanian G. Deletion mapping of the c3-c2 region of bacteriophage lambda. Virology. 1970 May;41(1):170–174. doi: 10.1016/0042-6822(70)90065-6. [DOI] [PubMed] [Google Scholar]
- Keppel F., Georgopoulos C. P., Eisen H. Host interference with expression of the lambda N gene product. Biochimie. 1974;56(11-12):1505–1509. doi: 10.1016/s0300-9084(75)80273-2. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Yanofsky C. Polarity suppressors defective in transcription termination at the attenuator of the tryptophan operon of Escherichia coli have altered rho factor. J Mol Biol. 1976 Sep 15;106(2):231–241. doi: 10.1016/0022-2836(76)90082-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liedke-Kulke M., Kaiser A. D. The c-region of coliphage 21. Virology. 1967 Jul;32(3):475–481. doi: 10.1016/0042-6822(67)90299-1. [DOI] [PubMed] [Google Scholar]
- Lozeron H. A., Anevski P. J., Apirion D. Antitermination and absence of processing of the leftward transcript of coliphage lambda in the RNAase III-deficient host. J Mol Biol. 1977 Jan 15;109(2):359–365. doi: 10.1016/s0022-2836(77)80039-9. [DOI] [PubMed] [Google Scholar]
- Oppenheim A. B., Katzir N., Oppenheim A. Regulation of protein synthesis in bacteriophage lambda. Restoration of gene expression in lambda N-strains by mutations in the cro gene. Virology. 1977 Jun 15;79(2):405–425. doi: 10.1016/0042-6822(77)90367-1. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B., Sauer R. T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976 Oct 8;194(4261):156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Ptashne M. ISOLATION OF THE lambda PHAGE REPRESSOR. Proc Natl Acad Sci U S A. 1967 Feb;57(2):306–313. doi: 10.1073/pnas.57.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P., Murialdo H. The role of gene Nu3 in bacteriophage lambda head morphogenesis. Virology. 1975 Mar;64(1):247–263. doi: 10.1016/0042-6822(75)90096-3. [DOI] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W., Mount D. W. Inactivation and proteolytic cleavage of phage lambda repressor in vitro in an ATP-dependent reaction. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2283–2287. doi: 10.1073/pnas.74.6.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W. Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc Natl Acad Sci U S A. 1975 Jan;72(1):147–151. doi: 10.1073/pnas.72.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- SUSSMAN R., JACOB F. [On a thermosensitive repression system in the Escherichia coli lambda bacteriophage]. C R Hebd Seances Acad Sci. 1962 Feb 19;254:1517–1519. [PubMed] [Google Scholar]
- Schwartz M. On the function of the N cistron in phage lambda. Virology. 1970 Jan;40(1):23–33. doi: 10.1016/0042-6822(70)90375-2. [DOI] [PubMed] [Google Scholar]
- Signer E. R., Manly K. F., Brunstetter M. A. Deletion mapping of the c-3-N region of bacteriophage. Virology. 1969 Sep;39(1):137–141. doi: 10.1016/0042-6822(69)90356-0. [DOI] [PubMed] [Google Scholar]
- Szybalski E. H., Szybalski W. Physical mapping of the att-N region of coliphage lambda: apparent oversaturation of coding capacity in the gam-ral segment. Biochimie. 1974;56(11-12):1497–1503. doi: 10.1016/s0300-9084(75)80272-0. [DOI] [PubMed] [Google Scholar]