Abstract
Lymphoproliferative disease virus (LPDV) is an exogenous oncogenic retrovirus that induces lymphoid tumors in some galliform species of birds. Historically, outbreaks of LPDV have been reported from Europe and Israel. Although the virus has previously never been detected in North America, herein we describe the widespread distribution, genetic diversity, pathogenesis, and evolution of LPDV in the United States. Characterization of the provirus genome of the index LPDV case from North America demonstrated an 88% nucleotide identity to the Israeli prototype strain. Although phylogenetic analysis indicated that the majority of viruses fell into a single North American lineage, a small subset of viruses from South Carolina were most closely related to the Israeli prototype. These results suggest that LPDV was transferred between continents to initiate outbreaks of disease. However, the direction (New World to Old World or vice versa), mechanism, and time frame of the transcontinental spread currently remain unknown.
Keywords: Lymphoproliferative disease virus (LPDV), alpharetrovirus, avian tumor viruses, exogenous retrovirus, oncogenesis, Order Galliformes
Introduction
Lymphoproliferative disease virus (LPDV) is an exogenous retrovirus that induces a neoplastic syndrome in domestic turkeys (Meleagris gallopavo), resulting in the formation of lymphoid tumors in multiple organs (Biggs et al., 1978). Although there is circumstantial evidence of LPDV-like outbreaks occurring in domestic turkeys in Europe prior to the 1970’s, the virus was not identified as an etiological agent of disease until 1972 in the United Kingdom (Biggs et al., 1974). Within a few years of its recognition, LPDV outbreaks were documented in additional European countries as well as Israel (Biggs, 1997; Ianconescu et al., 1983). These outbreaks remained confined to the European-Middle Eastern region and the virus has never been detected outside of this area, including North America. Although LPDV is characterized as an oncogenic retrovirus, it does not contain a putative viral oncogene, nor does it appear to exert its oncogenic effects through insertional mutagenesis of a cellular proto-oncogene; thus, the mechanism by which lymphoproliferative disease is induced is unknown (Chajut et al., 1991; Sarid et al., 1994; Gazit and Yaniv, 1999).
In domestic turkeys, disease due to LPDV infection is generally first noted around eight to 10 weeks of age, with a variable flock mortality that can reach 25% (Gazit and Yaniv, 1999; Biggs, 1997). The characteristic lesion of LPDV infection is a pleomorphic infiltrate of lymphocytes, including lymphoblasts intermixed with plasma cells and monocytic reticular cells in multiple tissues. The most commonly affected organs are spleen, thymus, pancreas, and liver, although smaller focal lesions can be present in a variety of other tissues (Biggs, 1997). LPDV has been demonstrated experimentally to infect both turkeys and chickens (Gallus gallus), although not ducks or geese (species not listed; Ianconescu et al., 1983), suggesting that the natural host range of the virus may be restricted to birds of the order Galliformes, which also includes other game birds such as pheasants, quail, and grouse; however, reports of natural LPDV infections have been limited to domestic turkeys. Under experimental conditions, viremia occurs approximately two weeks after infection and may last for up to 10 months (Zimber et al., 1983) and horizontal transmission can occur among birds in close contact (McDougall et al., 1978). How the virus is transmitted in nature (e.g., vector-borne, horizontal/vertical transmission) is uncertain.
Diagnostically, the identification of LPDV has been hampered by the inability to isolate and propagate the virus in an appropriate culture system, including embryonating eggs, primary cell cultures, and established cell lines (Gazit and Yaniv, 1999). However, inoculation of naive turkeys with lymphocytes transfected with an infectious clone resulted in the reproduction of disease and the detection of the virus in blood and tissues, indicating that LPDV is replication-competent (Gak et al., 1989; Gak et al., 1990). Thus, similar to ‘slowly-transforming’ exogenous strains of the related avian leukosis virus (ALV) (genus Alpharetrovirus), LPDV is a non-oncogene-containing, replication-competent, tumorigenic virus (Payne and Venugopal, 2000). Although a number of seminal studies conducted in Israel and the United Kingdom provided insights into the molecular genetics, pathogenicity, and cellular tropism of LPDV, the extent of its genetic diversity and source of emergence remains unknown (Chajut et al., 1992; Gak et al., 1989; Gak et al., 1991, Gazit et al., 1979; Gazit et al., 1982; Gazit et al., 1983; Gazit et al., 1986; McDougal et al., 1978; Yaniv et al., 1979; Zimber et al., 1983).
In January 2009, an adult female wild turkey in Columbia County, Arkansas, was found recumbent and moribund. The turkey died shortly thereafter and was submitted to the Southeastern Cooperative Wildlife Disease Study (SCWDS) at the University of Georgia for diagnostic evaluation. Grossly, this turkey had marked splenomegaly, multiple demarcated tan foci scattered throughout the liver, and a diffuse thickening of the proximal small intestinal wall. Microscopically, infiltrating neoplastic lymphoid cells were identified in various organs, including the spleen, liver, intestines, kidney, pancreas, and lung. Attempts to isolate endemic oncogenic avian viruses, including reticuloendotheliosis virus (REV), Gallid herpesvirus 2 (GaHV-2), and ALV, in a number of avian cell lines and specific-pathogen-free embryonating chicken eggs were unsuccessful. To further investigate a possible viral etiology for the lymphoid neoplasia, spleen, liver, and lung samples were screened for LPDV by PCR targeting a region spanning the p31 and capsid (CA) genes. Proviral LPDV DNA was detected in all three tissues, representing the first detection of LPDV outside of Europe and the Middle East, and the first natural LPDV infection in a wild avian host.
Following this initial identification, we attempted to determine whether LPDV may be a widespread, yet unrecognized, pathogen in the United States by screening select wild turkey diagnostic cases, as well as seasonally harvested wild turkeys, for the virus. In this report, we provide a description of natural LPDV infection in North American wild turkeys, including basic epidemiologic patterns, frequency and description of lesions associated with LPDV infection, the co-occurrence of LPDV with other common avian viral pathogens, and the prevalence of LPDV infection in apparently healthy birds. We conducted a comparative analysis of the proviral genome of North American LPDV and the Old World prototype strain from Israel and mapped the insertion site of LPDV into the host genome. We also performed a phylogenetic analysis of proviruses recovered from birds collected from 18 states between 2009-2012 with that of the Israeli prototype, and examined the evolutionary relationship of LPDV to other retroviruses.
Results
Detection of LPDV within the United States and lesions observed in diagnostic cases
Following the first recognition of LPDV in North America, we tested tissues from select clinically ill wild turkeys submitted to diagnostic laboratories throughout the eastern United States for LPDV proviral DNA. Including the prototype North American strain (12/AR/2009), LPDV was detected in 41 wild turkeys from 18 states, stretching from Maine to Louisiana and west to Colorado (Table 1), encompassing an area covering most of the natural geographic distribution of wild turkeys in the United States (Hatfield and Vance, 2009). The vast majority of LPDV-positive wild turkeys were adults (36/41; 87.8%), but five (12.2%) were hatch-year birds. There was a near even distribution of males (19/41; 46.3%) and females (18/41; 43.9%); sex was not determined for four wild turkeys. Tissues from two of the 41 LPDV-positive wild turkeys were not evaluated microscopically due to the poor post-mortem condition of the carcass. Of the remaining 39 LPDV-positive wild turkeys, only a small minority (6/39; 15.4%) had microscopic lesions consistent with LPDV infection in domestic turkeys (i.e., lymphoid neoplasia). However, as REV was also detected in two of these birds (196/NC/2012 and 453/NJ/2012), LPDV infection in the absence of any other detectable pathogens was diagnosed in only four birds with neoplasia-related mortality (12/AR/2009, 122/WV/2009, 152/GA/2011, 592/MS/2012; Table 1). The remaining 33 LPDV-positive wild turkeys were diagnosed with other causes of morbidity or mortality in the absence of lymphoid neoplasia, including fowlpox virus (FWPV) infection (16/39; 41.0%), bacterial infections, including systemic or skin infections (8/39; 20.5%), trauma (2/39; 5.1%), endoparasitism (1/39; 2.6%), toxicosis (1/39; 2.6%), or undetermined (5/39; 12.8%). In addition to the two birds with lymphoid neoplasia that were positive for both LPDV and REV, another 17 of the LPDV-positive turkeys (19/41; 46.3%) also tested positive for REV proviral DNA and 12 of 41 turkeys (29.3%) were simultaneously positive for LPDV, REV, and FWPV (Table 1).
Table 1.
LPDV strains recovered in the United States from 2009-2012 that were analyzed during the study. LPDV-positive birds that were also positive for reticuloendotheliosis virus (REV) and/or fowlpox virus (FWPV) are indicated.
| Virusa | County | State | Date | Collection methodb | GenBankIDc | FWPV | REV |
|---|---|---|---|---|---|---|---|
| 3/KS/2009 | Logan | Kansas | 2009 | Diagnostic case | KC801949 | ✕ | ✕ |
| 12/AR/2009 | Columbia | Arkansas | 2009 | Diagnostic case | KC802224 | ✕ | ✕ |
| 122/WV/2009 | Randolph | West Virginia | 2009 | Diagnostic case | KC801950 | ✕ | ✕ |
| 286/WV/2009 | Upshur | West Virginia | 2009 | Diagnostic case | KC801993 | ✕ | ✕ |
| 47/WV/2010 | Upshur | West Virginia | 2010 | Diagnostic case | KC801994 | ✕ | ✕ |
| 183/WV/2010 | Wayne | West Virginia | 2010 | Diagnostic case | KC801951 | ✕ | ✕ |
| 27h/SC/2011 | Barnwell | South Carolina | 2011 | Seasonal harvest | KC801981 | * | * |
| 28h/SC/2011 | Barnwell | South Carolina | 2011 | Seasonal harvest | KC802003 | * | * |
| 34h/SC/2011 | Barnwell | South Carolina | 2011 | Seasonal harvest | KC801982 | * | * |
| 35h/SC/2011 | Barnwell | South Carolina | 2011 | Seasonal harvest | KC801983 | * | * |
| 37h/SC/2011 | Barnwell | South Carolina | 2011 | Seasonal harvest | KC801984 | * | * |
| 38h/SC/2011 | Barnwell | South Carolina | 2011 | Seasonal harvest | KC801985 | * | * |
| 48h/SC/2011 | Aiken | South Carolina | 2011 | Seasonal harvest | KC801986 | * | * |
| 52h/SC/2011 | Aiken | South Carolina | 2011 | Seasonal harvest | KC801987 | * | * |
| 54h/SC/2011 | Aiken | South Carolina | 2011 | Seasonal harvest | KC801988 | * | * |
| 59h/SC/2011 | Aiken | South Carolina | 2011 | Seasonal harvest | KC801989 | * | * |
| 61h/SC/2011 | Aiken | South Carolina | 2011 | Seasonal harvest | KC802004 | * | * |
| 62h/SC/2011 | Aiken | South Carolina | 2011 | Seasonal harvest | KC801976 | * | * |
| 65h/SC/2011 | Aiken | South Carolina | 2011 | Seasonal harvest | KC801990 | * | * |
| 67h/SC/2011 | Dorchester | South Carolina | 2011 | Seasonal harvest | KC802005 | * | * |
| 68h/SC/2011 | Dorchester | South Carolina | 2011 | Seasonal harvest | KC801975 | * | * |
| 69h/SC/2011 | Dorchester | South Carolina | 2011 | Seasonal harvest | KC801991 | * | * |
| 70h/SC/2011 | Dorchester | South Carolina | 2011 | Seasonal harvest | KC801992 | * | * |
| 90/WV/2011 | Logan | West Virginia | 2011 | Diagnostic case | KC801995 | * | ✕ |
| 152/GA/2011 | Putnam | Georgia | 2011 | Diagnostic case | KC801952 | ✕ | ✕ |
| 141/CO/2012 | Las Animas | Colorado | 2012 | Diagnostic case | KC801953 | ✕ | ✕ |
| 71/MO/2012 | St. Clair | Missouri | 2012 | Diagnostic case | KC801954 | ✕ | ✕ |
| 95/WV/2012 | Morgan | West Virginia | 2012 | Diagnostic case | KC801955 | ☑ | ☑ |
| 159/PA/2012 | Clearfield | Pennsylvania | 2012 | Diagnostic case | KC801996 | ✕ | ☑ |
| 163-A/NY/2012 | Columbia | New York | 2012 | Diagnostic case | KC801997 | ✕ | ☑ |
| 163-B/NY/2012 | Columbia | New York | 2012 | Diagnostic case | KC801998 | ✕ | ☑ |
| 165-A/ME/2012 | Piscataquis | Maine | 2012 | Diagnostic case | KC801999 | ☑ | ☑ |
| 165-C/ME/2012 | Piscataquis | Maine | 2012 | Diagnostic case | KC802000 | ✕ | ✕ |
| 169/MD/2012 | Allegany | Maryland | 2012 | Diagnostic case | KC802001 | * | ☑ |
| 196/NC/2012 | Bertie | North Carolina | 2012 | Diagnostic case | KC802002 | ✕ | ☑ |
| 205/MO/2012 | Howard | Missouri | 2012 | Diagnostic case | KC801956 | * | ✕ |
| 216/PA/2012 | Lackawanna | Pennsylvania | 2012 | Diagnostic case | KC801957 | * | ✕ |
| 217/LA/2012 | East Feliciana | Louisiana | 2012 | Diagnostic case | KC801958 | * | ✕ |
| 453/NJ/2012 | Sussex | New Jersey | 2012 | Diagnostic case | KC801959 | ✕ | ☑ |
| 514/OH/2012 | Hancock | Ohio | 2012 | Diagnostic case | KC801977 | ✕ | ✕ |
| 517-B/ME/2012 | Cumberland | Maine | 2012 | Diagnostic case | KC801960 | ✕ | ☑ |
| 535/PA/2012 | Blair | Pennsylvania | 2012 | Diagnostic case | KC801961 | ☑ | ✕ |
| 560/PA/2012 | Huntingdon | Pennsylvania | 2012 | Diagnostic case | KC801962 | ☑ | ☑ |
| 592/MS/2012 | Monroe | Mississippi | 2012 | Diagnostic case | KC801963 | ✕ | ✕ |
| 604-A/NH/2012 | Concord | New Hampshire | 2012 | Diagnostic case | KC801964 | ☑ | ☑ |
| 604-B/NH/2012 | Sutton | New Hampshire | 2012 | Diagnostic case | KC801965 | ☑ | ☑ |
| 604-C/NH/2012 | Lisbon | New Hampshire | 2012 | Diagnostic case | KC801966 | ☑ | ☑ |
| 604-D/NH/2012 | Bradford | New Hampshire | 2012 | Diagnostc case | KC801967 | ☑ | ☑ |
| 604-E/NH/2012 | Hinsdale | New Hampshire | 2012 | Diagnostic case | KC801968 | ☑ | ☑ |
| 604-F/NH/2012 | Dublin | New Hampshire | 2012 | Diagnostic case | KC801969 | ☑ | ☑ |
| 621/NC/2012 | Hertford | North Carolina | 2012 | Diagnostic case | KC801978 | ☑ | ✕ |
| 640/IN/2012 | Orange | Indiana | 2012 | Diagnostic case | KC801979 | * | ✕ |
| 648/WV/2012 | Braxton | West Virginia | 2012 | Diagnostic case | KC801970 | * | ✕ |
| 652/NC/2012 | Camden | North Carolina | 2012 | Diagnostic case | KC801971 | ☑ | ☑ |
| 654/MS/2012 | Copiah | Mississippi | 2012 | Diagnostic case | KC801972 | ☑ | ✕ |
| 655/MS/2012 | Perry | Mississippi | 2012 | Diagnostic case | KC801973 | ✕ | ✕ |
| 660/SC/2012 | Williamsburg | South Carolina | 2012 | Diagnostic case | KC801980 | ☑ | ☑ |
| 667/MS/2012 | Stone | Mississippi | 2012 | Diagnostic case | KC801974 | ☑ | ☑ |
Virus identification: case number/state of collection/year of collection
Collection method: Diagnostic clinical case (entire bird) or seasonally harvested sample (liver only)
Other than the index case 12/AR/2009, GenBank accession numbers refer to p31/capsid sequences
FWPV = Fowlpox virus; REV = Reticuloendotheliosis virus; * = not tested; ✕ = negative; ☑ = positive
Of the six wild turkeys determined to have died from lymphoid neoplasia in which LPDV was detected, five were adults and one was a hatch-year bird. There was an even sex distribution among male and female birds. Each of the six cases were isolated disease events involving a single bird and all affected wild turkeys were found in a moribund condition exhibiting non-specific clinical signs including lethargy, disorientation, ataxia, and recumbency. Entire carcasses from five of the birds and tissues (skin, bone marrow, and liver) from one bird were submitted for diagnostic examination. All five of the examined wild turkey carcasses were emaciated. Other gross lesions included splenomegaly (3/5; 60%), tan nodules in the liver (2/5; 40%), proliferative growths in the unfeathered skin of the head and legs (2/5; 40%), and diffuse or nodular thickening of the intestinal wall (2/5; 40%) (Figure 1). Microscopically, the five wild turkey carcasses had infiltrates of neoplastic lymphoid cells in tissues from multiple organs, including liver (5/5; 100%), kidney (4/5; 80%), lung (4/5; 80%), spleen (3/5; 60%), gastrointestinal tract (2/5; 40%), skin (2/5; 40%), heart (2/5; 40%), brain (1/5; 20%), adrenal gland (1/5; 20%), pancreas (1/5; 20%), and skeletal muscle (1/5; 20%). The wild turkey for which only tissues were submitted had similar neoplastic infiltrates in the skin and bone marrow, but not in the liver. In all six cases, neoplastic round cells morphologically consistent with lymphocytes (often intermixed with variable numbers of plasma cells) formed densely cellular sheets that effaced and expanded the underlying parenchyma (Figure 2), concordant with lesions observed in LPDV-infected domestic turkeys (Biggs, 1997). In some cases, the neoplastic cells also formed coalescing nodules (most often in skin) and perivascular aggregates. Neoplastic cell populations were generally pleomorphic and cells had scant, eosinophilic cytoplasm and round, hyperchromatic nuclei with inapparent nucleoli and rare to occasional mitotic figures. Occasional necrotic foci were scattered among some of the more densely cellular areas in some tissues, most commonly in the liver and skin. The neoplastic lymphoid infiltrates were most reliably recognized in LPDV cases with little autolysis, as poor post-mortem condition limits detailed examination and interpretation of lesions and thus could hinder detection of cases. There were no obvious differences in the character, distribution, or frequency of lesions between the four LPDV-positive and two LPDV/REV-positive turkeys with lymphoid neoplasia.
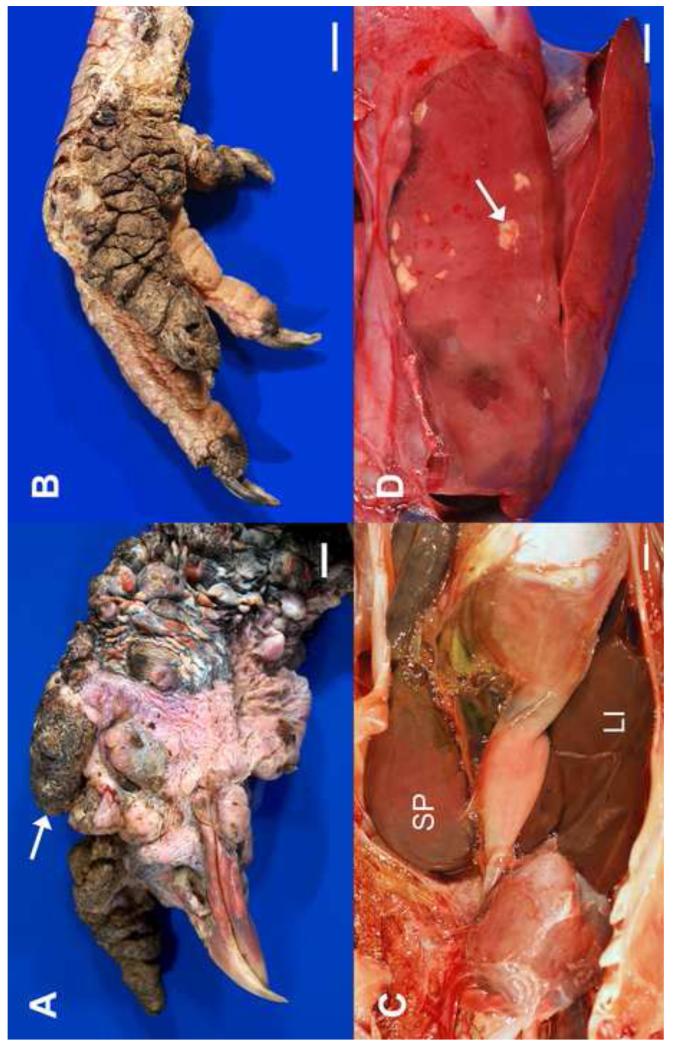
Figure 1.
Gross lesions in adult wild turkeys infected with LPDV. (A) The sparsely-feathered skin over much of the head and neck is covered by variably-sized, proliferative nodules, some of which contain superficial crusts (arrow); (B) The skin over two digits and the plantar aspect of the foot is severely thickened with multiple folds and is covered by dark, dry crusts; (C) The spleen (SP) is markedly enlarged (8.0 × 3.8 × 3.3 cm), a characteristic gross lesion also observed in domestic birds (see Gazit and Yaniv, 1999), (LI = liver); (D) The liver contains numerous, irregular, variably-sized pale foci, corresponding to aggregates of pleomorphic lymphoid cells (arrow). All scale bars = 1.0 cm.
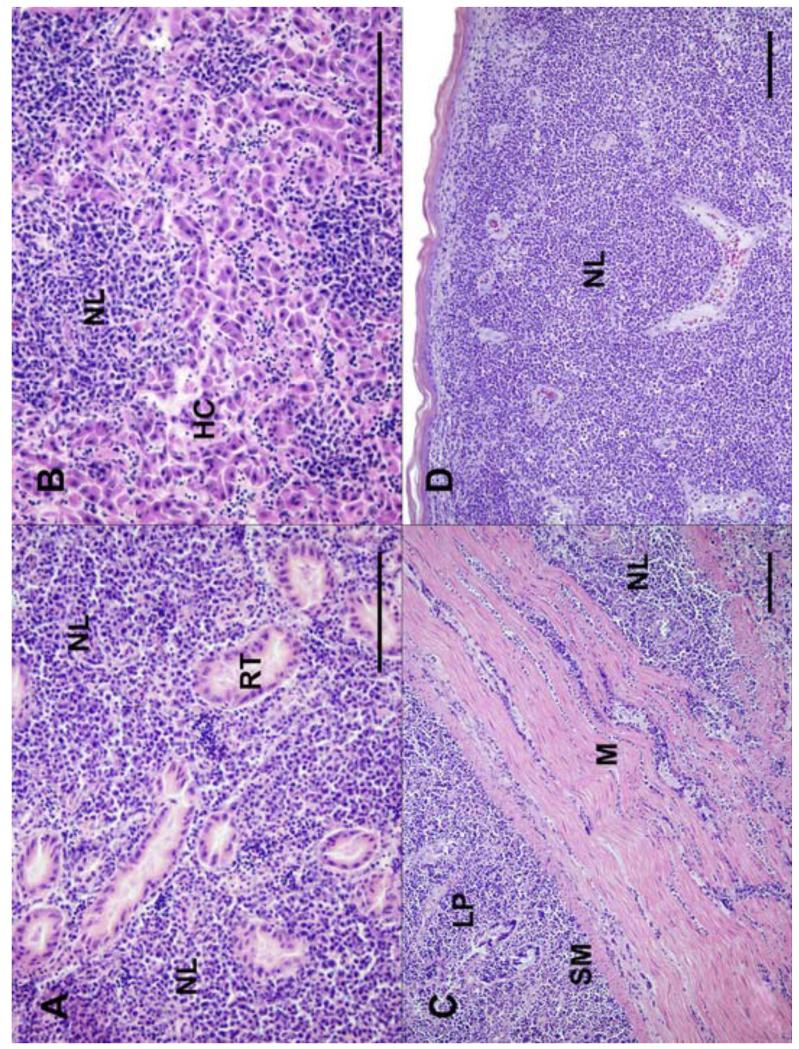
Figure 2.
Tissues from adult wild turkeys infected with LPDV. (A) Kidney: The interstitium is markedly expanded and effaced by sheets of neoplastic lymphocytes (NL). Remnant cross sections of renal tubules (RT) are completely surrounded by the neoplastic cells; (B) Liver: Most of the normal architecture is effaced by neoplastic lymphocytes. Remnant hepatic cords (HC) are compressed and distorted; (C) Intestine: There is transmural infiltration, marked expansion of the lamina propria (LP) and submucosa (SM), and multifocal perimysial expansion of underlying muscular tunics (M) by neoplastic lymphocytes; (D) Skin: The dermis is expanded and obscured by sheets of neoplastic lymphocytes that efface adnexa and there is mild orthokeratotic hyperkeratosis of the overlying epidermis. All scale bars = 100 μm. All images stained with hematoxylin and eosin.
Detection of LPDV in asymptomatic seasonally-harvested wild turkeys
To obtain preliminary insights into the prevalence of LPDV infection in apparently healthy wild turkeys, we tested livers from 74 hunter-collected turkeys from 10 different counties in South Carolina during 2010. Thirty-three (44.6%) of the birds tested were positive for LPDV proviral DNA by PCR. The remaining 41 (55.4%) were negative. Thus, nearly half of the birds tested in South Carolina were positive for LPDV infection, suggesting widespread prevalence within the state. None of the 74 wild turkeys were reportedly exhibiting overt signs of disease when harvested or had gross lesions identified during routine processing. However, microscopic examination of tissues for evidence of neoplasia was not performed on tissues from these birds.
Genomic analysis of the prototype North American LPDV
The complete proviral genome of the first LPDV case identified in North America (12/AR/2009) was PCR-amplified from liver tumor tissue. The proviral genome of 12/AR/2009 is 7,432 nucleotides in length (including both LTRs) and is flanked by two six-base pair repeats derived from the host (CCCAGC). The length of the LTRs, genes, and proteins are shown in Table 2. For 12/AR/2009, the tRNATrp primer binding site is 329-UGGUGACCCCGACGUGAU-346 and the polypurine tract is 7096-GGGGAGGGGGA-7106, identical to the Israeli strain of LPDV and very similar to other alpharetroviruses (Gak et al., 1991; Whitcomb et al., 1995). Comparison of the 12/AR/2009 genome to the prototype Israeli strain (GenBank accession U09568) demonstrated a nucleotide identity of 87.9%, with both viruses containing the same genomic organization of 5′-LTR-gag-pro-pol-env-LTR-3′. Nucleotide conservation among the major ORFs in the two prototypes was similar, with pol exhibiting the highest degree of identity (90.1%), followed by gag (88.3%), pro (88.3%), and env (86.6%). Although most genes were of similar length, the predicted SU protein of strain 12/AR/2009 was 47 amino acids longer than the Israeli prototype due to an extended N-terminus. In this context, the start codon for SU in 12/AR/2009 is positioned one nucleotide downstream from the previously identified splice acceptor site for env (Sarid et al., 1994).
Table 2.
Genomic characteristics of the North American prototype strain of LPDV.
| Gene/domain | Region | nt | aa | MW | pI | Position |
|---|---|---|---|---|---|---|
| U3 | 5′ LTR | 207 | * | * | * | 1-207 |
| R | 5′ LTR | 22 | * | * | * | 208-229 |
| U5 | 5′ LTR | 96 | * | * | * | 230-326 |
| Matrix (MA) | gag | 459 | 153 | 16.67 | 5.12 | 384-842 |
| p31 | gag | 708 | 236 | 25.78 | 5.14 | 843-1550 |
| Capsid (CA) | gag | 699 | 233 | 25.55 | 8.96 | 1551-2249 |
| Nucleocapsid (NC) | gag | 294 | 98 | 10.18 | 10.78 | 2250-2543 |
| Protease (PR) | pro | 444 | 148 | 15.98 | 8.87 | 2450-2893 |
| Reverse transcriptase (RT) | pol | 1743 | 581 | 65.34 | 6.75 | 2887-4629 |
| RT domain | pol | 834 | 278 | * | * | 2887-3720 |
| Tether (t) domain | pol | 519 | 173 | * | * | 3721-4239 |
| RNase H domain | pol | 390 | 130 | * | * | 4240-4629 |
| Integrase (IN) | pol | 810 | 270 | 30.10 | 9.92 | 4630-5439 |
| Surface (SU) | env | 972 | 324 | 35.99 | 9.20 | 5459-6430 |
| Transmembrane (TM) | env | 660 | 220 | 24.84 | 6.21 | 6431-7090 |
| U3 | 3′ LTR | 207 | * | * | * | 7107-7313 |
| R | 3′ LTR | 22 | * | * | * | 7314-7335 |
| U5 | 3′ LTR | 97 | * | * | * | 7336-7432 |
MW = molecular weight (kDa); pI = isoelectric point (pH); * = not applicable
Similar to members of the Alpharetrovirus genus, the pol polyprotein of LPDV is produced by ribosomal frameshifting (Sarid et al., 1994). However, in contrast to alpharetroviruses, the viral protease (pro) of LPDV is also produced by a -1 ribosomal frameshift initiated by a conserved 2447-GGGAAAC-2453 signal sequence and predicted stem-loop structure, thus being more similar to betaretroviruses in this instance (Sarid et al., 1994; Gruber et al., 2008; Stoye et al., 2011). In conjunction with pro not being expressed as part of gag as it is in other alpharetroviruses, the LPDV protease contains an Asp-Thr-Gly active site rather than the thermodynamically less stable and less active Asp-Ser-Gly triplet (Ingr et al., 2003). Comparison of the pol polyprotein [reverse transcriptase (RT) and integrase (IN) proteins; 850 amino acids] of the North American prototype of LPDV to other retroviruses demonstrated that it shared the highest amino acid identity to the Israeli LPDV prototype (92.9%), followed by Rous sarcoma virus (RSV) (44.0%) and ALV (43.7%), in accordance with the phylogenetic analysis of pol (see Evolutionary analysis of LPDV below). Other LPDV proteins that are generally more conserved among the retroviruses, such as the CA and matrix (MA) proteins, also shared similar levels of amino acid identity as pol to other alpharetroviruses, although the nucleocapsid (NC; 98 amino acids) of LPDV exhibits a moderate amino acid identity (38.7%) to simian immunodeficiency virus (SIV) from NC residues 6-96, including similar zinc finger (Cys-X2-Cys-X4-His-X4-Cys) motifs. Thus, although LPDV is most closely related to other avian oncogenic viruses of the genus Alpharetrovirus, it shares some identifiable characteristics with other closely related genera (i.e., Betaretrovirus frameshift strategies and Lentivirus NC homology).
A functional role for the p31 protein of LPDV (located between the MA and CA proteins) (Table 2) has not previously been ascribed (Sarid et al., 1994). However, two putative L domain motifs (PTAP and PPXY) separated by 11 amino acids were identified near the N-terminus, suggesting that p31 may recruit cellular factors that mediate budding of virions from infected cells (Freed, 2002; Pincetic and Leis, 2009). Additionally, the C-terminus of p31 in LPDV contains an LTDWGLVATEC motif similar to that of the RSV p10 protein that has been demonstrated to be critical for immature particle assembly, suggesting that p31 may also interact with CA (Phillips et al., 2008). The only large gap in the proviral genome sequence between the North American and Israeli LPDV prototypes was found in the U3 region of the LTR, where the approximate equivalent of nucleotide positions 21 to 49 (or 24 to 52) in the Israeli strain was missing in 12/AR/2009. Although this missing stretch of nucleotides is just upstream of direct repeat elements shown to be essential for efficient transcription, deletion of the distal 5′ end of U3 in LPDV (which includes parts of the missing region) has been shown to increase transcriptional activity, suggesting that this region may act as a negative regulatory element (Sarid et al., 1995).
LPDV integration
Mapping of the insertion site of the North American prototype strain (12/AR/2009) demonstrated that it was integrated, in reverse orientation, at nucleotide position 45,329,568 in chromosome 6 of the turkey genome (GenBank accessions NC_015016.1 and ADDD01042526), approximately 10.6 kB downstream of a C-C chemokine receptor type 4-like protein and 4.2 kB upstream of a beta-galactosidase-like protein. Although no known gene function has yet to be ascribed to the genomic region in which the provirus was integrated, BLAST analysis (using a Class Aves search set in the nucleotide collection database) of the integration site demonstrated the closest match was to a known oncogene, a serine/threonine kinase receptor-associated protein (GenBank accession XM_003202477), albeit only to the very 3′-terminus of the mRNA (Datta et al., 1998; Halder et al., 2006). Currently, it is unclear whether insertion at this site is related to the fatal oncogenesis observed in this case.
As both LPDV and FWPV were simultaneously detected in multiple birds (Table 1), we analyzed a subset of these cases to determine whether LPDV may be inserted into the genome of FWPV, similar to that observed in REV (Hertig et al., 1997). Although both LPDV and FWPV could be PCR amplified from original tissue samples, only FWPV could be detected after passage in avian cell culture, indicating that the isolated poxviruses did not contain LPDV sequences. This finding suggests that, in these cases, either the detection of both viruses is due to temporally distinct infections (e.g., LPDV integration into the host genome during a former infection followed by the diagnosis of an active FWPV infection) or that these were concomitant infections without recombination occurring between the two viruses.
Evolutionary analysis of LPDV
To determine the evolutionary relationships among the North American viruses, as well as to the prototype Israeli strain (GenBank accession U09568), we performed a phylogenetic analysis on the newly identified proviruses recovered from birds collected from 18 states between 2009-2012 (Table 1) using a portion of the gag polyprotein (partial p31/partial CA; 413 nt). A nucleotide and amino acid alignment of the sequences used to infer the p31/CA phylogeny is shown in Supplementary Figure 1. We also conducted a phylogenetic analysis of a subset of 15 proviruses using both a larger internal gene product [partial p31/full-length CA (977 nt)] and an external protein [partial surface (SU) glycoprotein; 530 nt] to determine if this provided greater resolution between the viruses or identified differences not observed in the partial p31/partial CA tree. Both the partial p31/full-length CA and SU trees had similar topologies to the 413 nt p31/CA phylogeny (data not shown; trees available from the authors on request). Sequences used in the inference of all three phylogenies have been deposited in GenBank under the accessions KC801949-KC802005 (p31/CA) and KC802006-KC802018 (SU).
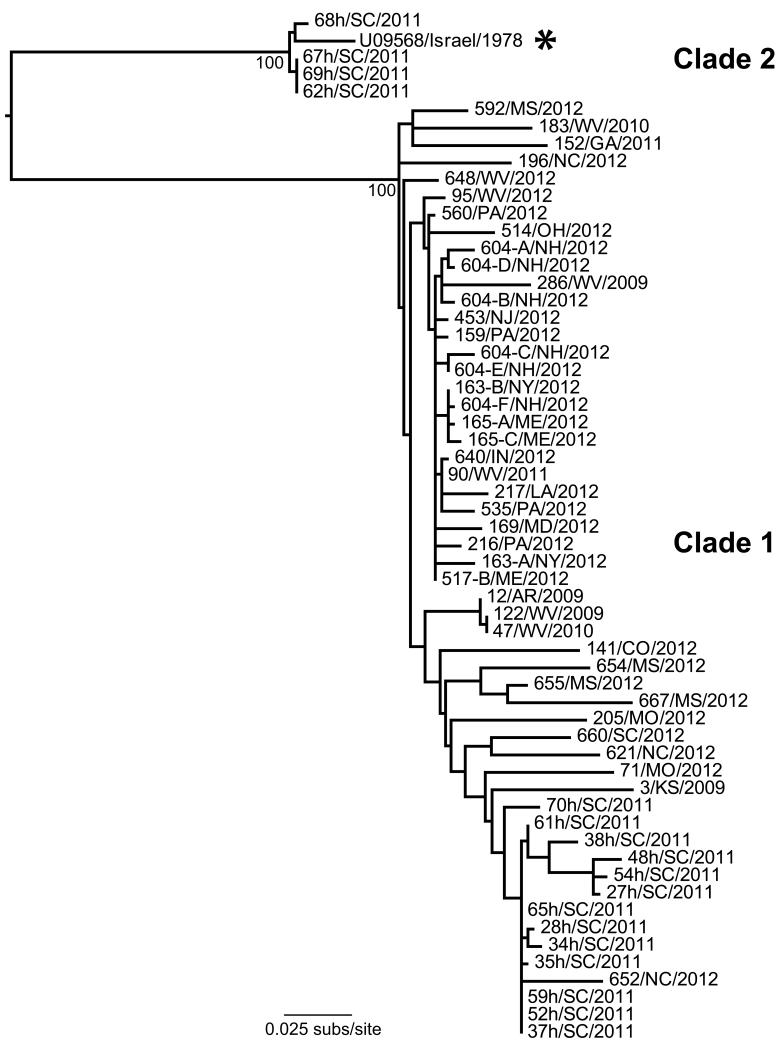
The p31/CA phylogeny revealed the proviruses from North America could be grouped into two major clades, termed clades 1 and 2 (Figure 3). A small subset of four viruses from South Carolina (62-, 67-, 68-, 69/SC/2011) was shown to cluster with the Israeli prototype strain of LPDV in clade 2. Remarkably, while these four viruses exhibited between 83-87% nucleotide identity in their p31/CA sequences to other viruses from the United States, they shared a 97% identity to the Israeli virus. In contrast, the majority of the proviruses sampled in the United States were divergent from the Israeli strain and fell into clade 1. Although this clade showed some evidence of geographical clustering, these groupings were weak, as there was little phylogenetic resolution among strains (i.e., low bootstrap support values), indicative of relatively widespread viral gene flow. The overall lack of temporal structure in the data precluded a molecular clock dating analysis.
Figure 3.
Phylogenetic tree of p31/capsid sequences of LPDV strains recovered from wild turkeys in the United States during 2009-2012, along with the prototype Israeli strain from 1978 (highlighted with an asterisk). Bootstrap support values are shown for key nodes and all horizontal branch lengths are drawn to a scale of nucleotide substitutions per site. The tree is mid-point rooted for purposes of clarity only. The 58 viruses used to infer the tree are designated as case number, followed by state and year of collection (as shown in Table 2).
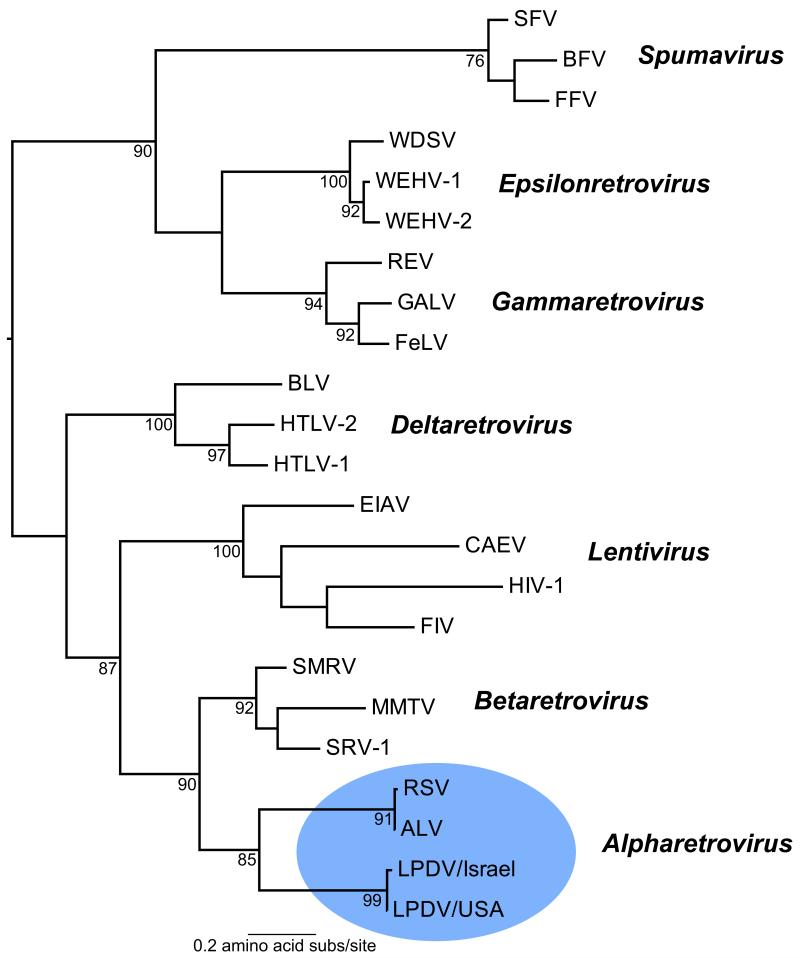
To better define the evolutionary relationship of LPDV to other retroviruses, we performed a phylogenetic analysis of LPDV with representative members from each of the six genera of Retroviridae, encompassing both Orthoretrovirinae and Spumaretrovirinae subfamilies, using conserved domains in RT within the pol polyprotein (Xiong and Eickbush, 1990). The amino acid RT phylogeny demonstrated that the North American and Israeli LPDV prototypes grouped with ALV and RSV within the Alpharetrovirus clade with strong (85% bootstrap) support (Figure 4), thereby demonstrating that LPDV is most closely related to other oncogenic alpharetroviruses. Recently, a wide diversity of lineages of avian endogenous retroviruses (ERV) has been identified in galliform (chicken, turkey) genomes that have properties characteristic of alpha-, beta-, and gammaretroviruses, with LPDV being similar to some avian beta-like ERVs (Bolisetty et al., 2012). However, as alpharetroviruses are postulated to have evolved from beta-like precursors (Bolisetty et al., 2012), that LPDV retains some betaretrovirus properties (i.e., frameshift strategies) while also possessing alpharetrovirus motifs (e.g., tRNATrp primer binding site) and clearly showing similar host associations (galliform birds) and a strong phylogenetic relationship with exogenous alpharetroviruses (ALV and RSV), suggests that its designation as an alpharetrovirus is the most appropriate given current data.
Figure 4.
Family-wide phylogeny of exogenous retroviruses based on conserved protein domains in reverse transcriptase. The clade containing LPDV and representing the Alpharetrovirus genus is shaded in blue. Bootstrap support values are shown for key nodes and all horizontal branch lengths are drawn to a scale of amino acid substitutions per site. The tree is mid-point rooted for purposes of clarity only. Viruses (and their GenBank accession numbers) used to infer the phylogeny were: ALV, Avian leukosis virus (ADO01000); BLV, Bovine leukemia virus (ACR15158); BFV, Bovine foamy virus (AAN08116); CAEV, Caprine arthritis encephalitis virus (AAG48629); EIAV, Equine infectious anemia virus (AAG02702); FeLV, Feline leukemia virus (BAM45657); FIV, Feline immunodeficiency virus (AAB49923); FFV, Feline foamy virus (NP_056914); GALV, Gibbon ape leukemia virus (AAC80264); HIV-1, Human immunodeficiency virus (BAF32463); HTLV-2, Human T-lymphotropic virus 2 (AAB59885); HTLV-1, Human T-lymphotropic virus 1 (AAB20769); LPDV, Lymphoproliferative disease virus [KC802224 (USA) and U09568 (Israel)]; MMTV, Mouse mammary tumor virus (P03365); REV, Reticuloendotheliosis virus (ABD46829); RSV, Rous sarcoma virus (CAA48535); SFV, Simian foamy virus (AFX98084); SMRV, Squirrel monkey retrovirus (NP_041261); SRV-1, Simian retrovirus 1 (ADI76909); WDSV, Walleye dermal sarcoma virus (NP_045937); WEHV-1, Walleye epidermal hyperplasia virus 1 (AAC59310); WEHV-2, Walleye epidermal hyperplasia virus 2 (AAD30054).
Discussion
This report is the first documentation of LPDV in the United States and provides the first description of the disease in naturally-infected wild birds, along with its genetic and phylogenetic relationship to viruses recovered in Israel in the 1990’s. Although the broad geographical distribution of the virus (Maine to Louisiana and west to Colorado), coupled with the non-migratory nature of wild turkeys, tentatively suggests that LPDV may have been endemic in the United States for a considerable time, it is unclear if management practices such as the extensive intra- and interstate translocation of wild turkeys through restoration projects (Tapley et al., 2005) may have inadvertently facilitated the more recent spread of the virus. Additionally, because the full extent of the host range of LPDV (which may include additional wild galliform birds or even avian species within other orders) and the route of virus transmission in nature (e.g., direct contact, arthropod-borne, vertical/horizontal transmission) have not been clearly defined, whether this apparent endemicity in the United States predates the emergence of LPDV (or recognition of LPDV-like disease) in Europe is uncertain. Nevertheless, our analysis shows that LPDV is widespread in the wild turkey population in the United States, is responsible for sporadic fatal tumorigenic disease, and has likely been missed as the etiological agent in undiagnosed cases of lymphoid hyperplasia and neoplasia.
Although fatal tumorigenesis was attributable to LPDV infection in only four of 40 LPDV-positive birds (10.0%), this may be a conservative estimate. Other birds had lesions that were consistent with LPDV infection (e.g., multicentric neoplasia, multi-organ lymphoplasmacytic cellular infiltrates), but concurrent infection with other agents (e.g., REV, FWPV, bacteria) and the observation of additional lesions in these birds precluded a definitive diagnosis of LPDV infection as the cause of death. Presently, in the dual LPDV/REV-positive cases in which fatal tumorigenesis were observed, it is unclear which virus was responsible for the neoplasia. However, as one of our current objectives is the production of recombinant LPDV proteins for use as antigen in the development of specific antisera for immunohistochemistry, this (in conjunction with REV-specific antisera) will likely aid in providing a more definitive diagnosis in such cases. The high prevalence of viral co-infections in the diagnostic cases tested (i.e., 46.3% and 36.6% of LPDV-positive birds were also positive for REV or FWPV, respectively) further demonstrates that screening for multiple pathogens should be performed to definitively assess the cause(s) of mortality. Although the integration of complete or partial REV genomes into FWPV (including vaccine strains) has been extensively demonstrated (Hertig et al., 1997; Liu et al., 2009), we did not detect LPDV incorporation within FWPV genomes in dually-infected turkeys, possibly suggesting that the association between the two viruses may be due to the persistence of LPDV DNA in previously infected but recovered birds, such that the two infections may commonly occur independently. However, the potential spread of LPDV in nature via integration into FWPV genomes, similar to that of REV, merits further investigation, as does the potential synergistic pathological effects of coinfection of LPDV with other pathogens.
The mechanism by which LPDV induces oncogenesis in its turkey host is unknown. Unlike RSV, LPDV does not carry an oncogene within its genome. However, LPDV is similar to the related ALV, as both viruses induce tumors in galliform birds independent of a transducing viral gene. In the case of ALV, although viral integration is a random process, insertion in the vicinity of the c-myc proto-oncogene has been demonstrated to lead to tumorigenesis (Neel et al., 1981; Hayward et al., 1981). Additionally, retroviral insertion into tumor-suppressor genes can be oncogenic, although this generally requires that both alleles be inactivated (e.g., Hicks and Mowat, 1988). Mapping of the insertion site of 12/AR/2009 demonstrated that it was integrated, in reverse orientation, into chromosome 6 of the turkey genome in a region of unknown function downstream of a C-C chemokine receptor type 4-like protein and upstream of a beta-galactosidase-like protein. Although LPDV integration into the turkey genome may be a random process, whether oncogenesis is triggered by insertion into a specific region, similar to ALV and c-myc, is currently unclear. As such, future research will be aimed at mapping the genomic integration site of a comprehensive set of proviruses in birds that were deemed to be unequivocal cases of LPDV-induced tumorigenesis, as well as other birds that died of apparently unrelated causes.
Other than the prototype Israeli strain, the lack of other sequences from historic LPDV outbreaks from Europe and Israel prohibits a detailed phylogenetic analysis of Old and New World viruses. However, it was striking that a subset of four proviruses sampled in South Carolina in 2011 (62-, 67-, 68-, 69/SC/2011) were more closely related genetically to the Israeli virus than to other North American viruses, forming a distinct monophyletic group in the p31/CA phylogeny (Clade 2 in Figure 3), as well as in the full-length CA or partial SU trees (data not shown). This grouping of South Carolina and Israeli isolates provides a direct link between viruses circulating in North America to those that first emerged in Europe, demonstrating that LPDV has been transferred between continents to result in outbreaks of disease. Despite their close evolutionary relationship, the route (North America to Europe or vice versa), approximate time frame, and mechanism by which the transcontinental spread of LPDV occurred is uncertain. However, if wild turkeys are the predominant natural host of the virus, and as their native range is exclusive to North America, it is possible that LPDV originated in North America and may have been inadvertently transferred to the Old World by the intercontinental movement of infected birds. Additionally, as no recent cases of LPDV have been detected in either Israel (personal communication, Irit Davidson, Kimron Veterinary Institute, Bet Dagan, Israel) or the United Kingdom (personal communication, Venugopal Nair, Pirbright Institute, Surrey, England), the sudden emergence of LPDV in the Old World followed by its apparent lack of endemic circulation or recurrent disease outbreaks is again compatible with the outbreak viruses originating from an exotic source. However, the evolutionary origins of LPDV and its precise transcontinental movements are currently unclear and will benefit from both retrospective and prospective analysis of additional viruses from the Middle East, Europe, and North America.
Whether the apparent widespread nature of LPDV in the United States is indicative of the recent identification of a long overlooked or misdiagnosed pathogen, or whether LPDV represents a newly emerging virus is uncertain. Tumorigenic diseases of unknown etiology have long been recognized in wild birds, including galliforms (Drew, 2007), suggesting that a long-standing failure to detect the virus is plausible. However, it is important to note that the population structure of wild turkeys within the United States over the last 50 years or so has undergone dramatic changes. Restoration projects, involving the movement of tens of thousands of birds across the United States to boost dwindling population numbers, have substantially altered the population demographics of turkeys by displacing birds into new territories (Tapley et al., 2005). Although there is no evidence that these restoration projects have facilitated the rapid spread of the virus, the recognition of the emergence of viral diseases into new areas after the inadvertent translocation of infected wildlife is not novel, as epitomized by the rapid expansion and movement of raccoon rabies throughout the northeastern United States in the 1980’-1990’s after the translocation of infected animals originating from the southeast during the 1970’s (Rupprecht and Smith, 1994).
LPDV is currently not listed as a recognized viral species by the ICTV (Stoye et al., 2011). However, based on the genetic and evolutionary data presented here (Figure 4), in addition to previous phylogenetic analyses of Israeli viruses (Chajut et al., 1992; Gazit and Yaniv, 1999), LPDV is clearly most closely related to avian leukosis-sarcoma viruses such as ALV and RSV. Accordingly, we suggest that it should be designated as a new species within the genus Alpharetrovirus, subfamily Orthoretrovirinae, family Retroviridae. Despite the apparent widespread geographic prevalence of the virus among wild turkeys, LPDV does not currently appear to be a serious threat to the North American poultry industry. However, this is conjectural until surveillance efforts have been made to screen commercial poultry for the presence of the virus and clinical trials are performed to definitively assess its pathogenic potential. As LPDV-associated lesions can superficially resemble other retroviral (e.g., reticuloendotheliosis) and non-retroviral (e.g. avian pox) diseases that are endemic to North America, in addition to our observation that dual infections with LPDV and these and other pathogens may be common, LPDV should be considered as a potential disease agent in both retrospective and prospective cases of leukocytic neoplastic or hyperplastic disease of unknown etiology.
Materials and Methods
Sample collection and virus testing
Following the first identification of LPDV in a North American wild turkey, we actively targeted the collection of tissues from clinically ill wild turkeys submitted to veterinary diagnostic laboratories throughout the eastern United States during 2009-2012. Samples were acquired from the diagnostic services at the SCWDS, the New York State Department of Environmental Conservation, the University of Maine Animal Health Laboratory, and the New Hampshire Veterinary Diagnostic Laboratory. These turkeys were submitted as entire carcasses and are designated as ‘diagnostic cases’. Where possible, we targeted wild turkeys with gross or microscopic lesions consistent with the historical description of LPDV infection in domestic turkeys (see McDougall et al., 1978; Zimber et al., 1983; Biggs, 1997) and/or proliferative skin lesions. Tissue samples acquired from diagnostic cases for LPDV testing varied, but when possible, efforts were made to test tissue collected directly from nodular lesions and/or lymphoid tissues. Unless stated otherwise, tissues from all diagnostic cases were examined by necropsy and fixed in 10% neutral-buffered formalin and processed for histopathological examination by standard hematoxylin and eosin staining. In certain cases, special histopathologic stains and ancillary diagnostic tests were performed as noted to assess for additional infectious agents such as bacteria, fungi, and other viruses. A diagnosis of LPDV infection as the sole cause of death was made when i) lymphoid infiltrates were detected in multiple tissues, ii) those tissues were PCR-positive for LPDV, and iii) other potential etiologies (e.g., bacteria, fungi, or other viruses) or alternative non-microbial causes of mortality (trauma, toxicosis) were not detected.
DNA was extracted from tissues in which lesions were observed using a QIAamp DNA mini kit (Qiagen, Valencia, CA) according to the manufacturer’s guidelines. LPDV PCR was performed using primers based on the Israeli prototype strain of LPDV that covered a region spanning the p31 and CA genes (5′-ATGAGGACTTGTTAGATTGGTTAC-3′, 5′-TGATGGCGTCAGGGCTATTTG-3). PCR products were amplified using a GoTaq® Flexi DNA polymerase system (Promega, Madison, WI) and purified products were labeled for sequencing using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA). All FWPV and REV cases were confirmed by PCR from tissue as previously described (Lüschow et al., 2004; Zavala et al., 2006) and also by histopathological examination of tissues. To determine if LPDV was incorporated into the genome of FWPV in birds where both viruses were detected, lesions that were PCR-positive for both viruses were mechanically homogenized in minimal essential media (Sigma-Aldrich, St. Louis, MO), clarified by centrifugation (6700 × g for 10 min), and an aliquot (100μl) was inoculated into DF-1 chicken fibroblast (CRL-12203), ConA-B1-VICK chicken T lymphocyte (CRL-12357), and/or Peking duck embryo fibroblast (CCL-141) cell cultures maintained at 37 °C in a 5% CO2 atmosphere. Cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA). Each case was serially passaged at weekly intervals and then DNA was extracted from cell culture supernatant and screened for both LPDV and FWPV as stated above.
In addition to diagnostic cases, we acquired liver samples from 74 adult wild turkeys harvested in South Carolina during the 2011 spring hunting season. The 74 samples were acquired from 10 counties (Aiken, Anderson, Barnwell, Chesterfield, Darlington, Dorchester, Florence, Hampton, Marion, and Orangeburg). Liver samples were processed and tested for LPDV as in the diagnostic cases. These turkeys are designated as ‘seasonally harvested’ birds.
Genomic sequencing
The complete proviral genome of the index North American case of LPDV (strain 12/AR/2009) was PCR amplified from lung tumor tissue (deposited as GenBank accession number KC802224). Primers for genomic sequencing were developed based on the Israeli prototype strain (GenBank accession number U09568). Gaps in the genome due to areas of divergence were filled in using primers based on newly acquired sequences when necessary. Primers are available from the authors upon request. PCR products were cloned using a PCR Cloning Kit (Qiagen) and contigs were assembled using Sequencher 4.1 (Gene Codes Corporation, Ann Arbor, MI), resulting in the full-length provirus. Mapping of the integration site of 12/AR/2009 into the turkey genome was performed by retroviral LTR arbitrarily primed PCR (RELAP-PCR) (Casper et al., 1998). RNA secondary structures were predicted using the RNAfold program (Gruber et al., 2008). Percent identities of genes and proteins among LPDV isolates and between different retroviruses were determined using the percent identity matrix in Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Theoretical molecular weights and isoelectric points of LPDV proteins were predicted using the Compute pI/MW tool in the ExPASy Bioinformatics Resource Portal (http://web.expasy.org/compute_pi/).
Phylogenetic analysis
We performed a phylogenetic analysis on the p31/capsid (CA) sequences (413 nt) of all LPDV strains recovered from wild turkeys in the United States during 2009-2012, combined with the prototype Israeli strain recovered in 1978. The 58 new proviruses used to infer the p31/CA tree were recovered from 58 individual birds (41 diagnostic cases and 17 seasonally harvested birds) and are designated as case number, followed by state and year of collection (as shown in Table 1). A maximum likelihood (ML) phylogeny was then estimated using PhyML 3.0 (Guindon et al., 2010) and employing the GTR+Γ4 model of nucleotide substitution (estimated Γ shape parameter of 0.549). The best tree was chosen after a combination of NNI and SPR branch-swapping. To infer support for individual nodes, a bootstrap analysis was conducted using 1000 replicate ML trees, again estimated using the GTR+Γ4 substitution model (optimized for each run) and with NNI branch-swapping. An equivalent phylogenetic analysis was performed on a subset of 15 proviruses using (i) partial p31/full-length CA (sequence alignment of 977 nt) and (ii) partial surface (SU) glycoprotein (sequence alignment of 530 nt). All analyses were undertaken using the same ML methods as described above.
To determine the evolutionary relationship of LPDV to other extant retroviruses, we conducted an additional phylogenetic analysis using conserved regions of the pol polyprotein [reverse transcriptase (RT)]. Accordingly, conserved domains in RT (Xiong and Eickbush, 1990) were compiled for 21 retroviruses representing the genera Alpharetrovirus, Betaretrovirus, Deltaretrovirus, Epsilonretrovirus, Gammaretrovirus, Lentivirus and Spumavirus. These sequences were combined with two LPDV sequences representing the Israeli (U09568) and North American (12/AR/2009) prototype strains and aligned using MUSCLE (Edgar, 2004). Because of the large number of insertions/deletions and alignment uncertainties, we used Gblocks (Talavera and Castresana, 2007) to purge all such regions prior to phylogenetic analysis. This resulted in a final sequence alignment of 23 taxa, 191 amino acid residues in length. Phylogenetic analyses of these data were again undertaken using the ML method available in PhyML 3.0, this time employing the WAG+Γ4 model of amino acid substitution (estimated Γ shape parameter of 1.110), and selecting the best tree after a combination of NNI and SPR branch-swapping. A bootstrap resampling analysis was conducted using 1000 replicate ML trees, again inferred using the WAG+Γ4 substitution model and NNI branch-swapping.
Supplementary Material
Highlights.
LPDV is responsible for periodic fatal tumorigenic disease in galliform birds
First description of LPDV in North America
First description of LPDV in wild birds
Phylogenetic analysis suggests transcontinental spread between New and Old World
LPDV has likely been the clandestine agent in many undiagnosed cases of neoplasia
Acknowledgments
We thank John Bryan, Elizabeth Bunting, Steven Kubiski, Joe Okoniewski, and Mark Ruder for diagnostic and field support, as well as other field biologists and veterinarians affiliated with the natural resource agency of each state for collection and submission of birds. Funding for this research was provided by Pittman-Robertson Federal Aid in Wildlife Restoration, the Arcadia Wildlife Preserve, Inc., and through the continued sponsorship of the SCWDS by member states and federal agencies. E.C.H. is supported by a National Health and Medical Research Council Australia Fellowship. Funding was additionally provided by a NRSA fellowship (F32AI100545) to A.B.A. from the National Institute for Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Biggs PM. Lymphoproliferative disease of turkeys. In: Calnek BW, Barnes HJ, Beard CW, McDougald LR, Saif YM, editors. Diseases of Poultry. 10th ed Iowa State University Press; Ames, IA: 1997. pp. 485–489. [Google Scholar]
- Biggs PM, McDougall JS, Frazier JA, Milne BS. Lymphoproliferative disease of turkeys. 1. Clinical aspects, Avian Pathol. 1978;7:131–139. doi: 10.1080/03079457808418265. [DOI] [PubMed] [Google Scholar]
- Biggs PM, Milne BS, Frazier JA, McDougall JS, Stuart JC. Proceedings 15th World Poultry Congress, World Poultry Science Association. Washington, D.C.: 1974. Lymphoproliferative disease in turkeys; pp. 55–56. [Google Scholar]
- Bolisetty M, Blomberg J, Benachenhou F, Sperber G, Beemon K. Unexpected diversity and expression of avian endogenous retroviruses. MBio. 2012;3(5):e00344–12. doi: 10.1128/mBio.00344-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper C, Leib-Mösch C, Salmons B, Günzburg WH, Baumann G, Höfler H, Erfle V, Atkinson MJ. Mapping of a mouse mammary tumor virus integration site by retroviral LTR-arbitrary polymerase chain reaction. Virus Res. 1998;54:207–215. doi: 10.1016/s0168-1702(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Chajut A, Yaniv A, Avivi L, Bar-Am I, Tronick SR, Gazit A. A novel approach for establishing common or random integration loci for retroviral genomes. Nucleic Acids Res. 1991;19:4299. doi: 10.1093/nar/19.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chajut A, Sarid R, Yaniv A, Smythers GW, Tronick SR, Gazit A. The lymphoproliferative disease virus of turkeys represents a distinct class of avian type-C retrovirus. Gene. 1992;122:349–354. doi: 10.1016/0378-1119(92)90225-e. [DOI] [PubMed] [Google Scholar]
- Datta PK, Chytil A, Gorska AE, Moses HL. Identification of STRAP, a novel WD domain protein in transforming growth factor-beta signaling. J. Biol. Chem. 1998;273:34671–34674. doi: 10.1074/jbc.273.52.34671. [DOI] [PubMed] [Google Scholar]
- Drew ML. Retroviral infections. In: Thomas NJ, Hunter DB, Atkinson CT, editors. Infectious Diseases of Wild Birds. Blackwell Publishing; Oxford, UK: 2007. pp. 216–235. [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO. Viral late domains. J. Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gak E, Yaniv A, Chajut A, Ianconescu M, Tronick SR, Gazit A. Molecular cloning of an oncogenic replication-competent virus that causes lymphoproliferative disease in turkeys. J. Virol. 1989;63:2877–2880. doi: 10.1128/jvi.63.6.2877-2880.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gak E, Yaniv A, Ianconescu M, Tronick SR, Gazit A. An in-vivo infectivity assay for cloned retroviruses lacking a susceptible cell culture. J. Virol. Methods. 1990;28:147–154. doi: 10.1016/0166-0934(90)90029-f. [DOI] [PubMed] [Google Scholar]
- Gak E, Yaniv A, Sherman L, Ianconescu M, Tronick SR, Gazit A. Lymphoproliferative disease virus of turkeys: sequence analysis and transcriptional activity of the long terminal repeat. Gene. 1991;99:157–162. doi: 10.1016/0378-1119(91)90122-r. [DOI] [PubMed] [Google Scholar]
- Gazit A, Yaniv A, Ianconescu M, Perk K, Aizenberg B, Zimber A. Molecular evidence for a type C retrovirus etiology of the lymphoproliferative disease of turkeys. J. Virol. 1979;31:639–644. doi: 10.1128/jvi.31.3.639-644.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit A, Schwarzbard Z, Yaniv A, Ianconescu M, Perk K, Zimber A. Organotropism of the lymphoproliferative disease virus (LPDV) of turkeys. Int. J. Cancer. 1982;29:599–604. doi: 10.1002/ijc.2910290518. [DOI] [PubMed] [Google Scholar]
- Gazit A, Yaniv A, Ilani A, Ianconescu M, Perk K, Zimber A. Genetic control of the organ specificity of lymphoproliferative disease virus (LPDV) of turkeys. Int. J. Cancer. 1983;31:351–356. doi: 10.1002/ijc.2910310316. [DOI] [PubMed] [Google Scholar]
- Gazit A, Yaniv A. Lymphoproliferative disease virus of turkeys (Retroviridae) In: Granoff A, Webster RG, editors. Encyclopedia of Virology. 2nd edition Academic Press; San Diego: 1999. pp. 911–915. [Google Scholar]
- Gazit A, Basri R, Ianconescu M, Perk K, Zimber A, Yaniv A. Analysis of structural polypeptides of the lymphoproliferative disease virus (LPDV) of turkeys. Int. J. Cancer. 1986;37:241–245. doi: 10.1002/ijc.2910370212. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–4. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Halder SK, Anumanthan G, Maddula R, Mann J, Chytil A, Gonzalez AL, Washington MK, Moses HL, Beauchamp RD, Datta PK. Oncogenic function of a novel WD-domain protein, STRAP, in human carcinogenesis. Cancer Res. 2006;66:6156–6166. doi: 10.1158/0008-5472.CAN-05-3261. [DOI] [PubMed] [Google Scholar]
- Hatfield MA, Vance S. Status and structure of the North American wild turkey management plan: an integrated approach to wildlife management. In: Rahm J, editor. Transactions of the Seventy-third North American Wildlife and Natural Resources Conference. Wildlife Management Institute; Washington, D.C.: 2009. pp. 44–49. [Google Scholar]
- Hayward WS, Neel BG, Astrin SM. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Hertig C, Coupar BE, Gould AR, Boyle DB. Field and vaccine strains of fowlpox virus carry integrated sequences from the avian retrovirus, reticuloendotheliosis virus. Virology. 1997;235:367–376. doi: 10.1006/viro.1997.8691. [DOI] [PubMed] [Google Scholar]
- Hicks GG, Mowat M. Integration of Friend murine leukemia virus into both alleles of the p53 oncogene in an erythroleukemic cell line. J. Virol. 1988;62:4752–4755. doi: 10.1128/jvi.62.12.4752-4755.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianconescu M, Yaniv A, Gazit A, Perk K, Zimber A. Susceptibility of domestic birds to lymphoproliferative disease virus (LPDV) of turkeys. Avian Pathol. 1983;12:291–302. doi: 10.1080/03079458308436172. [DOI] [PubMed] [Google Scholar]
- Ingr M, Uhlíková T, Strísovský K, Majerová E, Konvalinka J. Kinetics of the dimerization of retroviral proteases: the “fireman’s grip” and dimerization. Protein Sci. 2003;12:2173–2182. doi: 10.1110/ps.03171903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhao J, Su J, Pu J, Zhang G, Liu J. Full genome sequences of two reticuloendotheliosis viruses contaminating commercial vaccines. Avian Dis. 2009;53:341–346. doi: 10.1637/8579-010609-Reg.1. 2009. [DOI] [PubMed] [Google Scholar]
- Lüschow D, Hoffmann T, Hafez HM. Differentiation of avian poxvirus strains on the basis of nucleotide sequences of 4b gene fragment. Avian Dis. 2004;48:453–462. doi: 10.1637/7111. [DOI] [PubMed] [Google Scholar]
- McDougall JS, Biggs PM, Shilleto RW, Milne BS. Lymphoproliferative disease of turkeys. II. Experimental transmission and aetiology. Avian Pathol. 1978;7:141–55. doi: 10.1080/03079457808418266. [DOI] [PubMed] [Google Scholar]
- Neel BG, Hayward WS, Robinson HL, Fang J, Astrin SM. Avian leukosis virus-induced tumors have common proviral integration sites and synthesize discrete new RNAs: oncogenesis by promoter insertion. Cell. 1981;23:323–334. doi: 10.1016/0092-8674(81)90128-8. [DOI] [PubMed] [Google Scholar]
- Payne LN, Venugopal K. Neoplastic diseases: Marek’s disease, avian leukosis and reticuloendotheliosis. Rev. Sci. Tech. Off. Int. Epiz. 2000;19:544–564. doi: 10.20506/rst.19.2.1226. [DOI] [PubMed] [Google Scholar]
- Pincetic A, Leis J. The mechanism of budding of retroviruses from cell membranes. Adv. Virol. 2009:6239691–6239699. doi: 10.1155/2009/623969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips JM, Murray PS, Murray D, Vogt VM. A molecular switch required for retrovirus assembly participates in the hexagonal immature lattice. EMBO J. 2008;27:1411–1420. doi: 10.1038/emboj.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht CE, Smith JS. Raccoon rabies: the re-emergence of an epizootic in a densely populated area. Semin. Virology. 1994;5:155–164. [Google Scholar]
- Sarid R, Chajut A, Gak E, Kim Y, Hixson CV, Oroszlan S, Tronick SR, Gazit A, Yaniv A. Genome organization of a biologically active molecular clone of the lymphoproliferative disease virus of turkeys. Virology. 1994;204:680–691. doi: 10.1006/viro.1994.1584. [DOI] [PubMed] [Google Scholar]
- Sarid R, Gazit A, Tronick SR, Yaniv A. Identification of sequences in the long terminal repeat of the lymphoproliferative disease virus required for efficient transcription. Virology. 1995;208:789–794. doi: 10.1006/viro.1995.1213. [DOI] [PubMed] [Google Scholar]
- Stoye JP, Blomberg J, Coffin JM, Fan H, Hahn B, Neil J, Quackenbush S, Rethwilm A, Tristem M, King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego, California: 2011. Family Retroviridae; pp. 477–495. [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tapley JL, Abernethy RK, Kennamer JE. Status and distribution of the wild turkey in 2004. Proceedings of the National Wild Turkey Symposium. 2005;9:21–32. [Google Scholar]
- Whitcomb JM, Ortiz-Conde BA, Hughes SH. Replication of avian leukosis viruses with mutations at the primer binding site: use of alternate tRNAs as primers. J. Virol. 1995;69:6228–6238. doi: 10.1128/jvi.69.10.6228-6238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv A, Gazit A, Ianconescu M, Perk K, Aizenberg B, Zimber A. Biochemical characterization of the type C retrovirus associated with lymphoproliferative disease of turkeys. J. Virol. 1979;30:351–357. doi: 10.1128/jvi.30.1.351-357.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala G, Cheng S, Barbosa T, Haefele H. Enzootic reticuloendotheliosis in the endangered Attwater’s and greater prairie chickens. Avian Dis. 2006;50:520–525. doi: 10.1637/7655-052806R.1. [DOI] [PubMed] [Google Scholar]
- Zimber A, Perk K, Ianconescu M, Yegana Y, Gazit A, Yaniv A. Lymphoproliferative disease of turkeys: pathogenesis, viraemia and serum protein analysis following infection. Avian Pathol. 1983;12:101–116. doi: 10.1080/03079458308436152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.