Abstract
Recent metagenomic and mechanistic studies are consistent with a new model of periodontal pathogenesis. This model proposes that periodontal disease is initiated by a synergistic and dysbiotic microbial community rather than by a select few bacteria traditionally known as “periopathogens”. Low abundance bacteria with community-wide effects that are critical for the development of dysbiosis are now known as keystone pathogens, the best-documented example of which is Porphyromonas gingivalis. Here we review established mechanisms by which P. gingivalis interferes with host immunity and enables the emergence of dysbiotic communities. We integrate the role of P. gingivalis with that of other bacteria acting upstream and downstream in pathogenesis. Accessory pathogens act upstream to facilitate P. gingivalis colonization and coordinate metabolic activities, whereas commensals-turned-pathobionts act downstream and contribute to destructive inflammation. The recent concepts of keystone pathogens, along with polymicrobial synergy and dysbiosis (PSD), have profound implications for the development of therapeutic options for periodontal disease.
Keywords: dysbiosis, immune subversion, inflammation, P. gingivalis, periodontitis
Introduction
It is increasingly acknowledged that certain inflammatory diseases are associated with imbalances in the relative abundance or influence of microbial species within an ecosystem. This state is known as dysbiosis and leads to alterations in the host–microbe crosstalk that can potentially cause (or at least exacerbate) mucosal inflammatory disorders, such as inflammatory bowel disease, colorectal cancer, bacterial vaginosis, and periodontitis [1, 2]. The host-microbe homeostasis that characterizes a healthy mucosal tissue could be potentially destabilized by host-related factors such as diet, antibiotics, and immune deficiencies. Moreover, perturbations to the host-microbe ecosystem could also be precipitated by increased expression of microbial virulence factors that subvert the host immune response [3–5].
As a potential disease trigger, dysbiosis stands in stark contrast to the traditional view of a classic infection caused by a single or several select pathogens. An exemplar of this changing paradigm is periodontitis, a prevalent chronic inflammatory condition that leads to the destruction of the tooth-supporting tissues (periodontium) and potentially to systemic complications [6, 7]. Recent advances in this field are consistent with a new model of periodontal pathogenesis, according to which periodontitis is initiated by a synergistic and dysbiotic microbial community rather than by select ‘periodontal pathogens’ as traditionally thought [2, 8].
The mechanisms responsible for periodontal dysbiosis are currently poorly understood but likely include both microbial and host-related factors (e.g., congenital or acquired immunodeficiencies). Environmental factors (e.g., diet and smoking) can also manipulate the host-microbe balance unfavorably [9, 10]. From a microbe-centric perspective, the keystone-pathogen hypothesis holds that certain low-abundance microbes can orchestrate destructive periodontal inflammation by remodeling a normally symbiotic microbiota into a dysbiotic state [4]. Keystone or keystone-like pathogens may also be involved in polymicrobial inflammatory diseases occurring in other mucosal tissues [4, 5].
Porphyromonas gingivalis is a gram-negative asaccharolytic bacterium that has long been implicated in human periodontitis [11]. Recent evidence suggests that this bacterium contributes to periodontitis by functioning as a keystone pathogen [12, 13]. The objective of this review is to summarize and discuss the virulence credentials that qualify P. gingivalis as a ‘conductor’ in the orchestration of inflammatory bone loss in periodontitis.
The subgingival lifestyle of P. gingivalis
P. gingivalis resides in the subgingival crevice almost exclusively. Within this region, there are three distinct microenvironments for P. gingivalis: the complex sessile community on the root surface; the fluid phase of the gingival crevicular fluid (GCF); and in and on the gingival epithelial cells that line the crevice. Moreover, P. gingivalis can transition among these niches, each of which provides distinct opportunities and challenges for the organism. Adaption of P. gingivalis occurs on a global scale and indeed the organism differentially regulates around 30% of the expressed proteome according to community, planktonic or epithelial cell conditions [14, 15].
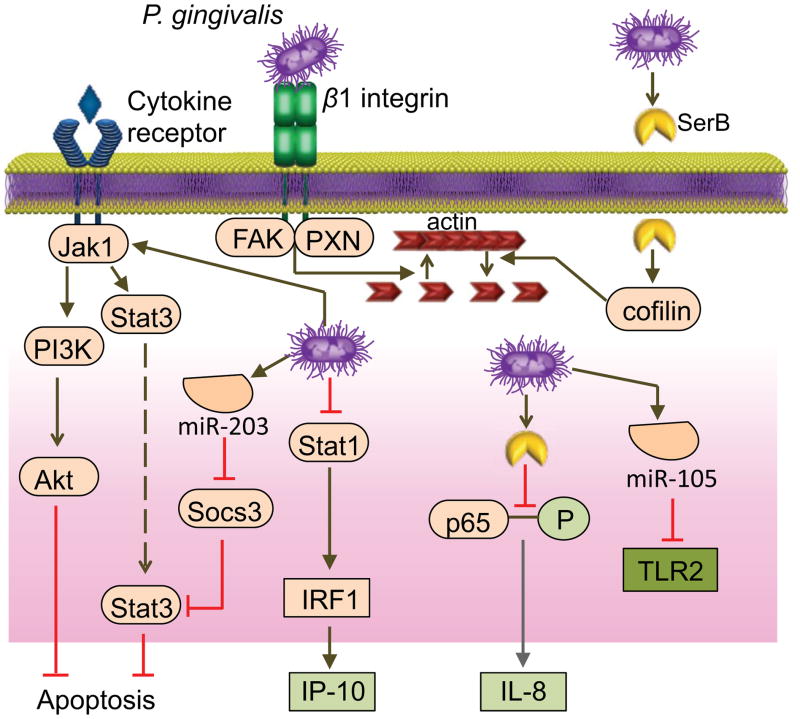
The gingival epithelial cells (GECs) of the subgingival crevice constitute both a physical barrier to microbial intrusion, and an interactive interface that signals microbial presence to the underlying cells of the immune system. P. gingivalis rapidly and abundantly invades GECs intracellularly, with both host cells and microbial interlopers remaining viable over the long term [16, 17]. The internalization process initiates with the FimA fimbrial mediated attachment of P. gingivalis to β1-integrin receptors on the GEC surface with the resultant recruitment and activation of the integrin focal adhesion complex (Fig. 1) [18]. Simultaneously, P. gingivalis secretes the functionally versatile serine phosphatase SerB, which can enter host cells and dephosphorylate and thus activate the actin depolymerizing molecule cofilin [19, 20]. The resulting transient and localized disruption of actin structure allows the organism to enter the interior of the cell. Integrin-dependent signaling also converges cytoskeletal remodeling and restores actin structure albeit in a condensed subcortical configuration [21]. P. gingivalis rapidly locates in the cell cytoplasm which is generally anoxic [22], although later may traffic through autophagosomes before spreading cell to cell [23, 24].
Figure 1. P. gingivalis interactions with gingival epithelial cells (GECs).
Internalization of P. gingivalis is initiated by binding and activation of β1-integrin receptors on the GEC surface mediated by the FimA fimbriae. P. gingivalis secretes the serine phosphatase SerB, which can enter host cells and activate the actin-depolymerizing molecule cofilin by dephosphorylating the Ser 3 residue. The resulting transient and localized disruption of actin structure allows entry of the organism into the cytoplasm. Integrin-dependent signaling through FAK and paxillin (PXN) also converges on the actin cytoskeleton, inducing a later and more long-lasting subcortical condensed microfilament arrangement. Intracellular P. gingivalis secretes SerB which dephosphorylates the Ser 536 residue of the p65 subunit of NF-κB, thus preventing nuclear translocation of p65 homodimers of NF-κB and suppressing CXCL8 (IL-8) production. Levels of Stat1 are diminished by P. gingivalis which reduce the activity of IFN regulatory factor 1 (IRF1) and downregulate CXCL10 (IP-10) synthesis. Decreased secretion of the neutrophil chemokine CXCL8 and the T-cell chemokine CXCL10 from GECs is called localized chemokine paralysis. Internalized P. gingivalis activates the JAK1-Stat3 and PI3K-Akt pathways thereby suppressing apoptosis. P. gingivalis increases the levels of a number of microRNAs within GECs. miR-105 suppresses TLR2 production, and miR-203 inhibits SOCS3 and elevates Stat3 production.
Internalized P. gingivalis bacteria waste little time before reprogramming host cell signal transduction and gene expression [25]. Infection of GEC results in acceleration through the cell cycle and suppression of apoptosis [26]. Anti-apoptotic pathways activated by P. gingivalis include those involving JAK-Stat and PI3K-Akt, which consequently suppress intrinsic mitochondrial-mediated cell death (Fig. 1) [16, 27]. In addition, ATP scavenging by a secreted nucleoside diphosphate kinase (Ndk) enzyme of P. gingivalis prevents apoptosis through the P2X7 receptor [28]. Ndk also contributes to intracellular persistence of P. gingivalis by increasing levels of glutathione which protect against reactive oxygen species [29]. Long-term cohabitation of P. gingivalis within GECs leads to an overall subtle and nuanced inter-kingdom interaction, which can affect innate immune status. For example, P. gingivalis induces the production of a variety of microRNAs in GECs: e.g., miR-105 which suppresses TLR2 production [30], and miR-203 which inhibits SOCS3 and SOCS6 production (see Fig. 1) [31]. Additional strategies employed by P. gingivalis to manipulate GEC innate immune function are discussed below.
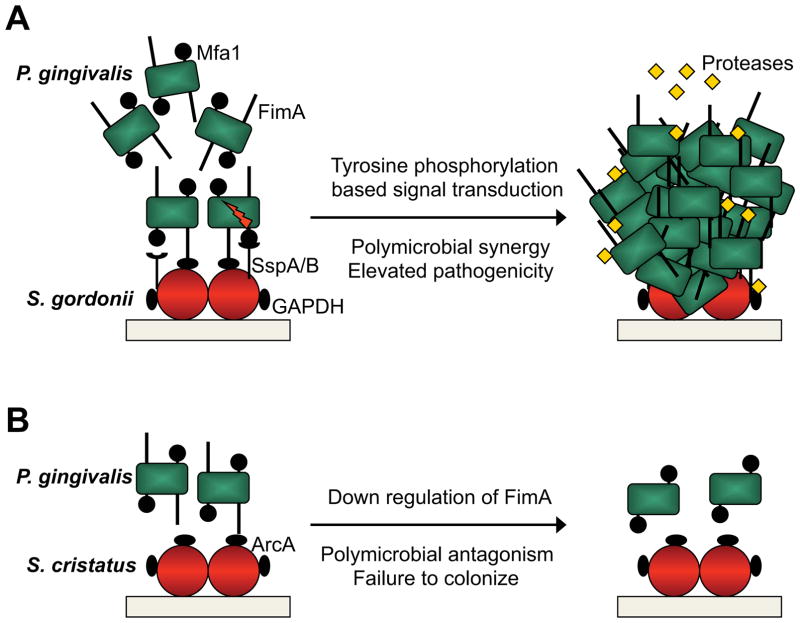
While oral epithelial cells can harbor several species of oral bacteria simultaneously [32], it is within the close confines of the multispecies biofilm on tooth surfaces that interbacterial communication becomes most relevant. As a strict anaerobe, P. gingivalis relies on antecedent colonizers such as streptococci and Fusobacterium nucleatum to reduce the oxygen tension and also provide metabolic support [33]. Coadhesion among these organisms facilitates nutritional and signaling interactions [34, 35]. P. gingivalis develops into heterotypic communities with S. gordonii following multimodal adhesion that involves both the FimA and Mfa1 component fimbriae of P. gingivalis that interact with streptococcal GAPDH and SspA/B surface proteins respectively (Fig. 2). Engagement of Mfa1 with SspA/B initiates a signal cascade within P. gingivalis. Increased expression of a protein tyrosine phosphatase (Ltp1) ultimately elevates the amount of the transcription factor CdhR, which suppresses production of Mfa1 and constrains further community development [33–36]. Moreover, tyrosine phosphorylation/dephosphorylation also regulates protease expression by P. gingivalis, thus influencing pathogenic potential [37]. The ability of S. gordonii to enhance P. gingivalis pathogenicity has also been established in vivo: oral co-infection of conventionally reared (specific pathogen-free) mice with both organisms induces more alveolar bone loss compared to infection with either species alone [38]. In the oral cavity, S. gordonii, hitherto considered as a commensal, would therefore be more accurately categorized as an accessory pathogen [34].
Figure 2. Synergistic and antagonistic interactions between P. gingivalis and oral streptococci.
(A) P. gingivalis fimbrial adhesins FimA and Mfa1 engage the S. gordonii adhesins GAPDH and SspA/B, respectively, and the two species accumulate into a heterotypic community. Mfa1 binding activates a signal transduction cascade within P. gingivalis based on protein tyrosine (de)phosphorylation. Ultimately expression of Mfa1 is downregulated and community development is constrained. Production and activity of P. gingivalis proteases is elevated and the heterotypic community has enhanced pathogencity in bone loss models in vivo. (B) Contact of the P. gingivalis FimA fimbriae with arginine deiminase (ArcA) on the surface of S. cristatus results in downregulation of FimA. Consequently P. gingivalis fails to adhere to S. cristatus, colonization is impeded, and P. gingivalis is inversely correlated with S. cristatus in vivo.
Not all interspecies interactions are synergistic, of course. Bacterial species compete for nutrients and attachment sites, and produce bacteriocins and toxic metabolites such as ROS. Interbacterial communication can also be antagonistic, for example arginine deiminase produced by Streptococcus cristatus represses synthesis of the FimA fimbrial adhesin in P. gingivalis [39]. Consequently, colonization and pathogenicity of P. gingivalis are impaired (Fig. 2). Indeed P. gingivalis and S. cristatus are negatively correlated in the subgingival biofilm [40,41]. The emerging perspective implicates the initial colonizers of dental biofilms in the pattern of subsequent microbial colonization. Distinct streptococcal species can determine the success or failure of keystone pathogen colonization and thus provide an additional level of control for the pathogenic potential of the entire community.
Within the fluid phase of the GCF host immune cells and effector molecules strive to minimize the impact of colonizing bacteria. Histological and electron microscopic observations reveal that gingival crevicular neutrophils form a ‘defense wall’ against the tooth-associated biofilm [42]. In periodontitis, however, the neutrophils largely fail to control the bacteria, despite maintaining viability and capacity to elicit immune responses, such as degranulation and release of ROS and extracellular DNA traps [42–45]. Although it is sometimes assumed that biofilms are intrinsically resistant to phagocytosis, recent studies have shown that neutrophils can be activated by biofilm matrix components or quorum-sensing molecules in ways that enable them to interfere with developing biofilms, specifically through phagocytosis, degranulation, and formation of extracellular traps [46–48]. In fact, depending on the nature and composition of biofilms, neutrophils can either move into a biofilm structure and phagocytose bacteria, or display a relatively immobile phenotype with limited capacity for phagocytosis, as shown in studies utilizing time-lapse video microscopy and confocal laser scanning microscopy [46, 47, 49, 50]. These findings suggest the operation of proactive microbial evasive mechanisms against neutrophils in the gingival crevice. Although P. gingivalis and other periodontal bacteria can endure oxidative stress [51–53], it is not known how they can resist the non-oxidative killing mechanisms of neutrophils. If the bacteria can disarm neutrophils in the gingival crevice, the subversive mechanism(s) involved should be appropriately targeted so as to not interfere with the host inflammatory response, which is essential for nutrient acquisition and the sustenance of dysbiotic microbial communities in periodontitis [4]. Accumulating evidence suggests that P. gingivalis can transiently interfere with the recruitment of neutrophils in the early stages of colonization and, moreover, has the potential to interfere with host immunity in a manner that enhances the survival of the entire microbial community (next section).
Manipulation of innate and adaptive immunity
Normal neutrophil recruitment is an important feature of the healthy periodontium. Genetic defects that compromise this function (e.g., leukocyte-adhesion deficiency) are associated with aggressive forms of periodontitis [54]. Adjacent to the tooth surface, the junctional gingival epithelium produces CXCL8 (IL-8) and generates a gradient for the recruitment of neutrophils to the gingival crevice [55]. GECs exposed to P. gingivalis fail to produce CXCL8 even when stimulated with other bacterial species that are otherwise potent inducers of this chemokine [56]. This ‘local chemokine paralysis’ depends upon the capacity of P. gingivalis to invade the epithelial cells [56] and secrete the serine phosphatase SerB, which specifically dephosphorylates S536 on NF-κBp65 (Fig. 1) [57]. P. gingivalis additionally acts on endothelial cells and inhibits the upregulation of E-selectin by other periodontal bacteria, thereby potentially interfering with the leukocyte adhesion and transmigration cascade [58]. In vivo studies in mice showed that the subversive effects of P. gingivalis on CXCL8 and E-selectin expression are transient [13], suggesting that P. gingivalis can only delay rather than block the recruitment of neutrophils. At least in principle, however, this mechanism could allow adequate time for P. gingivalis and other bacteria sharing the same niche to establish colonization in the relative absence of neutrophil defenses. Consistent with this notion, a SerB-deficient isogenic mutant of P. gingivalis induces enhanced neutrophil recruitment to the periodontium and is less virulent than the WT organism in terms of bone loss induction [59].
Studies in the oral gavage model of mouse periodontitis have shown that P. gingivalis can persist in the periodontium of both specific pathogen-free and germ-free mice [13]. This observation is consistent with the capacity of P. gingivalis to escape immune clearance through proactive manipulation of several leukocyte innate immune receptors and other defense mechanisms activated in concert, such as the complement cascade [60–62] (Fig. 3). Intriguingly, bystander bacterial species likely benefit from the ability of P. gingivalis to impair host defenses, since the colonization of P. gingivalis is associated with increased total counts and altered composition of the periodontal microbiota [13]. Although the precise mechanisms are uncertain, these dysbiotic alterations are required for periodontal pathogenesis as suggested by the failure of P. gingivalis to cause disease by itself in germ-free mice [13].
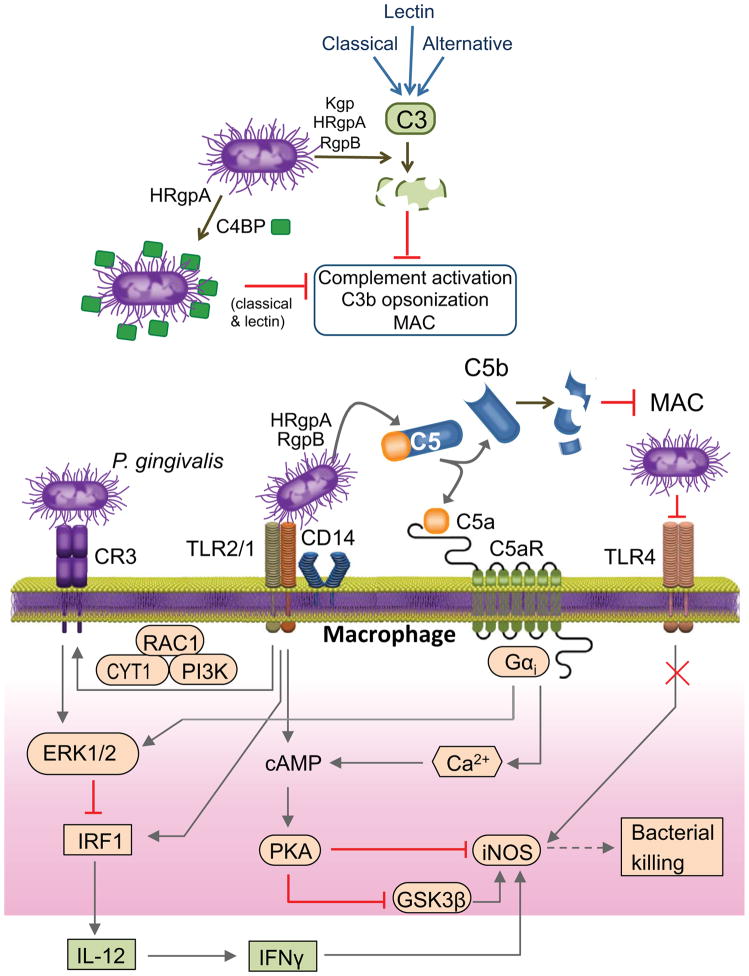
Figure 3. Manipulation of complement, TLRs, and their signaling crosstalk by P. gingivalis.
P. gingivalis gingipains (Kgp, HRgpA, RgpB) inhibit the classical, lectin, and alternative pathways of complement activation by degrading the central complement component C3. This prevents the deposition of C3b opsonin or the membrane attack complex (MAC) on the bacteria. P. gingivalis protects itself against complement also by using HRgpA to capture the circulating C4b-binding protein (C4BP), a negative regulator of the classical and lectin pathways. P. gingivalis interacts with TLR2 (specifically with the CD14–TLR2–TLR1 signaling complex) and with TLR4. TLR4 activation is prevented by the bacterium’s atypical LPS that can act as a TLR4 antagonist. A subset of TLR2 responses is subverted by P. gingivalis through instigation of signaling crosstalk with complement receptors. By means of its HRgpA and RgpB which release biologically active C5a from C5, P. gingivalis activates the C5a receptor (C5aR) in macrophages and induces intracellular Ca2+ signaling which synergistically enhances the otherwise weak cAMP responses induced by TLR2 activation alone. The resulting activation of protein kinase A (PKA) inactivates glycogen synthase kinase-3β (GSK3β) and inhibits iNOS-dependent intracellular killing (in macrophages). P. gingivalis-activated TLR2 also induces an inside-out signaling pathway, mediated by RAC1, PI3K and cytohesin-1 (CYT1), which transactivates complement receptor-3 (CR3). Activated CR3 binds P. gingivalis and induces ERK1/2 signaling, which in turn selectively downregulates IL-12 p35 and p40 mRNA expression through suppression of IFN regulatory factor 1 (IRF1). This ERK1/2 pathway is also induced downstream of C5aR. Inhibition of IL-12, and secondarily IFN-γ, results in defective immune clearance of P. gingivalis.
In the mouse model, subgingival dysbiosis and periodontitis require intact complement C5a receptor (C5aR) signaling. Indeed, P. gingivalis fails to colonize the periodontium of C5aR-deficient mice, whereas treatment of mice with a C5aR antagonist applied locally in the periodontium eliminates P. gingivalis, reverses dysbiosis, and inhibits development of periodontitis [13,63]. It is possible that P. gingivalis exploits C5aR signaling in several leukocyte types although this concept has thus far shown only in macrophages. In these cells, the C5aR-dependent subversive effects strictly require a crosstalk with TLR2, and both receptors can be activated in tandem by P. gingivalis [64]. Notably, P. gingivalis does not rely on immunological mechanisms for C5aR activation, since it can activate this complement receptor through C5a generated locally by its Arg-specific gingipains (HRgpA, RgpB) that have C5 convertase-like activity [64, 65]. P. gingivalis also expresses a number of potent TLR2 ligands including serine lipids and lipoproteins [66, 67].
At the molecular level, the P. gingivalis-induced C5aR-TLR2 crosstalk in macrophages leads to synergistic activation of cAMP-dependent protein kinase A for inhibition of glycogen synthase kinase-3β (GSK3β) and of iNOS-dependent intracellular bacterial killing [64] (Fig. 3). In the murine periodontal tissue, C5aR signaling synergizes with TLR2 to induce secretion of cytokines that promote periodontal inflammation and bone loss (TNF, IL-1β, IL-6, and IL-17A). This is likely to enhance the fitness of P. gingivalis and other periodontitis-associated bacteria which require an inflammatory environment to secure critical nutrients; that is, tissue breakdown products including peptides and hemin-derived iron. In stark contrast to the upregulation of bone-resorptive inflammatory cytokines, P. gingivalis-induced C5aR signaling in macrophages downregulates TLR2-induced IL-12 and hence inhibits IFN-γ production and cell-mediated immunity against the bacteria [63, 65]. The selective inhibition of bioactive IL-12 (IL-12p35/IL-12p40) associated with C5aR-TLR2 crosstalk involves ERK1/2 signaling-dependent suppression of the IFN regulatory factor-1 (IRF-1), a transcription factor that is crucial for the regulation of IL-12 p35 and p40 mRNA expression [65, 68]. Importantly, genetic ablation of C5aR or TLR2 promotes the killing of P. gingivalis in vivo [64, 69].
The inhibitory ERK1/2 pathway that regulates TLR2-induced IL-12 is also activated when P. gingivalis binds complement receptor 3 (CR3) on macrophages [70, 71] (Fig. 3). CR3 is a β2 integrin (CD11b/CD18) that can bind ligands when its high-affinity conformation is transactivated via inside-out signaling by other receptors such as chemokine receptors. P. gingivalis induces TLR2-mediated transactivation of CR3 through an inside-out pathway that involves RAC1, PI3K and cytohesin-1 [72, 73] (see Fig. 3). Upon binding CR3, P. gingivalis not only downregulates IL-12 but also enters macrophages in a relatively safe way [74], perhaps because CR3 is not linked to strong microbicidal mechanisms such as those activated by FcγR-mediated phagocytosis [75]. Indeed, P. gingivalis can persist intracellularly in WT macrophages for longer times than in CR3-deficient macrophages [74].
As alluded to above, P. gingivalis can activate C5aR signaling independently of the canonical activation of complement [64, 65]. In fact, P. gingivalis can block the canonical complement cascade regardless of the initiation pathway involved (classical, lectin, or alternative) since its gingipains readily degrade C3, the central complement component where all initiation pathways converge [76, 77] (Fig. 3). As a consequence, the deposition of C3b opsonin or the membrane attack complex on the bacterial surface is suppressed, whereas genetic or pharmacological ablation of the gingipains restores these complement functions [78,79]. It should be noted that although P. gingivalis generates biologically active C5a through direct C5 conversion, the resulting C5b fragment is readily degraded by the gingipains, ostensibly to prevent the formation of the membrane attack complex [80] (Fig. 3). All three gingipain enzymes mediate complement inactivation through C3 degradation, although HRgpA and RgpB are more potent than the Lys-specific gingipain (Kgp) [76].
P. gingivalis also employs degradation-independent mechanisms to interfere with complement activation. Specifically, P. gingivalis uses HRgpA to capture the circulating C4b-binding protein on its cell surface, thereby acquiring the ability to negatively regulate the classical and lectin pathways [81] (Fig. 3). All these mechanisms are consistent with the exquisite resistance of P. gingivalis to the lytic action of complement [76, 78]. Curiously, however, gingipain-deficient mutants appear to be as resistant as the WT organism after exposure to human serum, despite the deposition of active complement fragments on the bacterial surface of the mutants [78, 82]. This intrinsic resistance was attributed to an anionic polysaccharide structure anchored to the cell surface by lipid A (also known as A-LPS). An intriguing question, therefore, is why P. gingivalis has developed mechanisms to suppress an antimicrobial system that cannot kill it. As microbial evasive mechanisms seldom provide full protection, P. gingivalis may be using a number of different reinforcing mechanisms to maximize protection against complement. An alternative, though not mutually exclusive, interpretation is that inactivation of complement by P. gingivalis serves to protect otherwise complement-susceptible organisms in the same subgingival niche, in line with its role as a keystone pathogen.
The interactions of P. gingivalis with complement are quite complex in that its gingipains can exert dose-dependent biphasic effects on complement activation. At low concentrations, the gingipains not only cannot inhibit complement but actually activate the C1 complex and hence trigger the classical pathway [76]. It can be speculated that the diffusion of released gingipains away from the biofilm generate appropriate enzyme concentrations that activate complement and hence the flow of inflammatory exudate (gingival crevicular fluid), which, as discussed above, provides essential nutrients. Importantly, immunohistochemical studies have detected a concentration gradient of gingipains extending from the subgingival biofilm to the subjacent gingival connective tissue [83].
Besides TLR2, P. gingivalis can also interact with TLR4 by means of LPS, although in a rather unusual way. The organism can enzymatically modify the lipid A moiety of its LPS to either evade or antagonize TLR4 activation (Fig. 3), in contrast to the classical enterobacterial LPS which is a potent TLR4 agonist [55]. These modifications involve the generation of atypical LPS molecules with 5-acyl monophosphate lipid A structure (weak TLR4 agonist) or with 4-acyl monophosphate lipid A structure (potent TLR4 antagonist) [12, 55]. The atypical nature of P. gingivalis LPS molecules not only explains the failure of TLR4 to contribute to the host response against P. gingivalis in vivo [69] but additionally protect the organism against cationic antimicrobial peptides [84, 85].
P. gingivalis possesses a plethora of other mechanisms to manipulate innate immunity, possibly reflecting its ability to cope with diverse challenges or in different settings. For instance, through the use of distinct virulence factors, P. gingivalis is thought to exploit interactions with erythrocytes, DC, and aortic endothelial cells, which not only promote its fitness but also contribute to the pathogenesis of atherosclerosis [86–88]. Additional in vitro and animal model studies suggest that, through enzymatic modification of host proteins, P. gingivalis can breach immune tolerance in susceptible individuals and exacerbate rheumatoid arthritis [89]. The reader is referred to specialized reviews for additional information on systemic effects associated with P. gingivalis [62, 90–92].
Recent studies indicate that P. gingivalis can potentially also manipulate adaptive immunity by acting on APC GECs. Indeed, the interaction of P. gingivalis with DC induces a cytokine pattern that favors CD4+ T helper 17 (Th17) polarization at the expense of the Th1 lineage [93]. Specifically, P. gingivalis induces IL-1β, IL-6 and IL-23, but not IL-12, which moreover is particularly susceptible to proteolysis by the P. gingivalis gingipains [93]. GECs stimulated with P. gingivalis produce a potent admixture of pro- and anti- inflammatory cytokines and chemokines [17, 94]. For example, P. gingivalis infected GECs overexpress pro-IL-1β, although secretion requires an additional stimulus such as extracellular ATP, to activate the processing enzyme caspase-1 through the NLRP3 inflammasome [29, 95]. One major function of IL-1β is to enhance the antigen-driven proliferation of CD4+ T cells; however, P. gingivalis additionally inhibits GEC production of CXCL10 (IP-10) and other Th1 chemoattractants (CXCL9 and CXCL11) through downregulation of IRF-1 and Stat1 expression (Fig. 1) [96]. The inhibitory effect on CXCL10 is ‘dominant’ in that GECs exposed to P. gingivalis cannot express this chemokine in response to other oral bacteria that otherwise can readily induce CXCL10 [96]. In a related context, the ability of P. gingivalis to induce TLR2-dependent IL-10 production leads to inhibition of IFN-γ production by CD4+ and CD8+ T cells [97]. It could, therefore, be hypothesized that P. gingivalis modulate T-cell development and function in ways that promotes Th17-mediated inflammation over a Th1-dependent cell-mediated immune response, which is thought to promote clearance of P. gingivalis [60]. Numerous Th17 cells can be observed in periodontitis lesions [93] and can function as an osteoclastogenic subset that links T-cell activation to inflammatory bone loss [98, 99]. On the other hand, Th1 cells are thought to play a protective role in periodontitis [100] although some studies have attributed destructive effects to Th1 cells [101]. Overall, more research is warranted to better understand the roles of T-cell subsets in periodontitis and the biological relevance of their modulation by P. gingivalis in the context of its role as a keystone pathogen.
Keystone pathogens and models of disease
In inflammatory conditions associated with bacterial communities, traditional concepts of pathogen and commensal have become obsolete. This is well illustrated by periodontal disease where P. gingivalis can remain quiescent for long periods of time before (and after) expressing pathogenicity through manipulation of the host response and disruption of homeostasis. Conversely, organisms usually considered commensals, such as S. gordonii, can act as accessory pathogens and elevate the pathogenicity of P. gingivalis. Commensal organisms can also act as pathobionts, that is, following homeostasis breakdown and initiation of inflammation, these commensals-turned-pathogens can propagate and amplify destructive periodontal inflammation. In this regard, a recent study identified a bacterium (designated NI1060) in the murine oral cavity that selectively accumulates in damaged periodontal tissue and causes inflammatory bone loss by activating the intracellular PRR Nod1 [102]. NI1060 appears to thrive under inflammatory conditions, apparently because it can readily procure nutrients derived from tissue breakdown in an inflammatory environment. NI1060, moreover, contributes to the exacerbation of inflammation by inducing neutrophil-specific chemokines, thereby augmenting neutrophil infiltration in the periodontal tissue [102]. Other commensals (NI440 and NI968) dominate exclusively in healthy sites and do not behave as periodontal pathobionts [102]. The notion that there are pathobionts that can opportunistically contribute to periodontitis is consistent with recent metagenomic studies showing a strong association of previously underappreciated bacteria (including the Gram-positive Filifactor alocis and Peptostreptococcus stomatis and other species from the genera Prevotella, Megasphaera, Selenomonas, and Desulfobulbus) with periodontitis [8, 103, 104]. Moreover, as the bacterial biomass increases with increasing periodontal inflammation, the ecological shift from health to disease involves the emergence of newly-dominant community members as opposed to the appearance of novel species [8]. This finding further supports the idea that many periodontitis-associated bacteria are commensals in some circumstances, but under certain conditions can thrive beyond a threshold sufficient to cause or exacerbate periodontitis. This threshold could be numerical or physiological, or a combination of both. It therefore takes a ‘team effort’ to cause periodontitis in that the disease requires cooperative interactions among bacteria with different roles.
A recently formulated model that accommodates these concepts is called the Polymicrobial Synergy and Dysbiosis (PSD) model [2]. This model holds that physiologically compatible organisms assemble into heterotypic communities which exist in a controlled immuno-inflammatory state. While they are proinflammatory and can produce toxic products such as proteases, overgrowth and overt pathogenicity are controlled by the host response. The microbial constituents of the communities can vary among individuals, among sites, and over time. Colonization by keystone pathogens such as P. gingivalis elevates the virulence of the entire community following interactive communication with accessory pathogens. Initially, host immune surveillance is impaired and the dysbiotic community increases in number. Subsequently, the community proactively induces inflammation to sustain itself with derived nutrients, which will also shape a modified ‘inflammophilic’ community. The action of pathobionts in the community, in addition to overt pathogens, eventually leads to destruction of periodontal tissues. The PSD model reconciles a number of features of periodontal disease that were discordant with earlier concepts of pathogenicity. These include: the variable microbiota at disease sites, even within the same patient; the presence of pathogens in the absence of disease; the episodic nature of the disease; and the failure of P. gingivalis to cause periodontitis in the absence of the commensal microbiota [13].
Conclusions and future perspectives
Bacteria on human mucosal surfaces tend to accumulate into complex multispecies communities, a process controlled by a sophisticated series of interbacterial signaling and host response interactions. Within these communities, bacteria have specialized roles, such as provision of an essential enzyme for progressive nutrient metabolism. Bacteria that influence the pathogenicity of the entire community are keystone pathogens, the best-documented example of which is P. gingivalis. While P. gingivalis can affect gene and protein expression in other community members, the major keystone-related influence of the organism is likely through interference with host immunity. This is accomplished by a multipronged approach that compromises immune function on a number of levels (Fig. 1 and 3). It is important to bear in mind, however, that periodontitis is an inflammatory disease, and thus the timing, location and context of immune suppression by P. gingivalis will have major significance for the ultimate progression of disease. Upon initial colonization of the subgingival area, chemokine paralysis and immune evasion contribute to the persistence of P. gingivalis infection. As the reduced immune surveillance begins to benefit the entire biofilm community, local overgrowth of organisms may then overwhelm the structural integrity of the tissues, and cause inflammation to rebound. These host responses, however, may be insufficient to control P. gingivalis and, worse, contribute further to tissue damage and bone resorption. Tissue destruction also releases peptides and heme-containing compounds which stimulate the growth of P. gingivalis. Nutrients derived from inflammation and tissue degradation select for community members that are inflammophilic. Subsequently, however, the activities of P. gingivalis can be constrained, most likely due to a combination of host protective responses and the aggregate efforts of the bacterial community, and a controlled immuno-inflammatory state can be restored. This notion is consistent with the ‘burst model’ of periodontitis, according to which disease chronicity may not represent a constant pathologic process but rather a persistent series of acute insults (bursts) separated by periods of remission [105].
Recent concepts of keystone pathogens in a PSD model of periodontal disease have a profound impact on the development of therapeutic options for periodontal disease. Targeting of P. gingivalis directly, historically the strategy of choice, is no longer the most rational approach as it is difficult to completely eliminate the organism and P. gingivalis is effective keystone pathogen at low levels of abundance. The ability of P. gingivalis to survive inside epithelial cells also hinders elimination as intracellular P. gingivalis are protected from antibiotics and can serve as a source for recrudescence of infection [106, 107]. Rather, community manipulation has emerged as an option, albeit still theoretical. Elevating numbers of organisms that normally constrain P. gingivalis and reducing those that are synergistic with P. gingivalis would foster commensalism and prevent the transition to a pathogenic community. Targeting of host cell processes is another avenue worthy of exploration. This could involve anti-inflammatory approaches to inhibit destructive inflammation which indirectly would also exert antimicrobial effects (limitation of inflammatory exudate-derived nutrients) or the targeted blockade of immune evasion pathways. In this regard, antagonizing complement pathways in the gingival tissues could lock the host in a mode that is non-responsive to the subversive activities of P. gingivalis, and potentially to other keystone pathogens. Moreover, enhancing protective innate immunity in ways that counteract chemokine paralysis, TLR4 antagonism, and other bacterial strategies with community-wide impact may also help restore periodontal tissue homeostasis.
Acknowledgments
The authors’ research is supported by NIH/NIDCR grants; DE015254, DE017138, DE021580, DE021685 (GH) and DE011111, DE012505, DE016690, DE017921, DE022867 (RJL).
Footnotes
Conflicts of interest. The authors declare no financial or commercial conflict of interest.
References
- 1.Eloe-Fadrosh EA, Rasko DA. The human microbiome: from symbiosis to pathogenesis. Annu Rev Med. 2013;64:145–163. doi: 10.1146/annurev-med-010312-133513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: The Polymicrobial Synergy and Dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurashima Y, Goto Y, Kiyono H. Mucosal innate immune cells regulate both gut homeostasis and intestinal inflammation. Eur J Immunol. 2013 doi: 10.1002/eji.201343782.. [DOI] [PubMed] [Google Scholar]
- 4.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stecher B, Maier L, Hardt WD. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 6.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 7.Kebschull M, Demmer RT, Papapanou PN. “Gum bug leave my heart alone”-Epidemiologic and mechanistic evidence linking periodontal infections and atherosclerosis. J Dent ”Res. 2010;89:879–902. doi: 10.1177/0022034510375281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stabholz A, Soskolne WA, Shapira L. Genetic and environmental risk factors for chronic periodontitis and aggressive periodontitis. Periodontol 2000. 2010;53:138–153. doi: 10.1111/j.1600-0757.2010.00340.x. [DOI] [PubMed] [Google Scholar]
- 10.Divaris K, Monda KL, North KE, Olshan AF, Reynolds LM, Hsueh WC, Lange EM, et al. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Hum Mol Genet. 2013;22:2312–2324. doi: 10.1093/hmg/ddt065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 12.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91:816–820. doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuboniwa M, Hendrickson EL, Xia Q, Wang T, Xie H, Hackett M, Lamont RJ. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98. doi: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrickson EL, Xia Q, Wang T, Lamont RJ, Hackett M. Pathway analysis for intracellular Porphyromonas gingivalis using a strain ATCC 33277 specific database. BMC Microbiol. 2009;9:185. doi: 10.1186/1471-2180-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, Stavropoulos MF, et al. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9:1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol 2000. 2010;52:68–83. doi: 10.1111/j.1600-0757.2009.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yilmaz O, Watanabe K, Lamont RJ. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 2002;4:305–314. doi: 10.1046/j.1462-5822.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 19.Moffatt CE, Inaba H, Hirano T, Lamont RJ. Porphyromonas gingivalis SerB-mediated dephosphorylation of host cell cofilin modulates invasion efficiency. Cell Microbiol. 2012;14:577–588. doi: 10.1111/j.1462-5822.2011.01743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tribble GD, Mao S, James CE, Lamont RJ. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc Natl Acad Sci U S A. 2006;103:11027–11032. doi: 10.1073/pnas.0509813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yilmaz O, Young PA, Lamont RJ, Kenny GE. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology. 2003;149:2417–2426. doi: 10.1099/mic.0.26483-0. [DOI] [PubMed] [Google Scholar]
- 22.Riemer J, Bulleid N, Herrmann JM. Disulfide formation in the ER and mitochondria: two solutions to a common process. Science. 2009;324:1284–1287. doi: 10.1126/science.1170653. [DOI] [PubMed] [Google Scholar]
- 23.Yilmaz O, Verbeke P, Lamont RJ, Ojcius DM. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect Immun. 2006;74:703–710. doi: 10.1128/IAI.74.1.703-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi H, Furuta N, Morisaki I, Amano A. Exit of intracellular Porphyromonas gingivalis from gingival epithelial cells is mediated by endocytic recycling pathway. Cell Microbiol. 2011;13:677–691. doi: 10.1111/j.1462-5822.2010.01564.x. [DOI] [PubMed] [Google Scholar]
- 25.Handfield M, Mans JJ, Zheng G, Lopez MC, Mao S, Progulske-Fox A, Narasimhan G, et al. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell Microbiol. 2005;7:811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- 26.Kuboniwa M, Hasegawa Y, Mao S, Shizukuishi S, Amano A, Lamont RJ, Yilmaz OP. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10:122–128. doi: 10.1016/j.micinf.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao L, Jermanus C, Barbetta B, Choi C, Verbeke P, Ojcius DM, Yilmaz O. Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells. Mol Oral Microbiol. 2010;25:89–101. doi: 10.1111/j.2041-1014.2010.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yilmaz O, Yao L, Maeda K, Rose TM, Lewis EL, Duman M, Lamont RJ, Ojcius DM. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cell Microbiol. 2008;10:863–875. doi: 10.1111/j.1462-5822.2007.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi CH, Spooner R, DeGuzman J, Koutouzis T, Ojcius DM, Yilmaz O. Porphyromonas gingivalis-nucleoside-diphosphate-kinase inhibits ATP-induced reactive-oxygen-species via P2X7 receptor/NADPH-oxidase signalling and contributes to persistence. Cell Microbiol. 2013;15:961–976. doi: 10.1111/cmi.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, Stathopoulou P, et al. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J Biol Chem. 2009;284:23107–23115. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moffatt CE, Lamont RJ. Porphyromonas gingivalis induction of microRNA-203 expression controls suppressor of cytokine signaling 3 in gingival epithelial cells. Infect Immun. 2011;79:2632–2637. doi: 10.1128/IAI.00082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudney JD, Chen R, Sedgewick GJ. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J Dent Res. 2005;84:59–63. doi: 10.1177/154405910508400110. [DOI] [PubMed] [Google Scholar]
- 33.Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontol 2000. 2010;52:38–52. doi: 10.1111/j.1600-0757.2009.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitmore SE, Lamont RJ. The pathogenic persona of community-associated oral streptococci. Mol Microbiol. 2011;81:305–314. doi: 10.1111/j.1365-2958.2011.07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright CJ, Burns LH, Jack AA, Back CR, Dutton LC, Nobbs AH, Lamont RJ, Jenkinson HF. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitmore SE, Lamont RJ. Tyrosine phosphorylation and bacterial virulence. Int J Oral Sci. 2012;4:1–6. doi: 10.1038/ijos.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda K, Tribble GD, Tucker CM, Anaya C, Shizukuishi S, Lewis JP, Demuth DR, Lamont RJ. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69:1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin X, Lamont RJ, Wu J, Xie H. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J Bacteriol. 2008;190:4367–4371. doi: 10.1128/JB.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang BY, Wu J, Lamont RJ, Lin X, Xie H. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol. 2009;47:3902–3906. doi: 10.1128/JCM.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie H, Hong J, Sharma A, Wang BY. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. J Periodontal Res. 2012;47:578–583. doi: 10.1111/j.1600-0765.2012.01469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryder MI. Comparison of neutrophil functions in aggressive and chronic periodontitis. Periodontol 2000. 2010;53:124–137. doi: 10.1111/j.1600-0757.2009.00327.x. [DOI] [PubMed] [Google Scholar]
- 43.Delima AJ, Van Dyke TE. Origin and function of the cellular components in gingival crevice fluid. Periodontol 2000. 2003;31:55–76. doi: 10.1034/j.1600-0757.2003.03105.x. [DOI] [PubMed] [Google Scholar]
- 44.Schroeder HE, Listgarten MA. The gingival tissues: the architecture of periodontal protection. Periodontol 2000. 1997;13:91–120. doi: 10.1111/j.1600-0757.1997.tb00097.x. [DOI] [PubMed] [Google Scholar]
- 45.Vitkov L, Klappacher M, Hannig M, Krautgartner WD. Neutrophil fate in gingival crevicular fluid. Ultrastruct Pathol. 2010;34:25–30. doi: 10.3109/01913120903419989. [DOI] [PubMed] [Google Scholar]
- 46.Meyle E, Stroh P, Gunther F, Hoppy-Tichy T, Wagner C, Hansch GM. Destruction of bacterial biofilms by polymorphonuclear neutrophils: relative contribution of phagocytosis, DNA release, and degranulation. Int J Artif Organs. 2010;33:608–620. doi: 10.1177/039139881003300906. [DOI] [PubMed] [Google Scholar]
- 47.Wagner C, Zimmermann S, Brenner-Weiss G, Hug F, Prior B, Obst U, Hansch GM. The quorum-sensing molecule N-3-oxododecanoyl homoserine lactone (3OC12-HSL) enhances the host defence by activating human polymorphonuclear neutrophils (PMN) Anal Bioanal Chem. 2007;387:481–487. doi: 10.1007/s00216-006-0698-5. [DOI] [PubMed] [Google Scholar]
- 48.Meyle E, Brenner-Weiss G, Obst U, Prior B, Hansch GM. Immune defense against S. epidermidis biofilms: components of the extracellular polymeric substance activate distinct bactericidal mechanisms of phagocytic cells. Int J Artif Organs. 2012;35:700–712. doi: 10.5301/ijao.5000151. [DOI] [PubMed] [Google Scholar]
- 49.Guenther F, Stroh P, Wagner C, Obst U, Hansch GM. Phagocytosis of staphylococci biofilms by polymorphonuclear neutrophils: S. aureus and S. epidermidis differ with regard to their susceptibility towards the host defense. Int J Artif Organs. 2009;32:565–573. doi: 10.1177/039139880903200905. [DOI] [PubMed] [Google Scholar]
- 50.Gunther F, Wabnitz GH, Stroh P, Prior B, Obst U, Samstag Y, Wagner C, Hansch GM. Host defence against Staphylococcus aureus biofilms infection: phagocytosis of biofilms by polymorphonuclear neutrophils (PMN) Mol Immunol. 2009;46:1805–1813. doi: 10.1016/j.molimm.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 51.Mydel P, Takahashi Y, Yumoto H, Sztukowska M, Kubica M, Gibson FC, 3rd, Kurtz DM, Jr, et al. Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection. PLoS Pathog. 2006;2:e76, 712–725. doi: 10.1371/journal.ppat.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hajishengallis G, Wang M, Bagby GJ, Nelson S. Importance of TLR2 in early innate immune response to acute pulmonary infection with Porphyromonas gingivalis in mice. J Immunol. 2008;181:4141–4149. doi: 10.4049/jimmunol.181.6.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheets SM, Robles-Price AG, McKenzie RM, Casiano CA, Fletcher HM. Gingipain-dependent interactions with the host are important for survival of Porphyromonas gingivalis. Front Biosci. 2008;13:3215–3238. doi: 10.2741/2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hajishengallis E, Hajishengallis G. Neutrophil homeostasis and periodontal health in children and adults. J Dent Res. 2013:92. doi: 10.1177/0022034513507956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 56.Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeuchi H, Hirano T, Whitmore SE, Morisaki I, Amano A, Lamont RJ. The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-κB RelA/p65. PLoS Pathog. 2013;9:e1003326. doi: 10.1371/journal.ppat.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darveau RP, Cunningham MD, Bailey T, Seachord C, Ratcliffe K, Bainbridge B, Dietsch M, et al. Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect Immun. 1995;63:1311–1317. doi: 10.1128/iai.63.4.1311-1317.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bainbridge B, Verma RK, Eastman C, Yehia B, Rivera M, Moffatt C, Bhattacharyya I, et al. Role of Porphyromonas gingivalis phosphoserine phosphatase enzyme SerB in inflammation, immune response, and induction of alveolar bone resorption in rats. Infect Immun. 2010;78:4560–4569. doi: 10.1128/IAI.00703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol. 2011;11:187–200. doi: 10.1038/nri2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo Y, Nguyen KA, Potempa J. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000. 2010;54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 63.Abe T, Hosur KB, Hajishengallis E, Reis ES, Ricklin D, Lambris JD, Hajishengallis G. Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (CD88) antagonist. J Immunol. 2012;189:5442–5448. doi: 10.4049/jimmunol.1202339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, Triantafilou M, et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010;3:ra11. doi: 10.1126/scisignal.2000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, Li F, et al. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J Immunol. 2011;186:869–877. doi: 10.4049/jimmunol.1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jain S, Coats SR, Chang AM, Darveau RP. A novel class of lipoprotein lipase-sensitive molecules mediates Toll-like receptor 2 activation by Porphyromonas gingivalis. Infect Immun. 2013;81:1277–1286. doi: 10.1128/IAI.01036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clark RB, Cervantes JL, Maciejewski MW, Farrokhi V, Nemati R, Yao X, Anstadt E, et al. Serine lipids of Porphyromonas gingivalis are human and mouse Toll-like receptor 2 ligands. Infect Immun. 2013;81:3479–89. doi: 10.1128/IAI.00803-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hawlisch H, Belkaid Y, Baelder R, Hildeman D, Gerard C, Kohl J. C5a negatively regulates toll-like receptor 4-induced immune responses. Immunity. 2005;22:415–426. doi: 10.1016/j.immuni.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Burns E, Bachrach G, Shapira L, Nussbaum G. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: Activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J Immunol. 2006;177:8296–8300. doi: 10.4049/jimmunol.177.12.8296. [DOI] [PubMed] [Google Scholar]
- 70.Hajishengallis G, Shakhatreh MA, Wang M, Liang S. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J Immunol. 2007;179:2359–2367. doi: 10.4049/jimmunol.179.4.2359. [DOI] [PubMed] [Google Scholar]
- 71.Hajishengallis G, Lambris JD. Crosstalk pathways between Toll-like receptors and the complement system. Trends Immunol. 2010;31:154–163. doi: 10.1016/j.it.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harokopakis E, Hajishengallis G. Integrin activation by bacterial fimbriae through a pathway involving CD14, Toll-like receptor 2, and phosphatidylinositol-3-kinase. Eur J Immunol. 2005;35:1201–1210. doi: 10.1002/eji.200425883. [DOI] [PubMed] [Google Scholar]
- 73.Harokopakis E, Albzreh MH, Martin MH, Hajishengallis G. TLR2 transmodulates monocyte adhesion and transmigration via Rac1- and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J Immunol. 2006;176:7645–7656. doi: 10.4049/jimmunol.176.12.7645. [DOI] [PubMed] [Google Scholar]
- 74.Wang M, Shakhatreh MA, James D, Liang S, Nishiyama S, Yoshimura F, Demuth DR, Hajishengallis G. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J Immunol. 2007;179:2349–2358. doi: 10.4049/jimmunol.179.4.2349. [DOI] [PubMed] [Google Scholar]
- 75.Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11:637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. 2007;178:7242–7250. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- 77.Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slaney JM, Gallagher A, Aduse-Opoku J, Pell K, Curtis MA. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect Immun. 2006;74:5352–5361. doi: 10.1128/IAI.00304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schenkein HA. Complement factor D-like activity of Porphyromonas gingivalis W83. Oral Microbiol Immunol. 1991;6:216–220. doi: 10.1111/j.1399-302x.1991.tb00480.x. [DOI] [PubMed] [Google Scholar]
- 80.Wingrove JA, DiScipio RG, Chen Z, Potempa J, Travis J, Hugli TE. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J Biol Chem. 1992;267:18902–18907. [PubMed] [Google Scholar]
- 81.Potempa M, Potempa J, Okroj M, Popadiak K, Eick S, Nguyen KA, Riesbeck K, Blom AM. Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J Immunol. 2008;181:5537–5544. doi: 10.4049/jimmunol.181.8.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rangarajan M, Aduse-Opoku J, Paramonov N, Hashim A, Bostanci N, Fraser OP, Tarelli E, Curtis MA. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J Bacteriol. 2008;190:2920–2932. doi: 10.1128/JB.01868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Brien-Simpson NM, Pathirana RD, Walker GD, Reynolds EC. Porphyromonas gingivalis RgpA-Kgp proteinase-adhesin complexes penetrate gingival tissue and induce proinflammatory cytokines or apoptosis in a concentration-dependent manner. Infect Immun. 2009;77:1246–1261. doi: 10.1128/IAI.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, et al. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol. 2009;11:1587–1599. doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Curtis MA, Percival RS, Devine D, Darveau RP, Coats SR, Rangarajan M, Tarelli E, Marsh PD. Temperature dependent modulation of Porphyromonas gingivalis lipid A structure and interaction with the innate host defences. Infect Immun. 2011;79:1187–1193. doi: 10.1128/IAI.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Belstrom D, Holmstrup P, Damgaard C, Borch TS, Skjodt MO, Bendtzen K, Nielsen CH. The atherogenic bacterium Porphyromonas gingivalis evades circulating phagocytes by adhering to erythrocytes. Infect Immun. 2011;79:1559–1565. doi: 10.1128/IAI.01036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carrion J, Scisci E, Miles B, Sabino GJ, Zeituni AE, Gu Y, Bear A, et al. Microbial carriage state of peripheral blood dendritic cells (DCs) in chronic periodontitis influences DC differentiation, atherogenic potential. J Immunol. 2012;189:3178–3187. doi: 10.4049/jimmunol.1201053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Madrigal AG, Barth K, Papadopoulos G, Genco CA. Pathogen-mediated proteolysis of the cell death regulator RIPK1 and the host defense modulator RIPK2 in human aortic endothelial cells. PLoS Pathog. 2012;8:e1002723. doi: 10.1371/journal.ppat.1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maresz KJ, Hellvard A, Sroka A, Adamowicz K, Bielecka E, Koziel J, Gawron K, et al. Porphyromonas gingivalis facilitates the development and progression of destructive arthritis through its unique bacterial peptidylarginine deiminase (PAD) PLoS Pathog. 2013;9:e1003627. doi: 10.1371/journal.ppat.1003627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hayashi C, Gudino CV, Gibson FC, 3rd, Genco CA. Review: Pathogen-induced inflammation at sites distant from oral infection: bacterial persistence and induction of cell-specific innate immune inflammatory pathways. Mol Oral Microbiol. 2010;25:305–316. doi: 10.1111/j.2041-1014.2010.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zelkha SA, Freilich RW, Amar S. Periodontal innate immune mechanisms relevant to atherosclerosis and obesity. Periodontol 2000. 2010;54:207–221. doi: 10.1111/j.1600-0757.2010.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA-the citrullinated enolase connection. Nat Rev Rheumatol. 2010;6:727–730. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- 93.Moutsopoulos NM, Kling HM, Angelov N, Jin W, Palmer RJ, Nares S, Osorio M, Wahl SM. Porphyromonas gingivalis promotes Th17 inducing pathways in chronic periodontitis. J Autoimmun. 2012;39:294–303. doi: 10.1016/j.jaut.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. Epithelial cell pro-inflammatory cytokine response differs across dental plaque bacterial species. J Clin Periodontol. 2010;37:24–29. doi: 10.1111/j.1600-051X.2009.01505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yilmaz O, Sater AA, Yao L, Koutouzis T, Pettengill M, Ojcius DM. ATP-dependent activation of an inflammasome in primary gingival epithelial cells infected by Porphyromonas gingivalis. Cell Microbiol. 2010;12:188–198. doi: 10.1111/j.1462-5822.2009.01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jauregui CE, Wang Q, Wright CJ, Takeuchi H, Uriarte SM, Lamont RJ. Suppression of T-cell chemokines by Porphyromonas gingivalis. Infect Immun. 2013;81:2288–2295. doi: 10.1128/IAI.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gaddis DE, Maynard CL, Weaver CT, Michalek SM, Katz J. Role of TLR2 dependent-IL-10 production in the inhibition of the initial IFN-γ T cell response to Porphyromonas gingivalis. J Leukoc Biol. 2012;93:21–31. doi: 10.1189/jlb.0512220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11:763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 99.Gaffen SL, Hajishengallis G. A new inflammatory cytokine on the block: rethinking periodontal disease and the Th1/Th2 paradigm in the context of Th17 cells and IL-17. J Dent Res. 2008;87:817–828. doi: 10.1177/154405910808700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gemmell E, Yamazaki K, Seymour GJ. The role of T cells in periodontal disease: homeostasis and autoimmunity. Periodontol 2000. 2007;43:14–40. doi: 10.1111/j.1600-0757.2006.00173.x. [DOI] [PubMed] [Google Scholar]
- 101.Garlet GP. Destructive and protective roles of cytokines in periodontitis: A reappraisal from host defense and tissue destruction viewpoints. J Dent Res. 2010;89:1349–1363. doi: 10.1177/0022034510376402. [DOI] [PubMed] [Google Scholar]
- 102.Jiao Y, Darzi Y, Tawaratsumida K, Marchesan JT, Hasegawa M, Moon H, Chen GY, et al. Induction of bone loss by pathobiont-mediated nod1 signaling in the oral cavity. Cell Host Microbe. 2013;13:595–601. doi: 10.1016/j.chom.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Graves DT, Oates T, Garlet GP. Review of osteoimmunology and the host response in endodontic and periodontal lesions. J Oral Microbiol. 2011;3 doi: 10.3402/jom.v3403i3400.5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Johnson JD, Chen R, Lenton PA, Zhang G, Hinrichs JE, Rudney JD. Persistence of extracrevicular bacterial reservoirs after treatment of aggressive periodontitis. J Periodontol. 2008;79:2305–2312. doi: 10.1902/jop.2008.080254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eick S, Pfister W. Efficacy of antibiotics against periodontopathogenic bacteria within epithelial cells: an in vitro study. J Periodontol. 2004;75:1327–1334. doi: 10.1902/jop.2004.75.10.1327. [DOI] [PubMed] [Google Scholar]