Abstract
BACKGROUND
Obesity has a complicated metabolic pathology, and defining the underlying mechanisms of obesity requires integrative studies with molecular endpoints. Real time quantitative PCR (RT-qPCR) is a powerful tool that has been widely utilized. However, the importance of using carefully validated reference genes in RT-qPCR seems to be overlooked in obesity-related research. The objective of this study was to select a set of reference genes with stable expressions to be used for RT-qPCR normalization in rats under fasted vs. re-fed and chow vs. high fat diet (HFD) conditions.
DESIGN
Male Long-Evans rats were treated under four conditions, chow/fasted, chow/re-fed, HFD/fasted, and HFD/re-fed. Expression stabilities of the13 candidate reference genes were evaluated in the rat hypothalamus, duodenum, jejunum, and ileum using ReFinder software program. The optimal number of reference genes needed for RT-qPCR analyses was determined using geNorm.
RESULTS
Using geNorm analysis, we found that it was sufficient to use the two most stably expressed genes as references in RT-qPCR analyses for each tissue under specific experimental conditions. Unique subsets of reference genes out of the 13 candidate genes were identified, each of which is specific for one type of rat tissue (hypothalamus, duodenum, jejunum, or ileum) under a different combination of diet and feeding condition.
CONCLUSIONS
Our study demonstrates that gene expression levels of reference genes commonly used in obesity-related studies, such as ACTB, or RPS18, are altered by changes in acute or chronic energy status. These findings underline the importance of using reference genes that are stable in expression across experimental conditions when studying the rat hypothalamus and intestine, because these tissues play an integral role in regulation of energy homeostasis. It is our hope that this study will raise awareness among obesity researchers on the essential need for reference gene validation in gene expression studies.
Keywords: Obesity, reference gene, hypothalamus, intestine, RT-qPCR, rat
INTRODUCTION
The worldwide incidence of obesity has led to an increasing need for understanding the molecular mechanisms that drive this epidemic. Obesity is the combined consequence of genetic, behavioral, and environmental factors that drive an imbalance between energy intake and energy expenditure towards an increase in adiposity1. Hormones and peptides secreted from the adipose tissue and the gastrointestinal tract act as signals representing current energy status to the central nervous system (CNS)2. The hypothalamus is a particularly important CNS region that is responsive to these peripheral signals to initiate changes in food intake and/or energy expenditure in order to tightly regulate overall energy balance3. Gut to brain signaling is also thought to be important for lipid sensing4 and glucose sensing5. Due to the increase in frequency and success of bariatric surgery to treat obesity, attention has turned to understanding mechanisms underlying the gut-brain axis in regulating energy homeostasis6.
RT-qPCR has become a valuable method for gene expression analysis and is used extensively in obesity research. In order to avoid measuring the absolute amount of mRNA within a sample, data is analyzed relative to the control group. However, errors are introduced in the technique through the process of RNA isolation, reverse transcription, and real time PCR. To overcome these variables, a reference gene is used for normalizing gene expression. However, this assumes that the expression of the reference gene must remain constant in all cell/tissue types and under specific experimental conditions. Unfortunately, increasing data have shown that no single gene has constant expression across all cell types or under all physiological/pathological conditions7–9. Therefore, to obtain accurate gene expression information, it is imperative that stable reference genes are chosen for the specific type of tissue and experimental condition. Several algorithms, including the geNorm9, NormFinder10, BestKeep11, and comparative ΔCt (cycle thresholds) method12, have been developed for selection of suitable reference genes and are widely used. The use of at least three reference genes for the correct normalization of RT-qPCR data has been proposed by Vandesompele9 and is a recommended approach for normalizing RT-qPCR data9,13. Recently, ReFinder14, a web based program that integrates four mathematical programs, i.e. geNorm, NormFinder, BestKeeper, and comparative ΔCt method, was developed to provide a convenient and adequate means for reference gene evaluation12.
Rodents, such as mice and rats, have been commonly used in studying diet induced obesity and diabetes. Surprisingly, evaluation of reference genes in obesity research has received little attention15. In most gene expression studies, a single gene, including ACTB and RPS18, is still commonly used without any mentioning of whether these genes are affected by the experimental conditions16–19. In other fields, it has been realized that the expression stability of reference genes are influenced by conditions used in studies20–22. Therefore, we studied 13 commonly used reference genes in the rat hypothalamus, duodenum, jejunum, and ileum across different diets and feeding schemes typically used in obesity research, to determine which reference genes would be most appropriate for these particular investigations. We found that the expression levels of several commonly used reference genes, such as RPS18 in the hypothalamus or ACTB in the intestine, fluctuated across the varied diet and nutrition conditions. These data underscore the systematic validation of reference genes in obesity research.
MATERIALS AND METHODS
Animals
Male Long-Evans rats were single-housed under controlled conditions (12:12-hour light-dark cycle, 50–60% humidity, 25°C with free access to water and food except where noted) in the Metabolic Diseases Institute at University of Cincinnati. Rats were fed either a high fat diet (HFD) (n=12), (Research Diets, New Brunswick, NJ, D12451; 45% fat; 4.73kcal/g) or a standard chow diet (Harlan-Teklad, Indianapolis, IN) (n=12) for 8 weeks prior to experiments. All procedures for animal use were approved by the University of Cincinnati Institutional Animal Care and Use Committee.
Animal groups and tissue collection
Rats were assigned to one of the four treatment groups (n=6/group): chow/fasted, chow/re-fed, HFD/fasted, HFD/re-fed. All rats were fasted overnight with free access of water and then half of the rats were re-fed for 2 hours. At the end of 2-hr feeding in the dark cycle, all rats were individually placed in a CO2 chamber and then sacrificed by decapitation. Brain, duodenum, jejunum, and ileum were taken quickly and flash frozen in sub-zero 2-methylbutane and stored at −80°C until RNA isolation.
RNA isolation and cDNA synthesis
The hypothalamus was quickly dissected from a semi-frozen brain. Total RNA was isolated from the hypothalamus, duodenum, jejunum, and ileum using RNeasy mini-columns (Qiagen, Valencia, CA). Genomic DNA was eliminated by DNase I on column treatment with RNase-free DNase set (Qiagen, Valencia, CA). RNA integrity was confirmed by visualizing an approximate 2/1 ratio of 28S to 18S band on the 1% agarose gel stained by ethidium bromide. RNA purity was confirmed with absorbance ratio of >2.0 at 260nm/280nm using NanoVue (GE Healthcare, Piscataway, NJ). 1 μg of total RNA was used to generate cDNA using iScript following manufacturer's protocol (Bio-Rad, Hercules, CA). cDNA was made from all samples for each tissue at the same time to minimize experimental variations.
Primers and qPCR
TaqMan Gene Expression Assays [Applied Biosystems (ABI) (Carlsbad, CA)] were used for qPCR. Pre-optimized (with nearly 100% efficiency) primers/probes of 13 candidate reference genes (Table 1) were purchased from ABI. qPCR reactions were set at 10 μl with 5μl of the TaqMan Gene Expression Master Mix, 0.5μl primer/probe, 2μl of 6× diluted cDNA, and 2.5μl H2O. Plates were run on the ABI Prism 79000HT Fast Real-Time PCR System. All qPCRs were run in duplicates on the same thermal cycles (95°, 10min, 40 cycles of 0.01sec @95°C, 20sec @60°C). No amplification signal was detected in water or no-RT RNA samples. Each gene was run with all samples for each tissue on the same plate. The threshold value was manually set to 0.2 to guarantee the comparability between the Cts obtained from different genes and different runs.
Table 1.
Candidate reference genes and catalog numbers (ABI)
| Symbol | Gene name | Accession number | Function | Cat. No. |
|---|---|---|---|---|
| ACTB | Actin, beta | NM_031144.2 | Cytoskeletal structural protein | 4352931E |
| B2M | beta-2 microglobulin | NM_012512.2 | Assembly and surface expression of MHC class I molecules | Rn00560865_m1 |
| HMBS | Hydroxymethylbilane synthase | NM_013168.2 | Heme synthesis, porphyrin metabolism | Rn00565886_m1 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | NM_012583.2 | Generation of purine nucleotides through the purine salvage pathway | Rn01527840_m1 |
| PGK1 | Phosphoglycerate kinase I | NM_053291.3 | Phosphoprotein glycolyosis | Rn00821429_g1 |
| PPIB | Peptidylprolyl isomerase B | NM_022536.1 | Endoplasmic reticulum cyclosporine-binding protein | Rn00574762_m1 |
| RPLP0 | Ribosomal protein large, P0 | NM_022402.2 | Protein synthesis | Rn00821065_g1 |
| RPLP2 | Ribosomal protein large, P2 | NM_001030021.1 | Protein synthesis | Rn01479927_g1 |
| RPL32 | Ribosomal protein L32 | NM_013226.2 | Protein synthesis | Rn00820748_g1 |
| RPS18 | Ribosomal protein S18 | NM_213557.1 | Protein synthesis | Rn01428915_g1 |
| TBP | TATA box binding protein | NM_001004198.1 | General RNA polymerase II transcription factor | Rn01455648_m1 |
| UBC | Ubiquitin C | NM_017314.1 | Involved in muscle protein metabolism | Rn01789812_g1 |
| YWHAZ | Tyrosine 3-monoxygenase/tryptophan 5-monoxygenase activation protein, zeta polypeptide | NM_013011.3 | Belongs to the 14-3-3 family of proteins which mediate signal transduction by binding to phosphoserine-containing proteins | Rn00755072_m1 |
Data analysis and statistics
The gene expression stabilities of the 13 candidate reference genes from each tissue were determined by ReFinder. Upon the input of Ct values, ReFinder invokes four commonly used computational programs, geNorm, NormFinder, BestKeeper, and comparative ΔCt method, to process those data, respectively. The processed ranking results from each program were aggregated by ReFinder to generate gene expression stability rank orders. Based on their rankings from each program, ReFinder assigns an appropriate weight to each individual reference gene and then calculates the geometric mean of the weights for each gene to reach its overall final ranking, the comprehensive ranking order, among all 13 candidate genes. The details in the calculation procedures have been previously described23. We used the comprehensive rank order as our results.
The gene expression stabilities of the 13 candidate genes were analyzed under 4 separated conditions for each tissue: (1) fasted vs. re-fed under chow diet, (2) fasted vs. re-fed under HFD, (3) chow vs. HFD in fasted condition, and (4) chow vs. HFD in fasted then re-fed condition. 12 Cts from each individual gene were analyzed under four conditions described above, for each tissue. We also did an overall evaluation of gene expression stabilities in pooled conditions, in which 24 Ct values of each candidate gene were analyzed for each tissue from chow/fasted, chow/re-fed, HFD/fasted, and HFD/re-fed animals.
The optimal numbers of reference genes required for accurate normalization were determined by the pairwise variation (Vn/Vn+1) using geNorm. A Vn/n+1 value (n = the number of reference genes desired) represents a pairwise variation between two sets of reference genes with the second set containing an additional gene. A large variation means that the added gene has a significant effect and should be included in normalization analyses. To calculate V values, Cts from all candidate genes were input into geNorm excel based program. An arbitral cut-off value of 0.15 for Vn/n+1 is adopted9 to assist evaluation of optimal gene numbers. For example, if a V2/3 is 0.22 and a V3/4 is 0.12, then three reference genes are recommended for RT-qPCR analyses. Since V3/4value is 0.12 (< cutoff value), using four reference genes will not make a significant impact on RT-qPCR analyses.
RESULTS
Transcription profiles of 13 candidate reference genes
The expression level of 13 candidate genes (Table 1) was evaluated as threshold cycle (Ct) in rat hypothalamus, duodenum, jejunum, and ileum that were treated in chow/fasted, chow/re-fed, HFD/fasted, and HFD/re-fed conditions (Fig. 1, for each gene, n=6/group). These genes were selected from ABI rat endogenous control array gene card (Applied Biosystems, Carlsbad, CA). They were chosen because they were routinely used as reference genes for normalization and they are expected to have minimal differential expression across different tissues and experimental conditions.
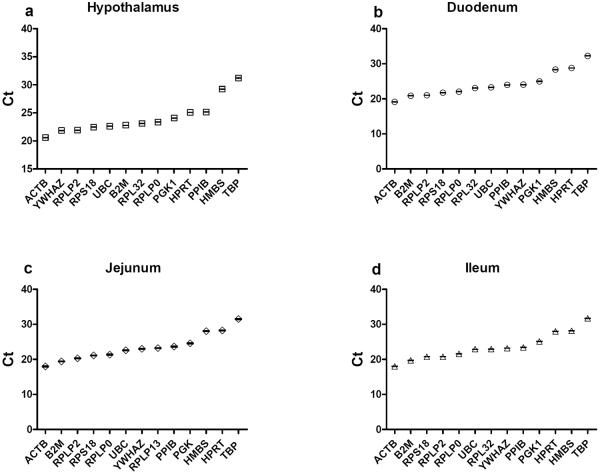
Figure 1. Distribution of threshold cycle (Ct) values of 13 candidate reference genes.
The boxes show the Ct of each gene in the hypothalamus (a), duodenum (b), jejunum (c), and ileum (d). Black center line indicates the median Ct. Samples were pooled from all four conditions: chow/fasted, chow/re-fed, HFD/fasted, and HFD/re-fed. Data are presented as mean±SEM (n=24)
The Cts across the candidate housekeeping genes ranged from 20.6 to 31.22 in the hypothalamus (Fig. 1a), 19.13 to 32.27 in the duodenum (Fig. 1b), 17.97 to 31.51 in the jejunum (Fig. 1c), and 17.98 to 31.67 in the ileum (Fig. 1d). The wide range of Ct values suggested that these candidate genes had different expression levels in the four tissues examined. Among the 13 candidate genes, ACTB mRNA was the most abundant whereas TBP mRNA was the least abundant in all four tissues.
Gene expression stability analysis of candidate reference genes
Hypothalamus
When comparing re-fed vs. fasted conditions, the most stably expressed reference genes within the hypothalamus were PGK1 and B2M in rats fed chow and PGK1 and HPRT in rats fed HFD. When comparing chow or HFD, the most stable reference genes were B2M and ACTB in fasted rats and RPLP2 and YWHAZ in re-fed rats. When the pooled conditions were considered, the reference genes that expressed consistently were B2M and RPLP0. The expression of TBP and PPIB genes were the most volatile across all conditions (Table 2).
Table 2.
Rank order of reference gene expression stability in the hypothalamus
| Rankinga, b | Chow fasted vs. re-fed | HFD fasted vs. re-fed | Fasted chow vs. HFD | Re-fed chow vs. HFD | Pooled conditions |
|---|---|---|---|---|---|
| 1 | PGK1 | PGK1 | B2M | RPLP2 | B2M |
| 2 | B2M | HPRT | ACTB | YWHAZ | RPLP0 |
| 3 | RPS18 | ACTB | UBC | B2M | ACTB |
| 4 | RPLP0 | RPS18 | RPLP0 | RPLP0 | HMBS |
| 5 | ACTB | RPL32 | HMBS | RPS18 | UBC |
| 6 | HMBS | B2M | HPRT | HPRT | YWHAZ |
| 7 | YWHAZ | UBC | YWHAZ | HMBS | HPRT |
| 8 | HPRT | HMBS | RPS18 | UBC | RPS18 |
| 9 | RPLP2 | YWHAZ | RPL32 | ACTB | RPLP2 |
| 10 | UBC | RPLP0 | PGK1 | PGK1 | PGK1 |
| 11 | RPL32 | RPLP2 | RPLP2 | RPL32 | RPL32 |
| 12 | PPIB | PPIB | PPIB | PPIB | PPIB |
| 13 | TBP | TBP | TBP | TBP | TBP |
The ranking order is the recommended comprehensive ranking by ReFinder. Gene expression stability decreases as the ranking order number increases.
Top two ranked genes are in bold.
Duodenum
In rat duodenum, when comparing fasting vs. re-feeding, the most stable genes were HPRT and RPLP2 in rats fed chow and HMBS and RPS18 in rats fed HFD. When comparing diets, the most stable genes were RPLP0 and RPS18 in fasted rats and HMBS and RPS18 in refed rats. Under the pooled conditions, RPS18 and HMBS were found to be the most stably expressed genes, ACTB and PGK1 were among the least stably expressed genes across all conditions (Table 3).
Table 3.
Rank order of reference gene expression stability in the duodenum
| Rankinga, b | Chow fasted vs. re-fed | HFD fasted vs. re-fed | Fasted chow vs. HFD | Re-fed chow vs. HFD | Pooled conditions |
|---|---|---|---|---|---|
| 1 | HPRT | HMBS | RPLP0 | HMBS | RPS18 |
| 2 | RPLP2 | RPS18 | RPS18 | RPS18 | HMBS |
| 3 | HMBS | RPLP2 | RPLP2 | RPLP2 | HPRT |
| 4 | RPS18 | RPLP0 | HMBS | PPIB | RPLP2 |
| 5 | RPLP0 | RPL32 | HPRT | RPL32 | PPIB |
| 6 | PPIB | B2M | YWHAZ | HPRT | YWHAZ |
| 7 | YWHAZ | HPRT | RPL32 | YWHAZ | RPL32 |
| 8 | UBC | YWHAZ | PPIB | UBC | RPLP0 |
| 9 | RPL32 | PPIB | UBC | B2M | UBC |
| 10 | TBP | UBC | B2M | TBP | B2M |
| 11 | PGK1 | PGK1 | ACTB | RPLP0 | ACTB |
| 12 | B2M | ACTB | PGK1 | ACTB | TBP |
| 13 | ACTB | TBP | TBP | PGK1 | PGK1 |
The ranking order is the recommended comprehensive ranking by ReFinder. Gene expression stability decreases as the ranking order number increases.
Top two ranked genes are in bold.
Jejunum
In the jejunum, when comparing fasted vs. re-fed conditions, the most stably expressed genes were identified as RPS18 and HPRT in rats fed chow and HMBS and YWHAZ in rats fed HFD. When comparing diets, RPLP0 and HMBS were identified as most stable genes in fasted rats, and RPLP2 and YWHAZ in fed rats. RPLP2 and RPLP0 were found to be the most stable genes under the pooled conditions. ACTB was among the genes with the least stable expressions across all conditions (Table 4).
Table 4.
Rank order of reference gene expression stability in the jejunum
| Rankinga, b | Chow fasted vs. re-fed | HFD fasted vs. re-fed | Fasted chow vs. HFD | Re-fed chow vs. HFD | Pooled conditions |
|---|---|---|---|---|---|
| 1 | RPS18 | HMBS | RPLP0 | RPLP2 | RPLP2 |
| 2 | HPRT | YWHAZ | HMBS | YWHAZ | RPLP0 |
| 3 | RPLP13 | RPLP0 | YWHAZ | HPRT | HPRT |
| 4 | RPLP2 | TBP | RPS18 | RPS18 | HMBS |
| 5 | UBC | HPRT | B2M | RPLP13 | RPS18 |
| 6 | HMBS | B2M | RPLP2 | HMBS | B2M |
| 7 | RPLP0 | RPLP2 | ACTB | PGK1 | YWHAZ |
| 8 | PGK1 | RPLP13 | RPLP13 | UBC | PGK1 |
| 9 | B2M | PGK1 | TBP | RPLP0 | RPLP13 |
| 10 | PPIB | RPS18 | PGK1 | B2M | UBC |
| 11 | YWHAZ | ACTB | HPRT | TBP | ACTB |
| 12 | TBP | UBC | UBC | ACTB | TBP |
| 13 | ACTB | PPIB | PPIB | PPIB | PPIB |
The ranking order is the recommended comprehensive ranking by ReFinder. Gene expression stability decreases as the ranking order number increases.
Top two ranked genes are in bold.
Ileum
In rat ileum, when comparing fasted vs. re-fed conditions, HMBS and RPLP2 were identified as the genes with most stable expressions in rats fed chow, and RPS18 and RPLP2 in rats fed HFD. However, when comparing diets, YWHAZ and RPS18 were found to be the most stable genes in fasted rats, and RPS18 and RPLP2 in re-fed rats. When the pooled conditions are considered, RPS18 and YWHAZ were found to be the most stably expressed genes. And again, ACTB was found among the least stable genes across all experimental conditions (Table 5).
Table 5.
Rank order of reference gene expression stability in the ileum
| Rankinga, b | Chow fasted vs. re-fed | HFD fasted vs. re-fed | Fasted chow vs. HFD | Re-fed chow vs. HFD | Pooled conditions |
|---|---|---|---|---|---|
| 1 | HMBS | RPS18 | YWHAZ | RPS18 | RPS18 |
| 2 | RPLP2 | RPLP2 | RPS18 | RPLP2 | YWHAZ |
| 3 | RPL32 | RPL32 | PPIB | YWHAZ | RPLP2 |
| 4 | UBC | HMBS | HMBS | RPL32 | RPL32 |
| 5 | RPS18 | UBC | RPLP2 | PPIB | UBC |
| 6 | YWHAZ | YWHAZ | PGK1 | UBC | PPIB |
| 7 | PGK1 | PGK1 | UBC | HMBS | HMBS |
| 8 | RPLP0 | PPIB | RPLP0 | HPRT | RPLP0 |
| 9 | PPIB | HPRT | RPL32 | RPLP0 | HPRT |
| 10 | HPRT | RPLP0 | ACTB | TBP | ACTB |
| 11 | ACTB | ACTB | HPRT | PGK1 | PGK1 |
| 12 | TBP | B2M | B2M | ACTB | TBP |
| 13 | B2M | TBP | TBP | B2M | B2M |
The ranking order is the recommended comprehensive ranking by ReFinder. Gene expression stability decreases as the ranking order number increases.
Top two ranked genes are in bold.
Optimal reference genes required for normalization
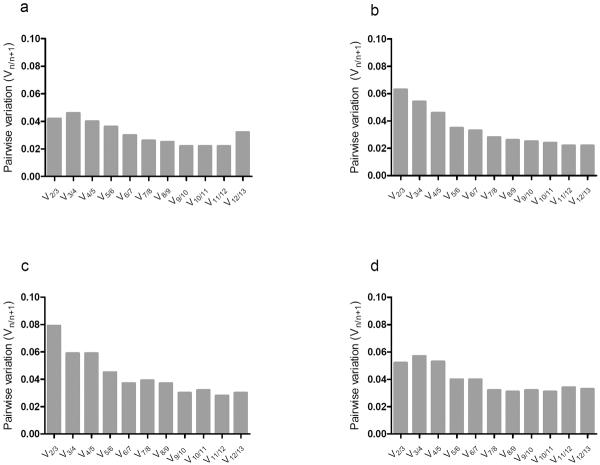
The optimal numbers of reference genes need for RT-qPCR analyses were determined in pooled conditions for each tissue by pairwise variation method (Vn/n+1) using geNorm software. Figures 2a–d showed that the V2/3 values were 0.042, 0.063, 0.079, and 0.052 for the hypothalamus, duodenum, jejunum, and ileum, respectively. Since they are all smaller than the recommended cut-off value of 0.15, it indicates that, under the pooled conditions used in this study, using two reference genes for normalization would be sufficient to obtain accurate data.
Figure 2. Pairwise variation (Vn/n+1) of 13 candidate reference genes.
To derive the numbers of reference genes needed for accurate RT-qPCR, Vn/n+1 were calculated by geNorm software by inputting the Ct of each candidate gene from each tissue, hypothalamus (a), duodenum (b), jejunum (c), and ileum (d). Vn/n+1, n = the number of reference genes desired, represents the pairwise variation between two sets of reference genes with the second set containing an additional gene. The cutoff value of V is 0.15.
DISCUSSION
The concept that reference genes used for normalization in RT-PCR analyses should be validated prior to use was initially suggested in 200224 and has been realized in various scientific research disciplines such as plant sciences23,25,26, cancer27–29, stem cell30–32, and cardiovascular research33–35. Some limited data have been published from metabolic research in rodents, which clearly show that different sets of reference genes are found only to be suitable for each experimental condition and for each tissue15,20–22. However, no previous research has identified the most stably expressed reference genes within the rat hypothalamus or small intestine under different dietary and feeding conditions for energy homeostasis studies. Therefore, our study is the first attempt in this area to provide first hand evidence on the necessity of reference gene optimization.
This study was designed (1) to evaluate gene expression stability across different dietary conditions among 13 commonly used endogenous control genes, and (2) to identify the reference genes most suitable for obesity studies that use RT-PCR analysis in the rat hypothalamus and intestine. Our data confirmed that expression of many of the 13 commonly used reference genes can be affected at different levels by both tissues and conditions used in experiments (Tables 2–5). As a result, the ranking order in gene expression stability among the 13 candidate genes from each tissue was found to vary across different experimental settings. For example, in the rat hypothalamus and jejunum, there was no single pair of reference genes that were stable across all conditions, suggesting that gene expression in these two tissues were more susceptible to the experimental conditions. Our studies also showed that TBP and PPIB were consistently found to be the least stable reference genes in the hypothalamus, while ACTB was regularly scored among the least stable reference genes in the rat intestine under all but three conditions (Table 3–5).
The lack of stability in expression by reference genes in each tissue across the various laboratory conditions exemplifies the complex physiological responses to changes in feeding conditions. In order to identify accurate gene expression during the course of obesity development, it is critical to carefully select reference genes used in RT-qPCR analyses. Unfortunately, in studies reported by several groups36–39, ACTB was frequently used as a reference gene for normalization in RT-qPCR analyses in the hypothalamus and intestine. Based on our findings in this study, the expression of ACTB is one of the least consistent in the intestine and can fluctuate quite widely in the hypothalamus (Tables 3–5). Therefore, depending on the experimental condition, the use of ACTB could lead to either an over- or underestimation of the role of the target gene.
In 2011, Lavin et al. published a study on the effect of HFD on anti-inflammation40. In an experiment with conditions similar to what we used in our study, they reported that in mice fed on a HFD for 10–12 weeks, no difference was observed in the expression of inflammation related genes, such as F4/80, CD11b, and IL1α, between fed vs. fasted conditions. However, in mice fed a low fat diet gene expressions of these inflammatory genes were down regulated in fasted vs. fed condition. Since their gene expression analyses were based on a single reference gene, ACTB which we now know can vary under the conditions that change energy homeostasis, their experimental design may be flawed.
It is recommended to use three reference genes in RT-qPCR normalization studies to ensure data quality9,13. However, due to the constraints in cost and time, it is not always feasible to include that many reference genes in an analysis. Our data analyses using geNorm (Fig. 2) indicated that two reference genes were adequate for RT-qPCR analyses under the experimental conditions we used. Therefore, in certain conditions, it would be sufficient to use two reference genes in RT-qPCR analyses.
In conclusion, our study demonstrated that expression of 13 candidate reference genes commonly used in obesity research was differentially affected by dietary and feeding conditions, as well as by tissue. From the 13 candidate reference genes, we have identified a subset of reference genes suitable for RT-qPCR normalization in the rat hypothalamus and intestine. The development of obesity is a complex event and subtle changes in gene expression during the development of obesity can impact a cascade of signaling pathways that further contribute to obesity and/or its comorbidities. Thus, in order to detect the small but significant changes in gene expression, normalization using highly stable candidate genes becomes critical. It is our hope that the reference genes identified here can be a resource for future obesity studies.
Footnotes
CONFLIC OF INTEREST The authors declare no conflict of interest.
REFERENCES
- 1.French SA, Story M, Jeffery RW. Environmental influences on eating and physical activity. Annual review of public health. 2001;22:309–35. doi: 10.1146/annurev.publhealth.22.1.309. [DOI] [PubMed] [Google Scholar]
- 2.Konturek SJ, Konturek JW, Pawlik T, Brzozowski T. Brain-gut axis and its role in the control of food intake. Journal of physiology and pharmacology : an official journal of the Polish Physiological Society. 2004;55:137–54. [PubMed] [Google Scholar]
- 3.Williams G, Harrold JA, Cutler DJ. The hypothalamus and the regulation of energy homeostasis: lifting the lid on a black box. The Proceedings of the Nutrition Society. 2000;59:385–96. doi: 10.1017/s0029665100000434. [DOI] [PubMed] [Google Scholar]
- 4.Breen DM, Yang CS, Lam TKT. Gut-brain signalling: how lipids can trigger the gut. Diabetes/metabolism research and reviews. 2011;27:113–9. doi: 10.1002/dmrr.1160. [DOI] [PubMed] [Google Scholar]
- 5.Burcelin R. The gut-brain axis: a major glucoregulatory player. Diabetes & metabolism. 2010;36(Suppl 3):S54–8. doi: 10.1016/S1262-3636(10)70468-7. [DOI] [PubMed] [Google Scholar]
- 6.O'Brien PE. Bariatric surgery: mechanisms, indications and outcomes. Journal of gastroenterology and hepatology. 2010;25:1358–65. doi: 10.1111/j.1440-1746.2010.06391.x. [DOI] [PubMed] [Google Scholar]
- 7.Bustin SA. Real-time, fluorescence-based quantitative PCR: a snapshot of current procedures and preferences. Expert review of molecular diagnostics. 2005;5:493–8. doi: 10.1586/14737159.5.4.493. [DOI] [PubMed] [Google Scholar]
- 8.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes and immunity. 2005;6:279–84. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 9.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer research. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 11.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnology letters. 2004;26:509–15. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 12.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC molecular biology. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.NdesoVampele J, Kubista M, Pfaffl MM. Reference gene validation software for improved normalization. Real-Time PCR: current technology and applications. 2009;4:47–64. [Google Scholar]
- 14.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant molecular biology. 2012 doi: 10.1007/s11103-012-9885-2. doi:10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 15.Cabiati M, et al. Tissue-specific selection of stable reference genes for real-time PCR normalization in an obese rat model. Journal of molecular endocrinology. 2012;48:251–60. doi: 10.1530/JME-12-0024. [DOI] [PubMed] [Google Scholar]
- 16.Auvinen HE, et al. The effects of high fat diet on the basal activity of the hypothalamus-pituitary-adrenal axis in mice. The Journal of endocrinology. 2012;214:191–7. doi: 10.1530/JOE-12-0056. [DOI] [PubMed] [Google Scholar]
- 17.Calegari VC, et al. Inflammation of the hypothalamus leads to defective pancreatic islet function. The Journal of biological chemistry. 2011;286:12870–80. doi: 10.1074/jbc.M110.173021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Furuta M, et al. Severe defect in proglucagon processing in islet A-cells of prohormone convertase 2 null mice. The Journal of biological chemistry. 2001;276:27197–202. doi: 10.1074/jbc.M103362200. [DOI] [PubMed] [Google Scholar]
- 19.Lam YY, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PloS one. 2012;7:e34233. doi: 10.1371/journal.pone.0034233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Djordjevic A, et al. Identification of Suitable Reference Genes for Gene Expression Studies in Tissues from Fructose-Fed Rats. nullnull, null.
- 21.Tanic N, Perovic M, Mladenovic A, Ruzdijic S, Kanazir S. Effects of aging, dietary restriction and glucocorticoid treatment on housekeeping gene expression in rat cortex and hippocampus-evaluation by real time RT-PCR. Journal of molecular neuroscience : MN. 2007;32:38–46. doi: 10.1007/s12031-007-0006-7. [DOI] [PubMed] [Google Scholar]
- 22.Xu L, et al. Selection of reference genes for qRT-PCR in high fat diet-induced hepatic steatosis mice model. Molecular biotechnology. 2011;48:255–62. doi: 10.1007/s12033-010-9366-2. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, Pan X, Xiao P, Farwell MA, Zhang B. Evaluation and identification of reliable reference genes for pharmacogenomics, toxicogenomics, and small RNA expression analysis. Journal of cellular physiology. 2011;226:2469–77. doi: 10.1002/jcp.22725. [DOI] [PubMed] [Google Scholar]
- 24.Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods (San Diego, Calif.) 2010;50:227–30. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Han X, et al. Selection of Reliable Reference Genes for Gene Expression Studies Using Real-Time PCR in Tung Tree during Seed Development. PloS one. 2012;7:e43084. doi: 10.1371/journal.pone.0043084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le DT, et al. Evaluation of Candidate Reference Genes for Normalization of Quantitative RT-PCR in Soybean Tissues under Various Abiotic Stress Conditions. PloS one. 2012;7:e46487. doi: 10.1371/journal.pone.0046487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frost K, et al. Inhibin/activin expression in human and rodent liver: subunits α and βB as new players in human hepatocellular carcinoma? British journal of cancer. 2011;104:1303–12. doi: 10.1038/bjc.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rienzo M, et al. Identification of valid reference housekeeping genes for gene expression analysis in tumor neovascularization studies. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2012 doi: 10.1007/s12094-012-0904-1. doi:10.1007/s12094-012-0904-1. [DOI] [PubMed] [Google Scholar]
- 29.Ma Y, Dai H, Kong X, Wang L. Impact of thawing on reference gene expression stability in renal cell carcinoma samples. Diagnostic molecular pathology : the American journal of surgical pathology, part B. 2012;21:157–63. doi: 10.1097/PDM.0b013e31824d3435. [DOI] [PubMed] [Google Scholar]
- 30.Farrokhi A, et al. Appropriate reference gene selection for real-time PCR data normalization during rat mesenchymal stem cell differentiation. Cellular and molecular biology (Noisy-le-Grand, France) 2012;(Suppl 58):OL1660–70. [PubMed] [Google Scholar]
- 31.Genovesi LA, Anderson D, Carter KW, Giles KM, Dallas PB. Identification of suitable endogenous control genes for microRNA expression profiling of childhood medulloblastoma and human neural stem cells. BMC research notes. 2012;5:507. doi: 10.1186/1756-0500-5-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chooi WH, Zhou R, Yeo SS, Zhang F, Wang D-A. Determination and Validation of Reference Gene Stability for qPCR Analysis in Polysaccharide Hydrogel-Based 3D Chondrocytes and Mesenchymal Stem Cell Cultural Models. Molecular biotechnology. 2012 doi: 10.1007/s12033-012-9604-x. doi:10.1007/s12033-012-9604-x. [DOI] [PubMed] [Google Scholar]
- 33.Brattelid T, et al. Normalization strategy is critical for the outcome of miRNA expression analyses in the rat heart. Physiological genomics. 2011;43:604–10. doi: 10.1152/physiolgenomics.00131.2010. [DOI] [PubMed] [Google Scholar]
- 34.Ellefsen S, et al. Per-unit-living tissue normalization of real-time RT-PCR data in ischemic rat hearts. Physiological genomics. 2012;44:651–6. doi: 10.1152/physiolgenomics.00004.2012. [DOI] [PubMed] [Google Scholar]
- 35.Tan SC, et al. Identification of valid housekeeping genes for quantitative RT-PCR analysis of cardiosphere-derived cells preconditioned under hypoxia or with prolyl-4-hydroxylase inhibitors. Molecular biology reports. 2012;39:4857–67. doi: 10.1007/s11033-011-1281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Althage MC, et al. Targeted ablation of glucose-dependent insulinotropic polypeptide-producing cells in transgenic mice reduces obesity and insulin resistance induced by a high fat diet. The Journal of biological chemistry. 2008;283:18365–76. doi: 10.1074/jbc.M710466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Funato H, Oda S, Yokofujita J, Igarashi H, Kuroda M. Fasting and high-fat diet alter histone deacetylase expression in the medial hypothalamus. PloS one. 2011;6:e18950. doi: 10.1371/journal.pone.0018950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rother E, et al. Hypothalamic JNK1 and IKKβ activation and impaired early postnatal glucose metabolism after maternal perinatal high-fat feeding. Endocrinology. 2012;153:770–81. doi: 10.1210/en.2011-1589. [DOI] [PubMed] [Google Scholar]
- 39.Tsuchida T, et al. MGAT2 deficiency ameliorates high-fat diet-induced obesity and insulin resistance by inhibiting intestinal fat absorption in mice. Lipids in health and disease. 2012;11:75. doi: 10.1186/1476-511X-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavin DN, et al. Fasting induces an anti-inflammatory effect on the neuroimmune system which a high-fat diet prevents. Obesity (Silver Spring, Md.) 2011;19:1586–94. doi: 10.1038/oby.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]