Abstract
Methyl groups placed on ϕXsB1 replicative form DNA by the Escherichia coli B modification enzyme are located in the overlap between fragments Mbo II-3 and Alu I-2, a 61-base-pair DNA segment. Mutations that led to loss of susceptibility to restriction by E. coli B occurred within this segment at three positions spanning 14 nucleotides. A sequence difference between ϕXsB1 and ϕXam3cs70, a ϕX174 strain not restricted by E. coli B, occurs at one of these positions. The site on simian virus 40 DNA methylated by the modification enzyme is located in the 115-base-pair overlap between fragments Hae III-I and Alu I-G. The sequences of these segments of ϕXsB1 and simian virus 40 DNA and two regions of phage f1 DNA recognized by the E. coli B restriction enzyme [Ravetch, J. V., Horiuchi, K. & Zinder, N. D. (1978) Proc. Natl. Acad. Sci. USA 75, 2266-2270] contain a homology of nine bases in the configuration:
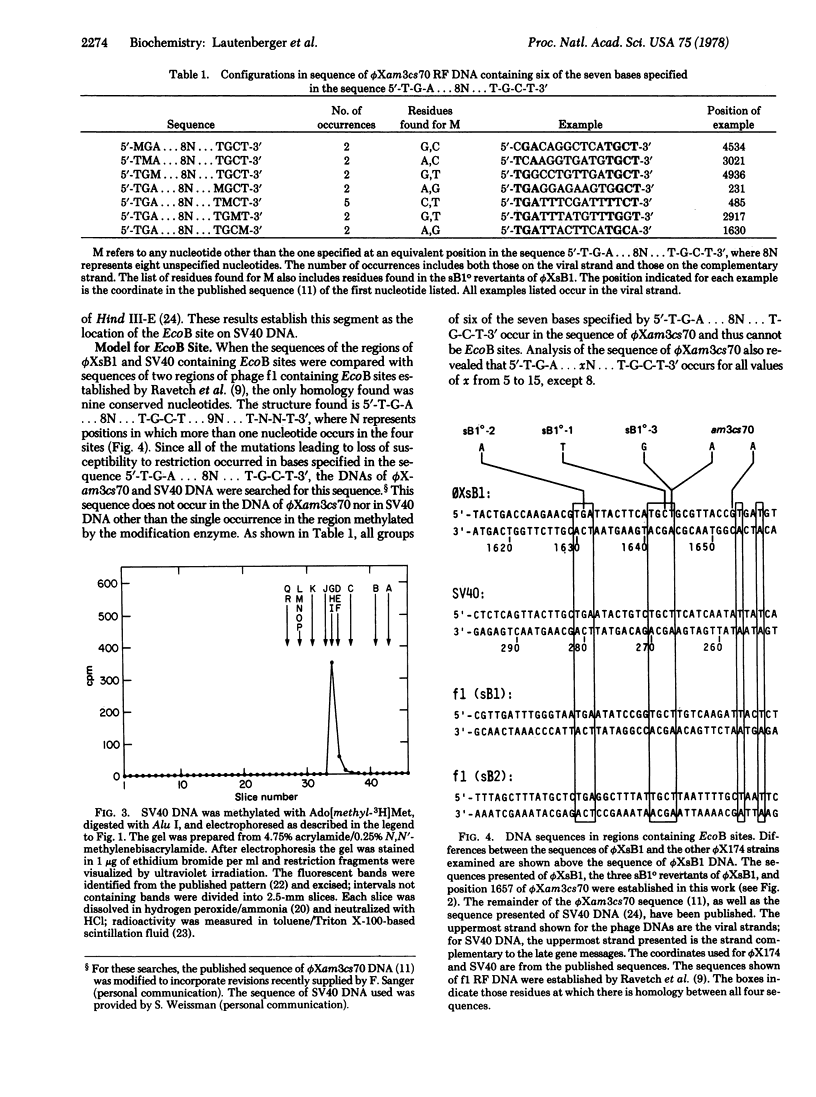
5′-T-G-A... 8N... T-G-C-T... 9N... T-N-N-T-3′.
The sequence 5′-T-G-A... 8N... T-G-C-T-3′ may constitute the restriction enzyme recognition site since it does not occur in ϕXam3cs70 DNA and occurs only once in simian virus 40 DNA, and since all observed mutations leading to loss of the site occur at one of the bases specified by this sequence. Analysis of the sequence of ϕXam3cs70 showed that if no other residues are recognized, all seven of these bases are essential for recognition and the interval between the two groups of specified bases must be precisely eight.
Keywords: DNA methylation, endonucleases, bacteriophage ϕX174
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W., Kühnlein U. Mutationeller Verlust B-spezifischer Restriktion des Bakteriophagen fd. Pathol Microbiol (Basel) 1967;30(6):946–952. [PubMed] [Google Scholar]
- Barnes W. M. DNA sequencing by partial ribosubstitution. J Mol Biol. 1978 Feb 15;119(1):83–99. doi: 10.1016/0022-2836(78)90271-1. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Air G. M., Hutchison C. A., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976 Nov 4;264(5581):34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Contreras R., Rogiers R., Van de Voorde A., Fiers W. Overlapping of the VP2-VP3 gene and the VP1 gene in the SV40 genome. Cell. 1977 Oct;12(2):529–538. doi: 10.1016/0092-8674(77)90129-5. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Kimball M., Linn S., Goodman H. M. Location and nucleotide sequence of the site on SV40 DNA methylated by the ECO B modification methylase. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1133–1140. doi: 10.1016/s0006-291x(74)80401-8. [DOI] [PubMed] [Google Scholar]
- Goldmark P. J., Linn S. Purification and properties of the recBC DNase of Escherichia coli K-12. J Biol Chem. 1972 Mar 25;247(6):1849–1860. [PubMed] [Google Scholar]
- Horiuchi K., Vovis G. F., Enea V., Zinder N. D. Cleavage map of bacteriophage f1: location of the Escherichia coli B-specific modification sites. J Mol Biol. 1975 Jun 25;95(2):147–165. doi: 10.1016/0022-2836(75)90388-5. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Zinder N. D. Cleavage of bacteriophage fl DNA by the restriction enzyme of Escherichia coli B. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3220–3224. doi: 10.1073/pnas.69.11.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. I. Purification, subunit structure, and catalytic properties of the modification methylase. J Biol Chem. 1972 Oct 10;247(19):6176–6182. [PubMed] [Google Scholar]
- Linn S., Lautenberger J. A., Eskin B., Lackey D. Host-controlled restriction and modification enzymes of Escherichia coli B. Fed Proc. 1974 May;33(5):1128–1134. [PubMed] [Google Scholar]
- Lyons L. B., Zinder N. D. The genetic map of the filamentous bacteriophage f1. Virology. 1972 Jul;49(1):45–60. doi: 10.1016/s0042-6822(72)80006-0. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum D., Smith M. Computer processing of DNA sequence data. J Mol Biol. 1977 Oct 15;116(1):29–30. doi: 10.1016/0022-2836(77)90116-4. [DOI] [PubMed] [Google Scholar]
- Middleton J. H., Edgell M. H., Hutchison C. A., 3rd Specific fragments of phi X174 deoxyribonucleic acid produced by a restriction enzyme from Haemophilus aegyptius, endonuclease Z. J Virol. 1972 Jul;10(1):42–50. doi: 10.1128/jvi.10.1.42-50.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panet A., van de Sande J. H., Loewen P. C., Khorana H. G., Raae A. J., Lillehaug J. R., Kleppe K. Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochemistry. 1973 Dec 4;12(25):5045–5050. doi: 10.1021/bi00749a003. [DOI] [PubMed] [Google Scholar]
- Pribnow D. Bacteriophage T7 early promoters: nucleotide sequences of two RNA polymerase binding sites. J Mol Biol. 1975 Dec 15;99(3):419–443. doi: 10.1016/s0022-2836(75)80136-7. [DOI] [PubMed] [Google Scholar]
- Ravetch J. V., Horiuchi K., Zinder N. D. Nucleotide sequence of the recognition site for the restriction-modification enzyme of Escherichia coli B. Proc Natl Acad Sci U S A. 1978 May;75(5):2266–2270. doi: 10.1073/pnas.75.5.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Schnegg B., Hofschneider P. H. Mutant of phi X174 accessible to host-controlled modification. J Virol. 1969 May;3(5):541–542. doi: 10.1128/jvi.3.5.541-542.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R. C., Van de Voorde A., Fiers W. Specific cleavage and physical mapping of simian-virus-40 DNA by the restriction endonuclease of Arthrobacter luteus. Eur J Biochem. 1976 Jan 2;61(1):119–138. doi: 10.1111/j.1432-1033.1976.tb10003.x. [DOI] [PubMed] [Google Scholar]