Abstract
Candidate gene and genome-wide association studies (GWAS) represent two complementary approaches to uncovering genetic contributions to common diseases. We systematically reviewed the contributions of these approaches to our knowledge of genetic associations with cancer risk by analyzing the data in the Cancer Genome-wide Association and Meta Analyses database (Cancer GAMAdb). The database catalogs studies published since January 1, 2000, by study and cancer type. In all, we found that meta-analyses and pooled analyses of candidate genes reported 349 statistically significant associations and GWAS reported 269, for a total of 577 unique associations. Only 41 (7.1%) associations were reported in both candidate gene meta-analyses and GWAS, usually with similar effect sizes. When considering only noteworthy associations (defined as those with false-positive report probabilities ≤0.2) and accounting for indirect overlap, we found 202 associations, with 27 of those appearing in both meta-analyses and GWAS. Our findings suggest that meta-analyses of well-conducted candidate gene studies may continue to add to our understanding of the genetic associations in the post-GWAS era.
Keywords: GWAS, candidate gene studies, meta-analysis, cancer
Introduction
Candidate gene association studies have been widely used to study genetic susceptibility to complex diseases, including cancer.1 Critics of candidate gene studies have pointed to non-replication of results, false positives, insufficient sample sizes, and limited prior knowledge of biologically relevant candidate genes.2 These concerns have prompted the use of systematic reviews, especially meta-analyses of multiple studies, to minimize false-positive associations and assess the credibility of findings.3 In recent years, genome-wide association studies (GWAS) have greatly accelerated the pace of discovery and found many novel genetic associations that were not anticipated by the candidate gene approach.4, 5 Associations discovered by GWAS raise additional questions, particularly because observed effects are typically very small.6 Furthermore, the implicated SNPs represent markers that require further investigation to identify causal variants,7 although this may become less of a problem as methods for fine-mapping associations improve.
A critical evaluation of a decade's worth of association studies is warranted as the next phase of cancer genetics research unfolds. In the present analysis, we used the data available in a 2008 paper published by Dong et al8 and in the Cancer Genome-wide Association and Meta Analyses database (Cancer GAMAdb)9 to complete a systematic review of genetic associations in cancer GWAS and meta-analyses and pooled analyses published over an 11-year period, from 2000–2011.
Materials and Methods
Cancer GAMAdb
To help consolidate the vast amount of information from both candidate gene and GWAS of cancer, the Centers for Disease Control and Prevention's (CDC) Office of Public Health Genomics and the National Cancer Institute's Division of Cancer Control and Population Sciences launched the Cancer GAMAdb in 2010.9 This continuously updated database catalogs published GWAS and meta-analyses and pooled analyses that have evaluated associations of genetic polymorphisms and cancer risk since January 1, 2000. Cancer GAMAdb builds on a published data set by Dong et al,8 which encompassed meta-analyses and pooled analyses of genetic polymorphisms, and cancer risk published until March 15, 2008. Associations in the database published after that date have been identified using the Human Genome Epidemiology (HuGE) Navigator database10 and the National Human Genome Research Institute (NHGRI) GWAS catalog.11 The Centers for Disease Control and Prevention's HuGE Navigator is a continuously updated knowledge base in HuGE.10 The NHGRI GWAS catalog extracts data from GWAS publications.11 Genetic associations with cancer are selected from these two databases for curation in the Cancer GAMAdb. Data describing the association(s)—including study population, minor allele frequencies, and effect sizes—are manually extracted from each article and entered into the Cancer GAMAdb. The current analysis is based on the data that were included in Cancer GAMAdb as of February 26, 2011.
Selection criteria
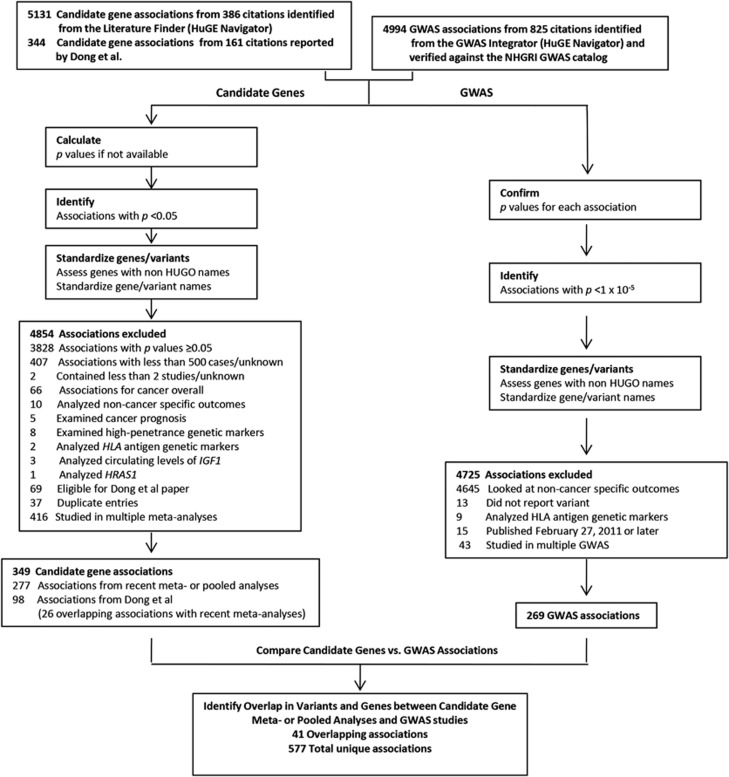
We selected genetic associations for our analysis according to the schema in Figure 1. We excluded meta-analyses and pooled analyses with P-values of odds ratios (OR) ≥0.05 (if P-values were not reported, we calculated P-values as described in Dong et al8). We excluded meta-analyses with fewer than 500 total cases or based on less than two studies for the meta-analyses and pooled analyses (or if either was unknown). We standardized the gene names with the Human Genome Organisation gene symbol and the National Center for Biotechnology Information Entrez Gene GeneID, as well as the RefSNP accession ID (rs numbers) for variant names where possible. To fill in the missing gene names, we searched by variant name using HuGE Navigator's Variant Name Mapper and the UCSC Genome Browser and collected region information if the variant was intergenic.
Figure 1.
Methodology used for inclusion of associations into the analyses.
Our analysis was limited to genetic associations with incident cancer of specified type; we excluded associations with other outcomes (eg, all cancers, precursor lesions, biomarkers, or survival). Associations with circulating levels of IGF1; human lymphocyte antigen (HLA) markers; high-penetrance genetic markers (eg, APC, BRCA1); and associations with HRAS1 (which have been questioned because of flawed genotyping methods) were also excluded.
When an association had been examined in multiple meta-analyses, we included only the most recent publication. We gave priority to the most recently reported overall association with a particular variant; however, if no significant overall association had been reported, we included the most recent subgroup-specific association. If a publication reported multiple significant contrasts (ie, results based on different genetic models) for the same variant, we included the contrast with the smallest P-value. When significant associations were found with the same variant in both meta-analyses and GWAS, we checked to be sure that they compared the same contrasts. Associations with combinations of two or more variants were considered unique, even if associations with the individual variants were also reported.
GWAS data were restricted to studies of incident cancer published before February 26, 2011. Studies were identified from the HuGE Navigator and checked against the NHGRI GWAS catalog to ensure completeness. In some instances, we checked the original publication for additional information; if we noticed data discrepancies between the GWAS catalog and the original paper, we used the data from the original paper. GWAS that included meta-analyses were classified as GWAS. Associations for which variants were not specified were excluded from analysis. When multiple GWAS reported the same association, we included only the most recently published study in our analysis.
Analysis strategy
Our analysis considered the extent to which associations reported in meta-analyses and GWAS overlapped. When both types of studies reported associations with the same variant, we called the overlap direct. When they reported associations with variants separated by less than 1 million base pairs, we called the overlap indirect. In an additional analysis, we also examined noteworthy associations, which we defined as those with false-positive report probabilities (FPRP) ≤0.2, a stringent threshold suggested by Wacholder et al,12 and used in the analysis by Dong et al.8 We calculated FPRPs at two levels of prior probability and at two levels of association (OR 1.5 and OR 1.2). As in the analysis by Dong et al,8 we chose to evaluate the associations using a low-prior probability of 0.001 (expected for a candidate gene) and a very low-prior probability of 0.000001 (expected for a random SNP). An association was considered noteworthy if it passed the FPRP threshold in one or more of these four categories.
Results
Significant associations are summarized in Table 1 by the cancer site and the study type.
Table 1. Number of significant associations (in variants and genes) reported in candidate gene meta-analysis and pooled analysis and GWAS, by cancer site.
| MAa | GWASb | |||
|---|---|---|---|---|
| Cancer Site | Variantsc | Genesd | Variantsc | Genesd |
| Bladder | 15 | 14 | 10 | 10 |
| Blood-related (ALL, MCL, NHL) | 1 | 1 | — | — |
| Breast | 80 | 59 | 36 | 30 |
| Cervical | 4 | 4 | — | — |
| Colorectal | 30 | 23 | 17 | 14 |
| Endometrial | 2 | 1 | — | — |
| Esophageal | 9 | 9 | 4 | 4 |
| Gastric | 21 | 17 | 2 | 2 |
| Genitourinary | 2 | 2 | — | — |
| Glioma | 18 | 13 | 9 | 8 |
| Head and neck | 14 | 11 | — | — |
| Hepatocellular | 8 | 4 | 4 | 6 |
| Hodgkin lymphoma | — | — | 4 | 3 |
| Laryngeal | 2 | 2 | — | — |
| Leukemia | 4 | 4 | 32 | 27 |
| Lung | 32 | 23 | 25 | 22 |
| Meningioma | 1 | 1 | — | — |
| Myeloproliferative | — | — | 1 | 1 |
| Nasopharyngeal | 4 | 3 | 6 | 6 |
| Neuroblastoma | — | — | 5 | 3 |
| Non-Hodgkin lymphoma | 10 | 8 | 2 | 2 |
| Oral | 1 | 1 | — | — |
| Ovarian | 14 | 12 | 10 | 10 |
| Pancreatic | — | — | 21 | 21 |
| Prostate | 53 | 40 | 56 | 35 |
| Renal cell | — | — | 3 | 3 |
| Skin | 20 | 8 | 8 | 7 |
| Testicular | — | — | 12 | 10 |
| Thyroid | — | — | 2 | 2 |
| Upper aero-digestive tract | 2 | 2 | — | — |
| Upper aero-digestive tract and lung | 1 | 1 | — | — |
| Urothelial | 1 | 1 | — | — |
| Total | 349 | 264 | 269 | 223 |
Abbreviations: ALL, adult lymphoblastic leukemia; GWAS, genome-wide association studies; MA, meta-analyses or pooled analyses; MCL, myeloid cell leukemia; NHL, non-Hodgkin lymphoma.
Total significant associations reported in previous systematic review of meta-analyses (Dong et al8) and meta-analyses and pooled data of individual studies published from 20 March 2008 through 26 February 2011. Meta-analyses were defined as those of candidate gene studies. Significance threshold was 0.05.
From GWAS catalog. Excludes variants that were not reported. GWAS with meta-analyses included were considered GWAS. Significance threshold was 1 × 10−5.
Some variants may be linked to one another due to proximity. Associations with combinations of two or more variants were considered unique, even if listed standalone variants were also reported.
Intergenic regions used if no gene provided by paper or associated with variant.
Meta- and pooled analyses
We identified 5131 gene-variant associations with incident cancer from 386 meta-analyses and pooled analyses published after the review by Dong et al review. We excluded 3828 (74.6%) associations because their reported P-values were ≥0.05; 1026 more were excluded for reasons listed in Figure 1. After applying all exclusion criteria, we found 277 significant associations; the review by Dong et al included 98 significant associations. Twenty-six (7.4%) of these were also found in meta-analyses published since the paper by Dong et al. Thus, there were 349 unique variant-cancer associations in all, involving 264 genes (76 with more than one associated variant) and spanning 25 different cancer types.
The largest number of candidate gene associations was found for breast cancer (n=80) followed by prostate cancer (n=53). Significant associations from meta-analyses and pooled analyses of candidate genes are listed in Supplementary Table 1.
Genome-wide association studies
We identified 4994 GWAS associations from 825 citations. We excluded 4645 associations with outcomes other than incident cancer and 80 for other reasons listed in Figure 1. In the end, there were 269 unique associations in 223 different genes with 21 different cancer types. The largest number of GWAS associations was found for prostate cancer (n=56) followed by breast cancer (n=36). Variants from GWAS are listed in Supplementary Table 2.
Combined
The combined results from candidate gene meta-analyses and GWAS included 577 unique associations of 446 different genes or chromosomal regions with 32 cancers. When we considered only direct overlap, we found 41 associations that had been reported in both meta-analyses and GWAS (Supplementary Table 3). The largest number of such associations was with prostate cancer (n=25), followed by breast cancer (n=8).
When we restricted our analysis to noteworthy associations (calculated FPRPs of ≤0.2 in either prior probability or OR) and allowed for direct and indirect overlap (both within and between study types), we found 202 unique associations in all. Of these, 66 were from candidate gene studies and 163 were from GWAS; 27 (13%) of these were found in both meta-analyses and GWAS (Table 2). We were unable to evaluate 38 GWAS associations for noteworthiness because the original publications did not report ORs and CIs. Allowing for indirect overlap, we found the largest numbers of noteworthy associations in leukemia (n=27), followed by prostate cancer (n=25). Noteworthy associations that were found only in meta-analyses (n=39) are listed in Table 3. All noteworthy associations are included in Supplementary Table 4.
Table 2. Number of noteworthy associations reported in candidate gene meta-analyses and pooled analyses and GWAS, accounting for direct and indirect overlap, by cancer site.
|
MAa |
||||||
|---|---|---|---|---|---|---|
| Cancer site | Dong, et alb | Subsequent MAc | Total | GWASd | Overlap | Total unique |
| Bladder | 2 | 2 | 3 | 10 | 3 | 10 |
| Blood-related | 0 | 1 | 1 | 0 | 0 | 1 |
| Breast | 3 | 13 | 15 | 14 | 7 | 22 |
| Colorectal | 1 | 5 | 6 | 14 | 1 | 19 |
| Endometrial | 0 | 1 | 1 | 0 | 0 | 1 |
| Esophageal | 0 | 5 | 5 | 3 | 1 | 7 |
| Gastric | 1 | 3 | 4 | 2 | 0 | 6 |
| Glioma | 0 | 0 | 0 | 5 | 0 | 5 |
| Head and neck | 0 | 1 | 1 | 0 | 0 | 1 |
| Hepatocellular | 0 | 1 | 1 | 4 | 0 | 5 |
| Hodgkin's lymphoma | 0 | 0 | 0 | 3 | 0 | 3 |
| Leukemia | 2 | 0 | 2 | 25 | 0 | 27 |
| Lung | 3 | 6 | 8 | 10 | 2 | 16 |
| Myeloproliferative | 0 | 0 | 0 | 1 | 0 | 1 |
| Nasopharyngeal | 0 | 1 | 1 | 4 | 0 | 5 |
| Neuroblastoma | 0 | 0 | 0 | 3 | 0 | 3 |
| Non-Hodgkin's lymphoma | 0 | 1 | 1 | 2 | 0 | 3 |
| Ovarian | 0 | 0 | 0 | 8 | 0 | 8 |
| Pancreatic | 0 | 0 | 0 | 16 | 0 | 16 |
| Prostate | 1 | 15 | 16 | 21 | 12 | 25 |
| Renal cell | 0 | 0 | 0 | 3 | 0 | 3 |
| Skin | 0 | 1 | 1 | 7 | 1 | 7 |
| Testicular | 0 | 0 | 0 | 6 | 0 | 6 |
| Thyroid | 0 | 0 | 0 | 2 | 0 | 2 |
| Total | 13 | 56 | 66 | 163 | 27 | 202 |
Abbreviations: GWAS, genome-wide association studies; MA, meta- or pooled analyses
Noteworthy associations from meta- and pooled analyses of candidate gene studies.
Reported in previous systematic review of meta-analyses (Dong et al8).
Reported in meta-analyses published from 20 March 2008 through 26 February 2011.
From GWAS catalog. Excludes variants that were not reported. GWAS with meta-analyses included were considered GWAS.
Table 3. Noteworthy associations only found in meta- and pooled analyses of candidate gene studies.
| Locus | Source/PMIDa | Gene/region | Variant | OR | 95% CI lower | 95% CI upper | P-valueb | FPRP at prior probability of 0.001 and OR 1.5 | FPRP at prior probability of 0.000001 and OR 1.5 | FPRP at prior probability of 0.001 and OR 1.2 | FPRP at prior probability of 0.000001 and OR 1.2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Blood-related cancer | |||||||||||
| 11q13.3 | 18843022 | CCND1 | rs17852153 | 1.62 | 1.28 | 2.05 | 0.0001 | 0.184 | 0.996 | 0.904 | 1.000 |
| Breast cancer | |||||||||||
| 2q13 | 20437198 | IL1B | rs1143627 | 1.4 | 1.17 | 1.67 | 1.8E-04 | 0.191 | 0.996 | 0.809 | 1.000 |
| 2q33.1 | Dong | CASP8 | rs1045485 | 0.89 | 0.85 | 0.94 | 5.7E-06 | 0.028 | 0.967 | 0.029 | 0.967 |
| 19629679 | 0.874 | 0.834 | 0.917 | 3.9E-08 | <0.001 | 0.037 | <0.001 | 0.038 | |||
| 2q33.2 | 20920330 | CTLA4 | rs231775 | 1.31 | 1.17 | 1.48 | 1.4E-05 | 0.014 | 0.936 | 0.153 | 0.995 |
| 5p12 | 21194473 | MRPS30 | rs10941679 | 1.12 | 1.09 | 1.15 | <1.e-30 | <0.001 | <0.001 | <0.001 | <0.001 |
| 17q22 | 21194473 | COX11/STXBP4 | rs6504950 | 0.95 | 0.92 | 0.97 | 1.4E-06 | 0.001 | 0.583 | 0.001 | 0.583 |
| 19q13.2 | Dong | TGFB1 | rs1800470 | 1.16 | 1.08 | 1.25 | 6.9E-05 | 0.090 | 0.990 | 0.108 | 0.992 |
| 20q13.2 | 19823929 | AURKA | rs2273535 | 1.23 | 1.1 | 1.37 | 1.7E-04 | 0.143 | 0.994 | 0.338 | 0.998 |
| 22q12.1 | Dong | CHEK2 | 1100delC | 2.4 | 1.8 | 3.2 | 2.5E-09 | 0.004 | 0.782 | 0.678 | 1.000 |
| Colorectal cancer | |||||||||||
| 1p26.22 | 19846566 | MTHFR | rs1801133 | 0.83 | 0.77 | 0.9 | 6.5E-06 | 0.006 | 0.866 | 0.014 | 0.933 |
| 11q22.2 | 19843588 | MMP1 | rs1799750 | 1.48 | 1.26 | 1.74 | 2.1E-06 | 0.004 | 0.785 | 0.270 | 0.997 |
| 12q15 | 20503107 | MDM2 | rs2279744 | 0.73 | 0.62 | 0.86 | 1.7E-04 | 0.163 | 0.995 | 0.747 | 1.000 |
| 19q13.2 | 20012233 | TGFB1 | rs1800469 | 1.62 | 1.3 | 2.02 | 1.8E-05 | 0.069 | 0.987 | 0.826 | 1.000 |
| 22q11.23 | Dong | GSTT1 | GSTT1 null | 1.37 | 1.17 | 1.6 | 8.1E-05 | 0.074 | 0.988 | 0.598 | 0.999 |
| Endometrial cancer | |||||||||||
| 15q21.2 | 19124504 | CYP19A1 | rs749292 | 1.3 | 1.17 | 1.45 | 2.5E-06 | 0.002 | 0.714 | 0.032 | 0.971 |
| Esophageal cancer | |||||||||||
| 1q31.1 | 21304218 | PTGS2 | rs20417 | 1.45 | 1.23 | 1.71 | 1.0E-05 | 0.015 | 0.939 | 0.451 | 0.999 |
| 4q23 | 20806441 | ADH1B | rs1229984 | 1.32 | 1.17 | 1.49 | 7.1E-06 | 0.007 | 0.878 | 0.103 | 0.991 |
| 16q12.2 | 20360147 | MMP2 | rs243865 | 0.7 | 0.59 | 0.82 | 9.9E-06 | 0.013 | 0.932 | 0.392 | 0.998 |
| 17p13.1 | 20827430 | TP53 | rs1042522 | 1.43 | 1.23 | 1.68 | 1.4E-05 | 0.018 | 0.950 | 0.451 | 0.999 |
| Gastric cancer | |||||||||||
| 1p36.22 | Dong | MTHFR | rs1801133 | 1.52 | 1.31 | 1.77 | 4.9E-08 | <0.001 | 0.140 | 0.057 | 0.984 |
| 2q13 | 20360147 | MMP7 | rs11568818 | 1.79 | 1.37 | 2.34 | 2.1E-05 | 0.173 | 0.995 | 0.923 | 1.000 |
| 4q13.3 | 19777350 | IL-8 | rs4073 | 1.363 | 1.199 | 1.527 | 9.2E-08 | <0.001 | 0.088 | 0.007 | 0.868 |
| 19q13.32 | 20981556 | ERCC2 | rs13181 | 0.3 | 0.21 | 0.44 | 7.2E-10 | 0.032 | 0.971 | 0.894 | 1.000 |
| Head and neck cancers | |||||||||||
| 11q22.2 | 19843588 | MMP1 | rs1799750 | 1.43 | 1.2 | 1.69 | 2.7E-05 | 0.037 | 0.974 | 0.577 | 0.999 |
| Hepatocellular carcinoma | |||||||||||
| 12q15 | 21240526 | MDM2 | rs2279744 | 1.57 | 1.36 | 1.8 | 1.0E-10 | <0.001 | <0.001 | 0.002 | 0.632 |
| Leukemia | |||||||||||
| 1p13.3 | Dong | GSTM1 | GSTM1 null | 1.2 | 1.14 | 1.25 | 8.6E-15 | <0.001 | <0.001 | <0.001 | <0.001 |
| 22q11.23 | Dong | GSTT1 | GSTT1 null | 1.19 | 1.14 | 1.29 | 3.5E-08 | 0.023 | 0.960 | 0.039 | 0.976 |
| Lung cancer | |||||||||||
| 1p13.3 and 22q11.23 | 19124497 | GSTM1, GSTT1 | GSTM1 and GSTT1 present | 0.71 | 0.62 | 0.82 | 3.2E-06 | 0.004 | 0.797 | 0.177 | 0.995 |
| 10q26.3 | 20031389 | CYP2E1 | rs2031920 | 0.8 | 0.72 | 0.89 | 4.1E-05 | 0.039 | 0.976 | 0.153 | 0.994 |
| 12q15 | Dong | MDM2 | rs2279744 | 1.27 | 1.12 | 1.44 | 2.E-04 | 0.162 | 0.995 | 0.505 | 0.999 |
| 16q12.2 | 20360147 | MMP2 | rs2285053 | 0.72 | 0.61 | 0.85 | 1.0E-04 | 0.113 | 0.992 | 0.713 | 1.000 |
| rs243865 | 0.55 | 0.48 | 0.63 | 6.2E-18 | <0.001 | <0.001 | <0.001 | <0.001 | |||
| 19q13.31 | Dong | XRCC1 | rs25487 | 1.34 | 1.16 | 1.54 | 5.E-05 | 0.038 | 0.975 | 0.383 | 0.998 |
| 19116388 | rs3213245 | 1.46 | 1.25 | 1.7 | 1.E-06 | 0.002 | 0.633 | 0.159 | 0.995 | ||
| 19q13.32 | Dong | ERCC2 | rs13181 | 1.3 | 1.13 | 1.49 | 2.E-04 | 0.143 | 0.994 | 0.566 | 0.999 |
| Nasopharyngeal cancer | |||||||||||
| 1p13.3 | 19338664 | GSTM1 | GSTM1 null | 1.42 | 1.21 | 1.66 | 1.1E-05 | 0.014 | 0.935 | 0.383 | 0.998 |
| Non-Hodgkin lymphoma | |||||||||||
| 4p14 | 19029192 | TLR | rs4833103 | 0.75 | 0.64 | 0.87 | 2.E-04 | 0.134 | 0.994 | 0.639 | 0.999 |
| Prostate cancer | |||||||||||
| 1q25.3 | Dong | RNASEL | rs627928 | 1.27 | 1.13 | 1.44 | 1.E-03 | 0.162 | 0.995 | 0.505 | 0.999 |
| 8p21.2 | 20564319 | NKX3-1 | rs1512268 | 1.17 | 1.12 | 1.23 | 2.E-12 | <0.001 | <0.001 | <0.001 | <0.001 |
| 17q24.3 | 20564319 | LOC124685 | rs1859962 | 1.21 | 1.12 | 1.3 | 8.E-07 | <0.001 | 0.161 | <0.001 | 0.318 |
| 19q13.2 | 20564319 | LOC644330 | rs887391 | 1.14 | 1.08 | 1.2 | 7.E-07 | <0.001 | 0.356 | <0.001 | 0.362 |
Abbreviations: CI, confidence intervals; FPRP, false-positive report probabilities; OR, odd ratios.
FPRPs in bold indicate values that are ≤0.2 and are therefore considered noteworthy.
Table does not include associations for which FPRPs could not be calculated due to missing ORs and CIs.
‘Dong' indicates variants reported in previous systematic review of meta-analyses (Dong et al8).
Italicized P-values indicate those derived from calculation using methods described in Wacholder et al.12 All other values are as reported in source noted.
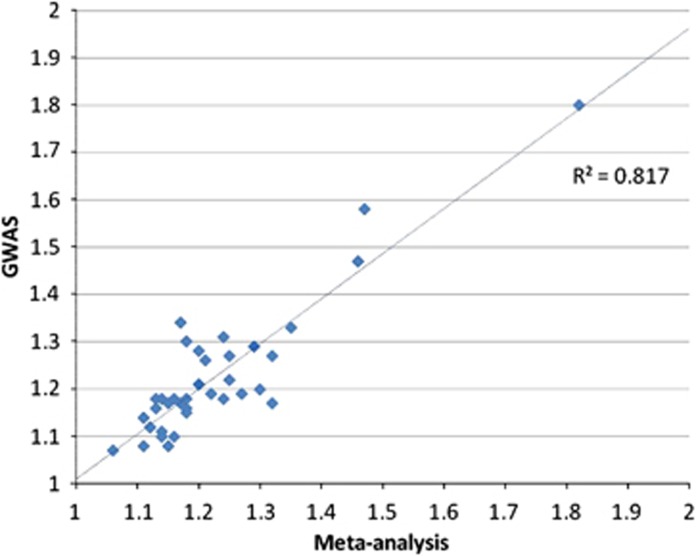
Meta-analyses and GWAS that examined the same variants (direct overlap) reported very similar ORs (Figure 2). All but three associations had ORs between 1.00 and 1.50. The largest effect sizes were observed for esophageal cancer and ALDH2 rs671 (heterozygous) in both meta-analysis (OR=2.52) and GWAS (OR=3.48).
Figure 2.
Odds ratios of variants common to candidate gene meta-analyses or pooled analyses (MA) and GWAS excluding ALDH2 in esophageal cancer (meta-analysis OR=2.52, GWAS OR=3.48).
Discussion
We summarized the principal findings from a decade of published genetic associations with incident cancer. We found that meta-analyses and pooled analyses of candidate gene studies had identified 349 statistically significant associations and GWAS identified 269. Very few associations were found in both groups; however, variant-cancer associations that were reported in both meta-analyses and GWAS had comparable effect sizes.
When we stratified on the basis of cancer type, there was considerable variation in the relative numbers of associations identified by meta-analyses and GWAS. For example, meta-analysis of candidate genes identified 80 breast cancer variants, versus 36 identified by GWAS. In contrast, meta-analysis found only four leukemia variants, compared with 32 identified by GWAS. The difference in the number of significant associations between the meta-analyses of candidate gene studies and GWAS could reflect variations in research interest, prevalence, or underlying knowledge of pathogenesis of different cancers.
Candidate gene studies and GWAS use different thresholds to define statistical significance. We used a P-value threshold of 0.05 for candidate gene studies and 1.0 × 10−5 for GWAS; the latter is used by the NHGRI GWAS Catalog, although 5 × 10−8 is more widely accepted in the literature today. These thresholds are consistent with those used in the original studies; however, it has been suggested that P=0.05 may be too lenient for candidate genes studies13 and P=5 × 10−8 may be too stringent for GWAS.14, 15 If this is true, then differences in the number of significant associations could also reflect an excess of false-positive findings from candidate gene studies13 and an excess of false negatives from GWAS.14 It has been suggested that in GWAS, where false negatives outnumber false positives, lowering the significance threshold to 10−7 would yield mostly genuine discoveries.15 Others have suggested that 10−7 be held as the criterion for early commercial genotyping arrays, but the standard 5 × 10−8 for current or merged commercial arrays.16 False-positive findings due to too lenient thresholds may be particularly pertinent for candidate gene studies that examine several variants and do not correct for multiple testing.
We identified noteworthy associations by calculating FPRPs as described by Wacholder et al.12 The FPRP for a genetic association takes into account not only the observed P-value but also the prior probability of the association and the statistical power of the test. We found 189 noteworthy associations in addition to the 13 previously reported by Dong et al.8 Most of these noteworthy associations were identified in GWAS; however, 39 were found exclusively in meta-analyses of candidate gene associations.
Meta-analysis of candidate gene association studies diminishes, but does not entirely exclude, random error and bias as causes of false-positive associations. GWAS also have challenges; in particular, the actual fraction of the genome interrogated in a GWAS varies with the genotyping platform and study population.17 Although imputation methods may help increase the genomic coverage, they are not perfect, especially for variants of lower frequency. For example, in an attempt to unify candidate gene and GWAS approaches in asthma, Michel et al18 found that GWAS coverage was insufficient for many asthma candidate genes.
More in-depth analysis in future studies could further elucidate why 39 candidate gene associations did not reproduce in GWAS. Insufficient power due to the limited ability of GWAS to detect rare variants may have a role. Candidate gene studies are not suited for the study of exceptionally rare variants either, not without incredibly large sample sizes. Uncommon variants, however, which include CHEK2, may still have frequencies too low to be detected through GWAS.4 The CHEK2 1100delC mutation, an established genetic risk factor for breast cancer, was found in 0.7% of cases and 0.4% of controls in a Swedish study population.19 Despite many GWAS conducted in breast cancer, CHEK2 has not passed the 1 × 10−5 threshold (as reported by the NHGRI GWAS Catalog).11 It is important to add, however, that the mutation was not discovered by candidate gene methods but by studying families with Li–Fraumeni syndrome.20 As in candidate gene studies, inadequate sample size should also be considered as a possible source of insufficient power. Significant positive correlations have been noted between the number of novel SNPs detected and the sample size of GWAS.21
In our study, the 41 associations common to both meta-analysis and GWAS had effect sizes that were generally similar and mostly small. A notable outlier is the association of ALDH2 rs671 risk for esophageal cancer, which has been described by three meta-analyses and one GWAS since 2000. ALDH2 encodes a key enzyme in the metabolism of consumed alcohol, which is a major epidemiologic risk factor for esophageal cancer. A 2009 paper by Khoury and Wacholder notes that very few association studies have considered gene–environment interactions, and that incorporating both genetic and environmental factors in the analysis may be one path to finding additional associations and larger effect sizes but may require extremely large sample sizes to achieve sufficient power.22 Other methodological challenges unique to genome-wide environmental interaction studies exist, which can perhaps explain the low number of publications in this field.23
Our analysis had some limitations. By considering only meta-analyses of candidate genetic associations, we could have left out some recent individual candidate gene studies with sufficiently large sample sizes to find noteworthy associations. By considering only the main associations in candidate gene meta-analyses, we could have overlooked important subgroup associations, such as some that seem to be race- or ethnicity-specific associations.24 We also did not use linkage disequilibrium between markers when defining indirect overlap but relied on physical distance. It is known that linkage disequilibrium and physical distance are correlated.25 Markers that are located close to each other generally exhibit higher linkage disequilibrium than those that are located further apart. Although a distance of 1 Mb may be considered large for identifying overlapping associations, at least one GWAS has traced an association to a causal variant located at roughly this distance away from the original signal.26 It should also be noted that reducing this distance would not change our conclusion that there is limited overlap between the two study types. Finally, we attempted to avoid duplication of cases by limiting our analysis to only the most recent meta-analysis and GWAS for each association. Nevertheless, this possibility cannot be completely excluded, especially because GWAS are often assembled from previously ascertained groups of cases and controls.
One criticism of candidate gene studies is that most genetic associations are not replicated in subsequent studies.2 Similar to GWAS, findings from the candidate gene studies must demonstrate replication to be considered valid. Meta-analysis of the published literature is an important tool in assessing the cumulative evidence on genetic associations.27 Consortia offer another approach to meta-analysis that may help protect against the effects of selective reporting and publication bias. In a study comparing meta-analyses of individual case-control studies with consortium analyses in breast cancer, the authors concluded that meta-analyses and consortia-wide analyses were complementary.28 Consortium-based analyses may be particularly useful for detecting variants modified by weak-to-moderate gene–environment interactions.29 Meta-analysis has also become increasingly popular in GWAS,30 where it can aid in exploring the heterogeneity across data sets and identifying more disease-related genes.31 In 2011, there were 173 publications on meta-analyses and pooled analyses of candidate genes in cancer, and 39 GWAS, of which 6 included a meta-analysis.9
In light of improved genetic sequencing technologies, some discussions on the future roles of GWAS and candidate gene studies are appropriate. One of the limitations of current GWAS technology is its limited ability to detect low-frequency variants. A study by Siu et al32 found that GWAS coverage of rare variants was still inadequate despite using chips designed to detect them. In addition, the quality of imputed low-frequency and, especially, rare variants in these studies is generally lower than that for common variants.33 Still, arrays and reference panels have improved much since the advent of GWAS, the most recent of which was not included in our analysis. It has been estimated that previous GWAS have detected less than 20% of all independent GWAS-detectable SNPs in chronic diseases, but future GWAS can potentially detect more SNPs through improved coverage and, especially, sample sizes.21
Studies that use recently developed arrays such as MetaboChip,34 ImmunoChip,35 and iCOGS array36 represent the latest reinvention of the candidate gene study. These chips can contain hundreds of thousands of SNPs that were chosen for replicating and fine-mapping loci identified from GWAS, as well as to cover the most promising candidate genes. A recent consortium-based meta-analysis that used the iCOGS array identified 23 new prostate cancer susceptibility loci.37 Next-generation sequencing is also increasingly helping to improve the understanding of genetic association studies.32 Projects such as ENCODE are likely to provide new insights into GWAS associations in non-coding regions of the genome.38 Together, these multiple approaches will help us identify additional genetic associations and understand their functional implications.
Acknowledgments
We would like to acknowledge the contributions of Camilla Marie Benedicto, for her role in the initiation of this analysis and Tram Kim Lam, Elizabeth M Gillanders, and Carolyn M Hutter for their thoughtful comments on the manuscript.
The authors declare no conflicts of interests. The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the Department of Health and Human Services.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet. 2002;70:124–141. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Gwinn M, Little J, et al. A road map for efficient and reliable human genome epidemiology. Nat Genet. 2006;38:3–5. doi: 10.1038/ng0106-3. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Gillanders EM, Manolio TA. Genetic architecture of cancer and other complex diseases: lessons learned and future directions. Carcinogenesis. 2011;32:945–954. doi: 10.1093/carcin/bgr056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte JS.Genome-wide association studies and beyond Ann Rev Public Health 2010319–20.4 p following 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ML, Monteiro AN, Gagther SA, et al. ‘Principles for the post-GWAS functional characterization of cancer risk loci.'. Nat Genet. 2011;43:513–518. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299:2423–2436. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schully SD, Yu W, McCallum V, et al. Cancer GAMAdb: database of cancer genetic associations from meta-analyses and genome-wide association studies. Eur J Hum Genet. 2011;19:928–930. doi: 10.1038/ejhg.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BK, Clyne M, Walsh M, et al. Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database. A J Epidemiol. 2006;164:1–4. doi: 10.1093/aje/kwj175. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoratou E, Montazeri Z, Hawken S, et al. Systematic meta-analyses and field synopsis of genetic association studies in colorectal cancer. J Natl Cancer Inst. 2012;104:1433–1457. doi: 10.1093/jnci/djs369. [DOI] [PubMed] [Google Scholar]
- Johansen CT, Wang J, McIntyre AD, et al. Excess of rare variants in non-genome-wide association study candidate genes in patients with hypertriglyceridemia. Circ Cardiovasc Genet. 2012;5:66–72. doi: 10.1161/CIRCGENETICS.111.960864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotou OA, Ioannidis JP. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol. 2012;41:273–286. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- Michel S, Liang L, Depner M, et al. Unifying candidate gene and GWAS approaches in asthma. PloS ONE. 2010;5:e13894. doi: 10.1371/journal.pone.0013894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsdottir K, Humphreys K, Bonnard C, et al. Linkage disequilibrium mapping of CHEK2: common variation and breast cancer risk. PLoS Med. 2006;3:e168. doi: 10.1371/journal.pmed.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DW, Varley JM, Szydlo TE, et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- Lindquist KJ, Jorgenson E, Hoffmann TJ, Witte JS. The impact of improved microarray coverage and larger sample sizes on future genome-wide association studies. Genet Epidemiol. 2013;37:383–392. doi: 10.1002/gepi.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ, Wacholder S.Invited commentary: from genome-wide association studies to gene-environment-wide interaction studies—challenges and opportunities Am J Epidemiol 2009169227–230.discussion 234–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H, Lutz S, Maus B, et al. Challenges and opportunities in genome-wide environmental interaction (GWEI) studies. Hum Genet. 2012;131:1591–1613. doi: 10.1007/s00439-012-1192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garte S. The role of ethnicity in cancer susceptibility gene polymorphisms: the example of CYP1A1. Carcinogenesis. 1998;19:1329–1332. doi: 10.1093/carcin/19.8.1329. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Noguchi E, Heinzmann A, et al. Extent and distribution of linkage disequilibrium in three genomic regions. Am J Hum Genet. 2001;68:191–197. doi: 10.1086/316944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M, Saut N, Oudot-Mellakh T, et al. Caution in interpreting results from imputation analysis when linkage disequilibrium extends over a large distance: a case study on venous thrombosis. PLoS ONE. 2012;7:e38538. doi: 10.1371/journal.pone.0038538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int Jl Epidemiol. 2008;37:120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- Janssens AC, Gonzalez-Zuloeta Ladd AM, Lopez-Leon S, et al. An empirical comparison of meta-analyses of published gene-disease associations versus consortium analyses. Genet Med. 2009;11:153–162. doi: 10.1097/GIM.0b013e3181929237. [DOI] [PubMed] [Google Scholar]
- Nickels S, Truong T, Hein R, et al. Evidence of gene–environment interactions between common breast cancer susceptibility loci and established environmental risk factors. PLoS Genet. 2013;9:e1003284. doi: 10.1371/journal.pgen.1003284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesupriya A, Yu W, Clyne M, Gwinn M, Khoury MJ. The continued need to synthesize the results of genetic associations across multiple studies. Genet Med. 2008;10:633–635. doi: 10.1097/gim.0b013e3181815360. [DOI] [PubMed] [Google Scholar]
- Zeggini E, Ioannidis JP. Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009;10:191–201. doi: 10.2217/14622416.10.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu H, Zhu Y, Jin L, Xiong M. Implication of next-generation sequencing on association studies. BMC Genomics. 2011;12:322. doi: 10.1186/1471-2164-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Chen C, Liang L, et al. One thousand genomes imputation in the national cancer institute breast and prostate cancer cohort consortium aggressive prostate cancer genome-wide association study. Prostate. 2012;73:677–689. doi: 10.1002/pros.22608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kang HM, Ding J, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A, Brown MA. Promise and pitfalls of the immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahcall OG. iCOGS collection provides a collaborative model. Nature Genet. 2013;45:343. doi: 10.1038/ng.2592. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Olama AA, Benlloch S, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nature Genet. 2013;45:385–391. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Barrett JC. How next-generation sequencing is transforming complex disease genetics. Trends Genet. 2013;29:23–30. doi: 10.1016/j.tig.2012.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.