Abstract
Data sharing is essential for the conduct of cutting-edge research and is increasingly required by funders concerned with maximising the scientific yield from research data collections. International research consortia are encouraged to share data intra-consortia, inter-consortia and with the wider scientific community. Little is reported regarding the factors that hinder or facilitate data sharing in these different situations. This paper provides results from a survey conducted in the European Network for Genetic and Genomic Epidemiology (ENGAGE) that collected information from its participating institutions about their data-sharing experiences. The questionnaire queried about potential hurdles to data sharing, concerns about data sharing, lessons learned and recommendations for future collaborations. Overall, the survey results reveal that data sharing functioned well in ENGAGE and highlight areas that posed the most frequent hurdles for data sharing. Further challenges arise for international data sharing beyond the consortium. These challenges are described and steps to help address these are outlined.
Keywords: biobanks, data sharing, consortia, genetic research
INTRODUCTION
The data-sharing landscape has changed considerably in the last decade due to several factors. First, data sharing is essential to study complex disease aetiology. This has catalysed the formation of international research consortia, each of which must tackle data-sharing issues. The more recent practice of inter-consortia collaborations adds yet another layer of complexity to data-sharing solutions and protocols. Second, data sharing is increasingly encouraged by the scientific community and by research funders1, 2, 3, 4, 5 in order to maximise the scientific returns from the data. Third, data sharing raises ethical and legal issues related to the privacy of research participants, which were often not foreseen when cohorts were established. Today, a typical consortium project in genomic sciences must develop solutions for data sharing that span intra-consortium, inter-consortium and sharing with scientific community at large. How easy is it for researchers and projects to fulfil the data-sharing requirements of today's science? Although the literature has emerged concerning the ethical and legal considerations surrounding data sharing,6, 7, 8, 9, 10 there is relatively little guidance for scientists on steps that could be undertaken to facilitate data sharing in consortia projects that need to negotiate this landscape.11, 12
This paper reports on the data-sharing experiences of the European Network for Genetic and Genomic Epidemiology (ENGAGE).13 The information is based on survey responses from ENGAGE partners who answered questions about situations encountered and provided recommendations to improve data sharing. We describe these results in the context of the data-sharing principles developed within ENGAGE and articulate other data-sharing challenges and solutions of the consortium, including data sharing post ENGAGE.
ENGAGE was established in 2008 with the main objective to share and analyse the wealth of data from a number of already-established cohort data sets.13 At project start, the consortium, funded by the 7th Framework Programme-Health Theme of the European Commission, comprised data from more than 80 000 genome-wide association scans and DNA and serum/plasma samples from over 600 000 individuals. During its 5-year duration (2008–2012), the 24 research organisations participating in ENGAGE have shared and analysed primarily Genome-wide association studies (GWAS) data to identify hundreds of genetic loci influencing dozens of medically-significant traits, ranging from type 2 diabetes and obesity, to smoking behaviour and birth weight. These discoveries resulted in approximately 170 publications as per August 2012, many of these in high-impact international journals, and more papers are under preparation.13
The ENGAGE data-sharing policy
An ENGAGE data-sharing policy was established early in the project by the Ethics core of the consortia in cooperation with ENGAGE members and the ENGAGE leadership.14 This policy was designed to fulfil three main objectives: (1) facilitate data sharing within the consortium, (2) facilitate data sharing between ENGAGE and other research consortia and (3) make the ENGAGE data widely available to the scientific community as required by the funders of the consortium. Practical tools were developed to support data sharing including data submission systems, sample availability systems and standard data access agreements (DAA) for the sharing of individual-level data.14, 15 Data-access catalogues describing the ENGAGE cohorts, data and specimens that could be made available to the wider scientific community were published on the ENGAGE website and in P3G's network catalogues.16, 17 ENGAGE partners were encouraged to deposit data produced by the consortium in the European Genome-Phenome Archive (EGA),18 a repository of genotype and phenotype data hosted by the European Bioinformatics Institute.19 The establishment of non-exclusive licensing protocols when research discoveries were made was encouraged to favour further uses by the wider scientific community. Although ENGAGE strongly endorsed principles of rapid data release to the scientific community, it had to take into consideration the specific requirements of each ENGAGE cohort in terms of, e.g., compliance with original consent, conformity with each cohort's confidentiality obligations and legal, ethical and security norms. In ENGAGE, data were shared only according to the rights and conditions for use determined by each data-generating partner.
Methods
An electronically-based questionnaire comprising 10 multiple choice questions and three open-ended questions was developed to collect information from data providers within the consortium about their data-sharing experiences and their recommendations for future collaborations. The questions applied to any type of ENGAGE collaborations, i.e., collaborations within ENGAGE or collaborations between ENGAGE and other research consortia, and covered the following areas: (1) technical, legal, ethical, administrative and financial hurdles encountered when sharing data; (2) concerns related to data privacy, confidentiality and use; (3) reasons for non-participation in ENGAGE studies; (4) usefulness of the ENGAGE data-sharing policy; (5) ease of collaboration in ENGAGE; (6) factors facilitating data sharing in ENGAGE; and (7) recommendations for improving data sharing in other consortia on the basis of the experiences from ENGAGE. Using a Likert-type scale, the survey participants were asked whether hurdles and concerns had been encountered: (a) never; (b) rarely; (c) a few times; or (d) many times. The questionnaire was sent in August 2012 to all ENGAGE principal investigators at each of the 24 ENGAGE partner institutions asking them to allocate at least one collaborator at their institution to fill in the questionnaire on behalf of their institution. To increase the chance of having all ENGAGE partner institutions represented, the questionnaire was in addition sent to all ENGAGE scientists as listed in the ENGAGE distribution list (215 subscribers). Two reminders were sent and the deadline for responding was extended once. The results were collected anonymously and with no indication of the respondent's affiliation.
RESULTS
Questionnaire results
In mid-September 2012, 26 replies had been collected. The survey participants reported to be primarily principal investigators, PostDoc researchers, senior researchers and PhD students who had a role of data analyst in the consortium.
Hurdles, concerns and reasons for non-participation in collaborations
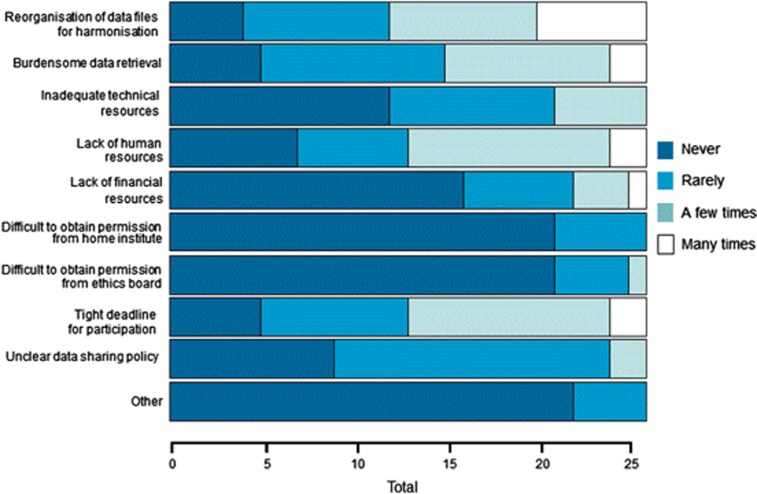
Overall, collaboration in ENGAGE was experienced as good. Seventy-three percent (n=19) of survey participants reported that they had encountered no difficulties when collaborating with other ENGAGE partners. However, 96% reported to have encountered at least one hurdle to data sharing while participating in ENGAGE and on average five hurdles were reported per respondent, although the frequency of these hurdles was generally low as can be seen in Figure 1 (hurdles). Most of the hurdles were either of technical nature (eg, lack of harmonisation of data sets) or organisational nature (eg, tight deadlines, burdensome procedures for data retrieval, lack of human resources). In comparison, the least-reported hurdles were related to obtaining permission from the one institution and the scientist's ethics board to participate in the collaboration, a result that confirms our previous investigation of the ENGAGE cohorts' ability to share data in ENGAGE.20
Figure 1.
Hurdles encountered when sharing data in ENGAGE.
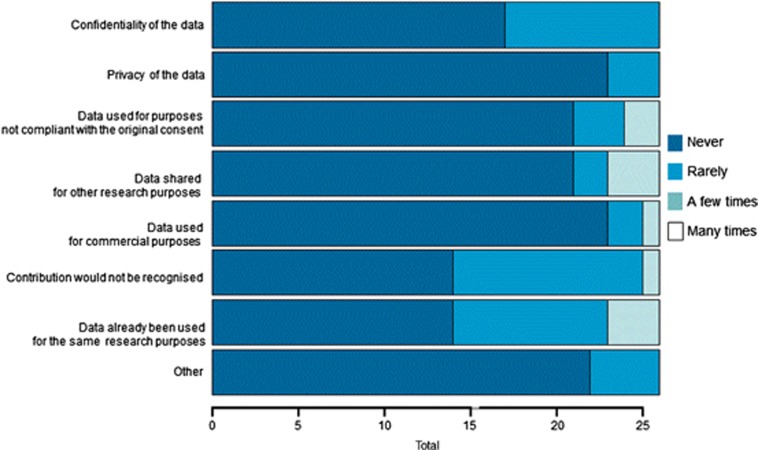
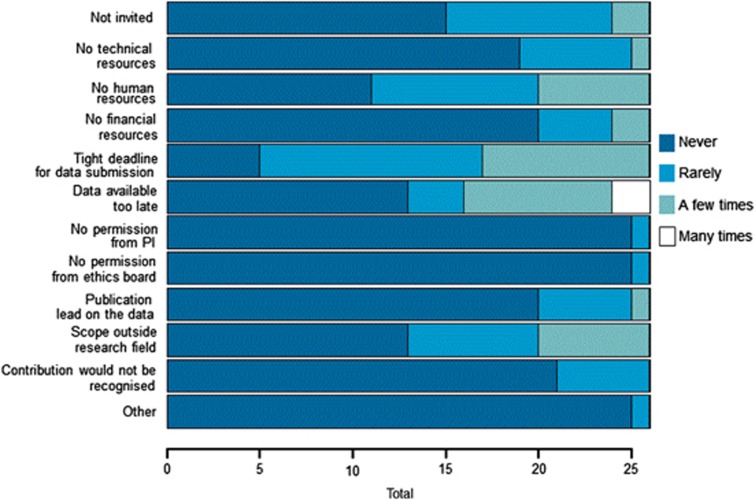
The number of concerns reported by survey participants ranged from 3–12, whereas seven participants responded that they had never encountered any concerns. Again, the frequency of these concerns was low as can be seen in Figure 2 (concerns). The most common concerns were that the data being shared might have already been used for other research purposes unknown to the survey participant, that the contribution from the survey participant may not have been recognised at the time of publication and that the confidentiality of the data may not have been protected well enough. Unfortunately, the questionnaire does not provide information as to whether the latter concern was related to the sharing of individual-level data, which was not very common in ENGAGE, or to the sharing of summary-level data. In comparison, the least-reported concerns were related to privacy (eg, risks of re-identification of the data) and the potential use of the data for commercial purposes. When asked whether there were ENGAGE collaborations for which their institution could have contributed data but did not, 11.5% of survey participants reported encountering this problem. There were four main reasons why they did not contribute data: the deadlines were too tight, the data that could have been used became available too late for use in ENGAGE, there were no human resources available to submit the data and the project was not within the scope of the participant's own research (see Figure 3). Again, obtaining permissions from the home institution and from the ethics board to participate in the collaboration was not an issue.
Figure 2.
Concerns encountered when sharing data in ENGAGE.
Figure 3.
Reasons for nonparticipation in an ENGAGE collaboration.
Usefulness and application of the ENGAGE data sharing policy
ENGAGE partners are encouraged to use Data Access Agreements (DAA) when sharing individual-level data in the consortium. Fifty-four percent (n=14) of study respondents reported that they did not share individual data and therefore did not use any DAA. Twenty-three percent (n=6) did not know whether DAAs had been established or not and only 14% (n=4) could report that they had systematically established DAAs. Similarly, only 19% (n=5) of study participants reported that they had deposited data (either, genotypic, phenotypic or omics) in the EGA and 38% (n=10) did not know whether data had been deposited or not. Forty-two percent (n=11) reported that they did not deposit data and some of the reasons evoked are that the data could not be deposited due to (1) participation in another consortium, (2) because it was not legally possible, (3) because it was seen as unnecessary and time-consuming and (4) because there were no incentives to deposit such data. Finally, 81% (n=21) of the survey participants could report that they did not implement licensing policies when sharing data in ENGAGE collaborations.
Factors facilitating data sharing in ENGAGE and recommendations
Factors that facilitated data sharing in ENGAGE included good collaboration (77%), good technical solutions (38%) and clarity of the data-sharing policy (31%). Data-sharing experiences in ENGAGE were largely seen as comparable to experiences from other consortia (61.5%). Half of the respondents provided suggestions for recommendations to facilitate data sharing in future collaborations. These are primarily related to the development of organisational, technical and governance tools as summarised in Table 1. Important points are the need for: (1) good information flow, (2) transparent publication policies and mechanisms for author recognition, (3) harmonised data-sharing policies across countries and funders, (4) simplified procedures for data sharing and (5) the development of good technical tools that provide easy access to the data.
Table 1. Recommendations for future research collaborations involving data sharing.
| Organisational tools |
| Provide systematic information about collaboration projects (plans, deadlines, reminders) |
| Implement transparent publication policies/mechanisms for author recognition |
| Governance tools |
| Use harmonised/unified data-sharing policies across countries and funders |
| Implement simplified Material Transfer Agreements (MTAs) and/or Data Access Agreements (DAAs) |
| Simplify ethics approval |
| Delegate harmonising issues to people in charge of developing policies |
| Technical tools |
| Develop systems for systematic information about data availability (without access to the data) and multiple files upload |
| Further develop tools for data harmonisation across cohorts |
| Use more efficient web servers |
Discussion
Several lessons can be learned from the ENGAGE data-sharing experience. First, the ENGAGE experience confirms that good collaboration is a key element to successful data sharing as research groups who enjoy working together are more inclined to share data.21 Second, results from our survey illustrate that hurdles to data sharing that are encountered in large research consortia such as ENGAGE are often primarily related to technical and organisational issues for which solutions can be developed, and such solutions are essential to facilitate data sharing in a consortium.22 Third, in agreement with reports from other consortia,10, 22, 23 our results highlight that bottlenecks in data sharing occur due to the need to harmonise data. Harmonisation initiatives have been set up to facilitate data harmonisation and should be developed further.24, 25, 26 Fourth, the lack of coordinated rules for data sharing across consortia creates hurdles to data sharing. Even when a consortium develops a comprehensive data-sharing policy, it usually applies to data sharing within the consortium and will have limited applicability in cross-consortia collaborations. This was the case for ENGAGE where most research was based on inter-project collaborations. Because of the complexities of regulating and structuring data sharing across consortia, it is difficult to develop a straightforward data-sharing policy. The degree to which this hampers or slows down research is difficult to ascertain, but most certainly this has a role. Fifth, incentive mechanisms should be developed to encourage researchers to make their data widely available to the scientific community.7 Such mechanisms may include requiring that publications acknowledge a wide number of data producers as authors,7 or that quantitative parameters to acknowledge the use of bioresources be developed.27 In addition, consortia should, at an early stage, set up plans for data sharing with the wider community, which includes the allocation of financial and human resources to realise the data-sharing plan, even after the project ends. With no such mechanisms in place, data deposit in repositories such as the European Genome-Phenome Archive (EGA) will not be actualised even if the importance is recognised.
Data sharing post ENGAGE
ENGAGE as a consortium officially ended in December 2012. At the project start, the European Commission, which funded the project, had requested that solutions for the release and sharing of ENGAGE data to the wider scientific community be established. Such solutions were provided through the development of the ENGAGE Data Access Catalogues.16, 17 Towards the end of the project, the General Assembly of ENGAGE also discussed plans to make ENGAGE data accessible to bona-fide researchers outside of the consortia after the project funding was over, with no preferential access for ENGAGE researchers. A proposal was set up to use the European Genome-Phenome Archive at the EBI for data archiving.18 The establishment of an ENGAGE Data Access Committee (DAC) that would evaluate and approve access to ENGAGE data was preferred. Internal agreement was reached among the project partners with respect to which data would be archived at the EBI and how. However, in March 2013, the establishment of an ENGAGE DAC was still pending due to a lack of funds to cover the administrative costs of such a committee. In the meantime, the recommendation from ENGAGE to its member institutions is to deposit data at the EBI with sufficient contact details on the EBI's project webpage. External access requests should be handled on a case-by-case basis by the main data providers.
Conclusion
Although data sharing and wider access to research data are essential to address questions of complex aetiology, current data-sharing procedures still place considerable demands on scientists and research consortia. Further steps are needed to fully enable wide data sharing as envisioned by funders and the scientific community. Key elements include harmonising the ethical and legal landscapes of data contributors, developing technological and organisational tools for secure data sharing and developing mechanisms for the recognition of data holders' contribution. Several initiatives17, 24, 28 are working on the development of procedures and data-sharing tools that are made freely available to the scientific community and can easily be integrated and adapted for data-sharing needs within consortia. Such tools include the IDAC (International Data Access Clearing house),17 which offers a one-stop policy interoperability and data access screening service via a ‘consent filter' the DataSHaPER (Data Schema and Harmonization Platform for Epidemiological Research), which aims at providing a toolbox for prospective harmonisation of emerging biobanks;26 the PhenX toolkit, which provides standard measures related to complex diseases, phenotypic traits and environmental exposures;25 DataSHIELD (Data Aggregation Through Anonymous Summary Statistics from Harmonised Individual levEL Databases),29 a statistical tool that allows pooled data analysis without physically sharing the data; BRIF (Bioresource Research Impact Factor),30 a quantitative parameter that allows the use of a bioresource to be traced; and ORCID (Open Researcher and Contributor ID), a coding system that permits to uniquely identify scientific and academic authors.31 Increased use of these solutions will streamline the data-sharing routines, increase incentives for data sharing, reduce duplicative efforts and data-sharing burdens that many consortia currently experience and accelerate the science.
Acknowledgments
This research was supported through funds from The European Community's Seventh Framework Programme (FP7/2007-2013), ENGAGE Consortium, grant agreement HEALTH-F4-2007- 201413. We thank Inga Prokopenko, University of Oxford; Nancy Pedersen and Erik Ingelsson, Karolinska Institute; Dorret Boomsma, VU University; Anil Jugessur, Norwegian Institute of Public Health and Jaakko Kaprio, University of Helsinki, for their comments to the questionnaire template. We thank all ENGAGE scientists who participated in the survey.
The authors declare no conflict of interest.
References
- Birney E, Hudson TJ, Green ED, et al. Prepublication data sharing. Nature. 2009;461:168–170. doi: 10.1038/461168a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppers BM, Harris JR, Tasse AM, et al. Towards a data sharing Code of Conduct for international genomic research. Genome Med. 2011;3:46. doi: 10.1186/gm262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health NIH Data Sharing Policy,2007
- Organisation for Economic Co-operation and Development OECD principles and guidelines for access to research data from public funding,2007
- Wellcome Trust Wellcome Trust Data Sharing Policy,2013
- Im HK, Gamazon ER, Nicolae DL, Cox NJ. On sharing quantitative trait GWAS results in an era of multiple-omics data and the limits of genomic privacy. Am J Hum Genet. 2012;90:591–598. doi: 10.1016/j.ajhg.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J, Heeney C, Hawkins N, de VJ, Boddington P. Data sharing in genomics–re-shaping scientific practice. Nat Rev Genet. 2009;10:331–335. doi: 10.1038/nrg2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. The Tension Between Data Sharing and the Protection of Privacy in Genomics Research. Annu Rev Genomics Hum Genet. 2012;13:415–431. doi: 10.1146/annurev-genom-082410-101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppers BM, Dove ES, Litton JE, Nietfeld JJ. Questioning the limits of genomic privacy. Am J Hum Genet. 2012;91:577–578. doi: 10.1016/j.ajhg.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AL, Basford M, Dressler LG, et al. Ethical and practical challenges of sharing data from genome-wide association studies: the eMERGE Consortium experience. Genome Res. 2011;21:1001–1007. doi: 10.1101/gr.120329.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord P, MacDonald A, Sinnott R, Ecklund D, Westhead M, Jones A.Large-scale data sharing in the life sciences: Data standards, incentives, barriers and funding models (The "Joint Data Standards Study"). 2005. UK e-Science Technical Report Series,.
- Rodriguez H, Snyder M, Uhlen M, et al. Recommendations from the 2008 International Summit on Proteomics Data Release and Sharing Policy: the Amsterdam principles. J Proteome Res. 2009;8:3689–3692. doi: 10.1021/pr900023z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Network for Genetic and Genomic Epidemiology ENGAGE,2012
- ENGAGE Principles for Data Sharing, Data Release and Intellectual Property,2009
- Gostev M, Fernandez-Banet J, Rung J, et al. SAIL—a software system for sample and phenotype availability across biobanks and cohorts. Bioinformatics. 2011;27:589–591. doi: 10.1093/bioinformatics/btq693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENGAGE Data Access Catalogue,2013
- Public Population Projects in Genomics and Society (P3G),2011
- European Genome-Phenome Archive (EGA),2012
- European Bioinformatics Institute (EMBL-EBI),2012
- Tasse AM, Budin-Ljosne I, Knoppers BM, Harris JR. Retrospective access to data: the ENGAGE consent experience. Eur J Hum Genet. 2010;18:741–745. doi: 10.1038/ejhg.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce N, Smith AH. Data sharing: not as simple as it seems. Environ Health. 2011;10:107. doi: 10.1186/1476-069X-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SN, Caporaso N, Fitzpatrick AL, et al. Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience. Genet Epidemiol. 2011;35:159–173. doi: 10.1002/gepi.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly Y, Dove ES, Knoppers BM, Bobrow M, Chalmers D. Data sharing in the post-genomic world: the experience of the International Cancer Genome Consortium (ICGC) Data Access Compliance Office (DACO) PLoS Comput Biol. 2012;8:e1002549. doi: 10.1371/journal.pcbi.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BioSHaRE-EU (Biobank Standardisation and Harmonisation for Research Excellence in the European Union),2012
- PhenX Toolkit,2013
- Data Schema and Harmonization Platform for Epidemiological Research (DataSHaPER),2012
- Cambon-Thomsen A, Thorisson GA, Mabile L, et al. The role of a Bioresource Research Impact Factor as an incentive to share human bioresources. Nat Genet. 2011;43:503–504. doi: 10.1038/ng.831. [DOI] [PubMed] [Google Scholar]
- BBMRI-LPC (Large Prospectives Cohorts),2013
- Wolfson M, Wallace SE, Masca N, et al. DataSHIELD: resolving a conflict in contemporary bioscience—performing a pooled analysis of individual-level data without sharing the data. Int J Epidemiol. 2010;39:1372–1382. doi: 10.1093/ije/dyq111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bioresource Research Impact Factor (BRIF),2012
- Open Researcher and Contributor ID (ORCID),2012