1. Disease characteristics
1.1 Name of the disease (synonyms)
Alagille syndrome (ALGS); Alagille syndrome 1 (ALGS1); Alagille syndrome 2 (ALGS2). Other synonyms include Alagille–Watson Syndrome (AWS); cholestasis with peripheral pulmonary stenosis; arteriohepatic dysplasia (AHD); hepatic ductular hypoplasia, syndrome; Miller–Watson syndrome.1, 2, 3, 4
1.2 OMIM# of the disease
ALGS1: 118450; ALGS2: 610205.
1.3 Name of the analysed genes or DNA/chromosome segments
JAG1 (Jagged1 gene; locus 20p12.2; disease ALGS1); NOTCH2 (Notch2 gene; locus 1p12-p11; disease ALGS2).
1.4 OMIM# of the gene(s)
JAG1 (601920); NOTCH2 (600275).
1.5 Mutational spectrum
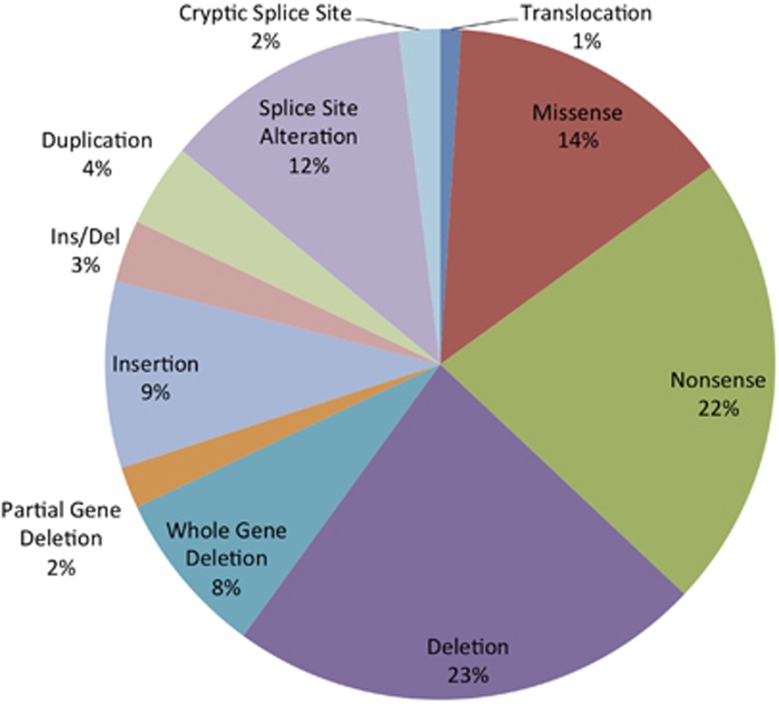
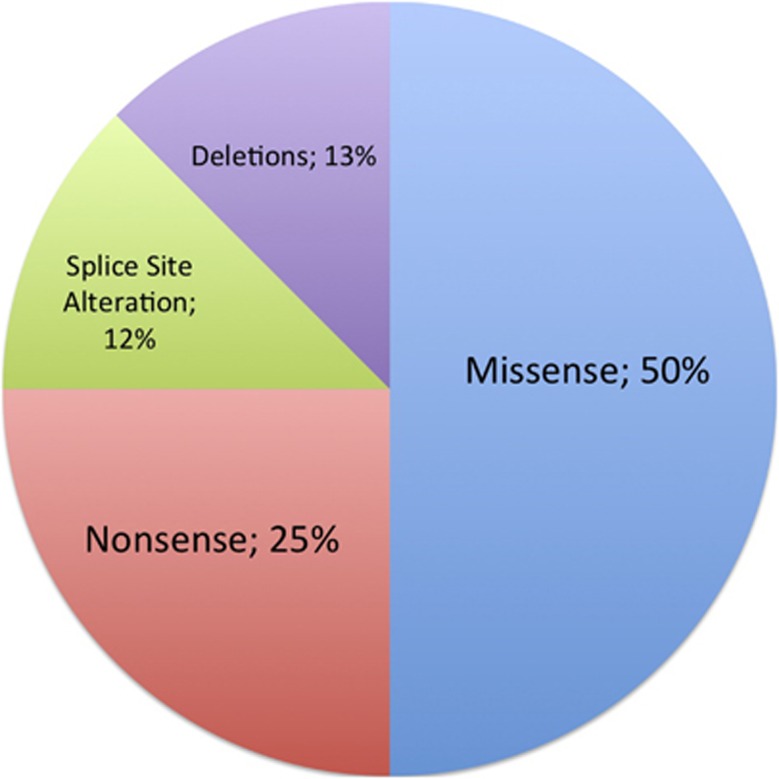
JAG1 mutations are identified in roughly 94% of patients with clinically diagnosed Alagille syndrome (ALGS) and NOTCH2 mutations are identified in about 1.5% of patients with clinically diagnosed ALGS. The distribution of mutation types is shown below for JAG1 (Figure 1) and NOTCH2 (Figure 2).5, 6, 7
Figure 1.
Prevalence of JAG1 mutations by mutation type (n=422 unrelated probands).5
Figure 2.
Prevalence of NOTCH2 mutations by mutation type (n=8 unrelated probands).5, 6
1.6 Analytical methods
Genomic sequencing of Jagged1 coding regions and Jagged1 multiplex ligation-dependent probe amplification (MLPA) assays are utilized to detect mutations or deletions/duplications within the gene. Gene-sequencing panels including Jagged1 have also recently become available. Clinical testing for NOTCH2 is currently offered in Germany, Austria and England. For a full list of laboratories that offer JAG1 or NOTCH2 testing, please refer to GeneTests.8
1.7 Analytical validation
Possible mutations detected during initial bidirectional Sanger sequencing can be re-sequenced in normal, ethnically matched controls to exclude possible polymorphisms. Due to the variable expressivity of Alagille syndrome, it is not recommended that unaffected family members be used as controls. Family members carrying the mutation may present with mild or subclinical disease features.9, 10
1.8 Estimated frequency of the disease (incidence at birth (‘birth prevalence') or population prevalence)
In 1977, the incidence of ALGS was estimated to be 1 in 70 000 live births.11 However, given that not all patients with ALGS present with neonatal cholestasis, as the original estimate assumes, this figure is thought to be an underestimate. Based on the work by Kamath et al.10 in 2003 we estimate that the incidence of ALGS is 1 in 30 000–50 000 live births, but due to the variable phenotype it is likely to remain underdiagnosed.
1.9 If applicable, prevalence in the ethnic group of the investigated person
Not applicable.
1.10 Diagnostic setting

Comment:
Mutation testing for ALGS is used mainly to confirm a suspected clinical diagnosis or to carry out screening in the relatives of an affected individual. A list of laboratories that perform clinical and research testing of the ALGS genes can be found on the GeneTests website.8 Prenatal testing is available for families with known mutations.
2. Test characteristics

2.1 Analytical sensitivity
(proportion of positive tests if the genotype is present)
The sensitivity for genomic sequencing approaches 100% for mutation detection. Deletions of exons or whole gene deletions are detected by MLPA and may be cross-checked with homozygous polymorphisms in deleted regions via Sanger sequencing.5, 6, 9, 10, 11 Additionally, mutations outside coding exons in promoters or enhancers are likely to be missed.7
**When screening parents for their child's known mutation, it is important to note that a negative result does not rule out the possibility of germline mosaicism.12
2.2 Analytical specificity
(proportion of negative tests if the genotype is not present)
Analytical specificity is nearly 100%. False positives are rare. In some cases, missense mutations may be reported as disease-causing or likely disease-causing by protein prediction software but later shown to be benign polymorphisms based on functional protein assays.
2.3 Clinical sensitivity
(proportion of positive tests if the disease is present)
The clinical sensitivity can be dependent on variable factors such as age or family history. In such cases a general statement should be given, even if quantification can only be made case by case.
In patients clinically consistent with ALGS, JAG1 mutations are identified in ∼94%, NOTCH2 mutations in ∼1.5% and no mutation in ∼4.5% of patients. Therefore, the clinical sensitivity is ∼95% and is estimated to increase as we learn more about the cause of disease in the 4.5% of patients who are clinically consistent with ALGS but have no identifiable mutation in Jagged1 or Notch2.5
2.4 Clinical specificity
(proportion of negative tests if the disease is not present)
The clinical specificity can be dependent on variable factors such as age or family history. In such cases a general statement should be given, even if quantification can only be made case by case.
Although clinical specificity cannot be predicted, it is not 100%. Due to highly variable expressivity, many individuals who do not meet the full diagnostic criteria for ALGS or have mild features of the disease may still carry mutations in Jagged1 or Notch2. Family members of affected individuals may carry the same genetic mutation but have mild clinical manifestations such as slightly elevated liver enzymes.9, 10
2.5 Positive clinical predictive value
(life-time risk to develop the disease if the test is positive)
ALGS is an autosomal dominant disorder and therefore a single mutation on one allele is sufficient to cause disease. Although non-penetrance is rare, ALGS has variable expressivity and severity of disease cannot be predicted based on the presence of a mutation. Both mild and severe ALGS have been described and there is no apparent link between mutation type or location and severity of disease.13, 14, 15, 16
2.6 Negative clinical predictive value
(probability of not developing the disease if the test is negative)
Assume an increased risk based on family history for a non-affected person. Allelic and locus heterogeneity may need to be considered.
Index case in that family had been tested:
Nearly 100%.
Index case in that family had not been tested:
Genetic heterogeneity with undiscovered genes means that ∼5% of individuals who test negative for both Jagged1 and Notch2 genes may still have the disease.14
3. Clinical Utility
3.1 (Differential) diagnostics: The tested person is clinically affected
(To be answered if in 1.10 ‘A' was marked)
3.1.1 Can a diagnosis be made other than through a genetic test?

The five primary diagnostic criteria are cholestatic liver disease (characterized by bile duct paucity), cardiac disease, skeletal abnormalities (butterfly vertebrae), eye abnormalities (posterior embryotoxon) and characteristic facial features. Secondary diagnostic criteria include renal and vascular abnormalities. The presence of three of the five primary diagnostic criteria is sufficient for diagnosis of ALGS.10, 17, 18, 19
3.1.2 Describe the burden of alternative diagnostic methods to the patient
The majority of clinical diagnostic tests (biochemical blood tests) are performed as a part of routine clinical care and therefore do not add additional risk or burden. A liver biopsy is not always necessary for the diagnosis of ALGS if initial evaluation of liver function tests is sufficient. Genetic evaluations of affected individuals and their parents have the largest impact if the family wishes to address the chances of future children being affected with Alagille syndrome.
3.1.3 How is the cost effectiveness of alternative diagnostic methods to be judged?
Although an accurate clinical assessment can confirm the diagnosis of ALGS and therefore establish the need for appropriate monitoring and management, genetic testing remains useful for genetic counselling and prenatal testing.
3.1.4 Will disease management be influenced by the result of a genetic test?

3.2 Predictive Setting: The tested person is clinically unaffected but carries an increased risk based on family history
(To be answered if in 1.10 ‘B' was marked).
3.2.1 Will the result of a genetic test influence lifestyle and prevention?
If the test result is positive (please describe):
If a clinically unaffected person undergoes genetic testing and is found to have a disease causing mutation, he/she should be screened for clinical features of the disease and then referred to the appropriate specialists.
If the test result is negative (please describe):
If a clinically unaffected person undergoes genetic testing and does not have a disease-causing mutation identified, the likelihood of developing the disease approaches 0% and therefore there will be no changes in lifestyle.
3.2.2 Which options in view of lifestyle and prevention does a person at-risk have if no genetic test has been done (please describe)?
We recommend that individuals who are at risk (have a significant family history of ALGS) undergo genetic testing and follow the guidelines in section 3.2.1.
3.3 Genetic risk assessment in family members of a diseased person
(To be answered if in 1.10 ‘C' was marked).
3.3.1 Does the result of a genetic test resolve the genetic situation in that family?
Yes.
3.3.2 Can a genetic test in the index patient save genetic or other tests in family members?
No—if testing is positive for ALGS in the index patient, it can reduce the need for testing for other genetic conditions in family members by providing a diagnosis. However, testing for ALGS may still be suggested in clinically affected or unaffected family members.
3.3.3 Does a positive genetic test result in the index patient enable a predictive test in a family member?
Yes. Given the variable expressivity of ALGS, a positive genetic test result in the index patient provides a molecular diagnosis of ALGS that can inform the diagnosis of other mildly affected family members.9, 10
3.4 Prenatal diagnosis
(To be answered if in 1.10 ‘D' was marked).
3.4.1 Does a positive genetic test result in the index patient enable a prenatal diagnosis?
Yes. Prenatal diagnosis can be performed if the index patient has a positive genetic test result. Testing for the known familial mutation can be performed on fetal DNA.
4. IF APPLICABLE, FURTHER CONSEQUENCES OF TESTING
Please assume that the result of a genetic test has no immediate medical consequences. Is there any evidence that a genetic test is nevertheless useful for the patient or his/her relatives? (Please describe).
Genetic tests can provide families knowledge about the genetics and inheritance pattern of ALGS. Genetic test results may also address questions about the risk of future children inheriting the disease.
Acknowledgments
This work was supported by EuroGentest2 (Unit 2: ‘Genetic testing as part of health care'), a Coordination Action under FP7 (Grant Agreement Number 261469) and the European Society of Human Genetics. This work was also supported by the NIDDK (R01-DK81702 and U01-DK062481).
The authors declare no conflict of interest.
References
- Alagille D, Borde J, Habib EC, Thomassin N. Surgical attempts in atresia of the intrahepatic bile ducts with permeable extrahepatic bile duct. Study of 14 cases in children. Arch Fr Pediatr. 1969;26:51–71. [PubMed] [Google Scholar]
- Riely CA, Cotlier E, Jensen PS, Klatskin G. Arteriohepatic dysplasia: a benign syndrome of intrahepatic cholestasis with multiple organ involvement. Ann Intern Med. 1979;91:520–527. doi: 10.7326/0003-4819-91-4-520. [DOI] [PubMed] [Google Scholar]
- Watson GH, Miller V. Arteriohepatic dysplasia: familial pulmonary arterial stenosis with neonatal liver disease. Arch Dis Child. 1973;48:459–466. doi: 10.1136/adc.48.6.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagille D, Estrada A, Hadchouel M, Gautier M, Odievre M, Dommergues JP. Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): review of 80 cases. J Pediatr. 1987;110:195–200. doi: 10.1016/s0022-3476(87)80153-1. [DOI] [PubMed] [Google Scholar]
- Warthen DM, Moore EC, Kamath BM, et al. Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum Mutat. 2006;27:436–443. doi: 10.1002/humu.20310. [DOI] [PubMed] [Google Scholar]
- Kamath BM, Bauer RC, Loomes KM, Chao G, Gerfen J, Hutchinson A, et al. NOTCH2 mutations in Alagille syndrome. J Med Genet. 2011;49:138–144. doi: 10.1136/jmedgenet-2011-100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Warthen DM, Sanchez-Lara PA, et al. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the Notch signaling pathway. Am J Hum Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeneTests Medical Genetics Information Resource (database online) Copyright, University of Washington, Seattle, WA, USA, 1993–2013. Available at http://www.genetests.org (accessed 16 May 2013).
- Guegan K, Stals K, Day M, Turnpenny P, Ellard S. JAG1 mutations are found in approximately one third of patients presenting with only one or two clinical features of Alagille syndrome. Clin Genet. 2011;82:33–40. doi: 10.1111/j.1399-0004.2011.01749.x. [DOI] [PubMed] [Google Scholar]
- Kamath BM, Bason L, Piccoli DA, Krantz ID, Spinner NB. Consequences of JAG1 mutations. J Med Genet. 2003;40:891–895. doi: 10.1136/jmg.40.12.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danks DM, Campbell PE, Jack I, Rogers J, Smith AL. Studies of the aetiology of neonatal hepatitis and biliary atresia. Arch Dis Child. 1977;52:360–367. doi: 10.1136/adc.52.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakudis J, Ropke A, Kujat A, et al. Paternal mosaicism of JAG1 mutations in families with Alagille syndrome. Eur J Hum Genet. 2001;9:209–216. doi: 10.1038/sj.ejhg.5200613. [DOI] [PubMed] [Google Scholar]
- Krantz ID, Colliton RP, Genin A, et al. Spectrum and frequency of jagged1 (JAG1) mutations in Alagille syndrome patients and their families. Am J Hum Genet. 1998;62:1361–1369. doi: 10.1086/301875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Driancourt C, Raynaud N, et al. Mutations in JAGGED1 gene are predominantly sporadic in Alagille syndrome. Gastroenterology. 1999;116:1141–1148. doi: 10.1016/s0016-5085(99)70017-x. [DOI] [PubMed] [Google Scholar]
- Spinner NB, Colliton RP, Crosnier C, Krantz ID, Hadchouel M, Meunier-Rotival M. Jagged1 mutations in Alagillesyndrome. Hum Mutat. 2001;17:18–33. doi: 10.1002/1098-1004(2001)17:1<18::AID-HUMU3>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- McElhinney DB, Krantz ID, Bason L, et al. Analysis of cardiovascular phenotype and genotype-phenotype correlation in individuals with a JAG1 mutation and/or Alagille syndrome. Circulation. 2002;106:2567–2574. doi: 10.1161/01.cir.0000037221.45902.69. [DOI] [PubMed] [Google Scholar]
- Emerick KM, Rand EB, Goldmuntz E, Krantz ID, Spinner NB, Piccoli DA. Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology. 1999;29:822–829. doi: 10.1002/hep.510290331. [DOI] [PubMed] [Google Scholar]
- Kamath BM, Podkameni G, Hutchinson AL, et al. Renal anomalies in Alagille syndrome: a disease-defining feature. Am J Med Genet. 2012;158A:85–89. doi: 10.1002/ajmg.a.34369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath BM, Spinner NB, Emerick KM, et al. Vascular anomalies in Alagille syndrome: a significant cause of morbidity and mortality. Circulation. 2004;109:1354–1358. doi: 10.1161/01.CIR.0000121361.01862.A4. [DOI] [PubMed] [Google Scholar]