Abstract
We have done conformational study of hNck2 SH3 domain by means of far-ultraviolet (far-UV) circular dichroism (CD) and X-ray solution scattering (XSS). The results indicated that the following: (1) hNck2 SH3 domain protein exhibited concentration dependent monomer–dimer transition at neutral pH, while the secondary structure of this protein was independent of the protein concentration. (2) The hNck2 SH3 domain also exhibited pH dependent monomer–dimer transition. This monomer–dimer transition was accompanied with helix-β transition of the secondary structural change. Moreover, the acid-induced conformation, which was previously studied by Liu and Song by CD and nuclear magnetic resonance (NMR), was found to be not compact, but the conformation of the protein at acidic pH was similar to the cold denatured state (C-state) reported by Yamada et al. for equine β-lactoglobulin. We calculated that a structure of the equilibrium helix-rich intermediate of the hNck2 SH3 domain by DAMMIF program.
Keywords: Equilibrium helix-rich intermediate, Monomer–dimer transition, C-state
1. Introduction
Src homology domain 3 (SH3) is a small protein (50–70 amino acids), which is involved in the signal transduction in the cell. SH3 domains comprise huge protein family [1]. The SH3 domains take the same protein fold topology: two small orthogonal three-stranded β-sheets with an associated irregular two-stranded-sheet packing against each other in a sandwich structure [2,3].
Although no α-helices are involved in the native structure, src SH3 domain is folded through an α-helix-rich intermediate [2–6]. The α-helix-rich intermediate was also found on the folding pathway of Fyn SH3 domain [4]. These α-helix-rich folding intermediate is rather common, as found in many other proteins [4–22]. The folding pathways via such α-helix-rich intermediate have been also thoroughly investigated in many β-rich proteins such as bovine and equine β-lactoglobulins [9,17–22].
The equilibrium α-helix-rich intermediates were also found in several proteins [5,14–16].
Particularly, a mutant of src SH3 domain, A45G, took an equilibrium α-helix-rich intermediate at acidic pH [5]. The equilibrium intermediate has the same α-helix content and the same Rg with the folding α-helix-rich intermediate within the experimental error [4,5]. The equilibrium α-helix-rich intermediate was also found in equine β-lactoglobulin in various conditions; pH 1.5 and pH 4 with 4 M urea at 25 °C [14–16]. Fujiwara et al. reported that the equilibrium intermediate at pH 4 with 4 M urea was a molten globule state, and was the same with the kinetically-obtained intermediate [15]. Another α-helix-rich intermediate at lower temperature (as low as −15 °C) was reported by Yamada et al. [23]. This intermediate is rich in α-helix, but no peak was observed in Kratky plot, indicating that this state is not compact. They ascribed this state as “C-state” (cold-denatured state) [23,24].
Liu and Song recently reported that hNck2 SH3 domain protein took helix-rich intermediate at acidic pH though it takes the native β-structure at neutral pH as is the case for src SH3 domain [2–4,25–27]. They also found a 4AlaMut mutant, L25A/W26A/L27A/L28A, that took helix-rich conformation even at neutral pH and reported that these equilibrium intermediates of hNck2 SH3 domain protein seems to be similar to A45G mutant of src SH3 domain at acidic pH [5,25].
We have then started studying the conformational features of hNck2 SH3 domain using CD and XSS methods. As mentioned above, the equilibrium helix-rich intermediate at acidic pH and the native structure at neutral pH of hNck2 SH3 domain were previously studied by NMR [25–27]. However, the conformational features, such as the whole molecular size and shape in solution, were yet unknown.
XSS method is a powerful technique for characterizing the structural feature in solution such as the whole molecular size and shape. Such structural information will be useful in structural biology, biological functions and folding research field.
In the present study, structural properties of the hNck2 SH3 domain in solution have been investigated over a wide range of pH values in conjunction with far-UV CD and XSS methods. As the results, we found that (1) protein concentration dependent monomer–dimer transition occurred at neutral pH. (2) Monomer–dimer transition also occurred in a pH-dependent manner. The equilibrium helix-rich intermediate was observed at acidic pH, as Liu and Song found [25]. However, in contrast to Liu and Song’s speculation, the equilibrium intermediate is not compact globule state. This indicates that the equilibrium helix-rich intermediate at acidic pH is not molten globule state, but should be ascribed to C-state found in the work with equine β-lactoglobulin [23,24].
From XSS results, we show proposed structures of the dimer at pH 8 and the equilibrium helix-rich intermediate at pH 2 of the hNck2 SH3 domain calculated by DAMMIF [28] and DAMAVER [29] programs.
2. Materials and methods
2.1. Materials
The hNck2 SH3 domain was expressed in Escherichia coli from the plasmid pET32a [25]. The cells were cultured at 37 °C to reach an optimal density at 600 nm of ≃0.4, and then isopropyl β-D-1-thiogalactopyranoside was added to a final concentration of 0.4 mM to induce the expression of the recombinant protein at ≃20 °C. The recombinant protein was purified by Chelating Sepharose Fast Flow gel (GE Healthcare). Purity of the protein was confirmed by Tris–tricine PAGE [30].
The temperature was controlled within ±1 °C by a temperature controller ULT-80 of NESLAB. On far UV–CD and XSS measurements, proteins were dissolved in the corresponding solvents for a few hours prior to each measurement or right before measurements. All solutions were filtrated with 0.45 μm filter (Millipore or IWAKI Inc). Protein concentration was determined spectrophotometrically after each measurement with an extinction coefficient ε280 = 27,880 M−1*cm−1 [31]. After measurements, used protein solutions were collected, dialyzed and lyophilized except the solutions used for X-ray irradiation. Reversibility of the protein solution was then checked, and no significant changes were observed.
2.2. Far-UV CD measurements at equilibrium
The samples of hNck2 SH3 domain were prepared in 50 mM sodium phosphate buffer at various pHs. The concentration of the protein was less than 1.1 mg/ml. Far-UV CD measurements were performed at 12 °C with a spectropolarimeter specially designed by Unisoku Inc. [4–6,13,17–22]. Cuvettes of 1 mm path-length were used for all measurements.
Resulting spectra did not show any significant time-dependent changes, indicating each state is sufficiently equilibrated.
2.3. XSS measurements
XSS experiments were done at the beamline of Bio-SAXS in Stanford Synchrotron Radiation Light Source (SSRL) for concentration dependence at pH 6, and at the beamline of 6 A at Photon Factory at KEK (Tsukuba, Japan) for pH dependent measurements [32–34].
At SSRL, XSS measurements were done, keeping the sample-to-detector-distance at ca. 1.7 m with a CCD-based X-ray detector. The X-ray wavelength of λ= 1.127 Å was used for data collection.
At Photon Factory, XSS measurements were done, keeping the sample-to-detector-distance at ca. 1.3 m with a CCD-based X-ray detector (Hamamatsu Photonics, C7300). The X-ray wavelength was λ= 1.504 Å with the CCD detector [35,36]. The obtained data were corrected for image distortion, non-uniformity of sensitivity, and the contrast reduction on X-ray image intensifier [35,36]. Equilibrium experiments were performed using a static-flow cell at 12 °C for preventing radiation damage of the proteins [37]. Helium gas was flowed in and around the observation cell in order to prevent frost from forming on the window. Measurements were repeated at least two times for confirming data reproducibility. This procedure also confirmed each state is sufficiently equilibrated.
X-ray scattering data were obtained from protein and the corresponding buffers. The scattering data of the buffers were subtracted from those of the protein solutions. X-ray scattering data were analyzed by Guinier approximation, as assuming an exponential dependence of the scattering intensity on h2, where h = 4πsinθ/ λ and θ is half of the scattering angle [38].
The linear Guinier region was selected to obtain a radius of gyration (Rg) and zero angle intensity {I(0)}, where the value of Rg *h is ≤1.3. For the analysis of Guinier plots, both single and double exponential analyses were used [20–22,39,40]. The double exponential analysis gives us two components, a larger I(0)(L) and Rg(L), and a smaller I(0)(S) and Rg(S). The larger I(0)(L) and Rg(L) usually give values of aggregated or oligomeric states. In contrast, the smaller I(0)(S) and Rg(S) give values of monomeric state. In case of a mixed solution of the monomer and aggregates or oligomers, we can get both I(0) and Rg for the monomer by using double exponential analysis for Guinier plots, reproductively [20–22,39,40]. When aggregated or oligomers component was included in the protein solution, the double exponential analysis is very convenient and useful analysis method as are the cases of previous reported by other proteins [20–22,39,40]. Bovine β-lactoglobulin was employed for scaling scattering intensities between different setups at both synchrotrons.
Concentrations of hNck2 SH3 domain used in the XSS experiments were less than 4.0 mg/ml.
Structural calculations were done for the scattering data at pH 2, 6 and 8 by using DAMMIF program [28]. 10 times calculations were done for each scattering data. Obtained models were averaged and filtered using DAMAVER program suite to obtain most probable structure [29].
3. Results
3.1. Protein concentration dependence of hNck2 SH3 domain at pH 6
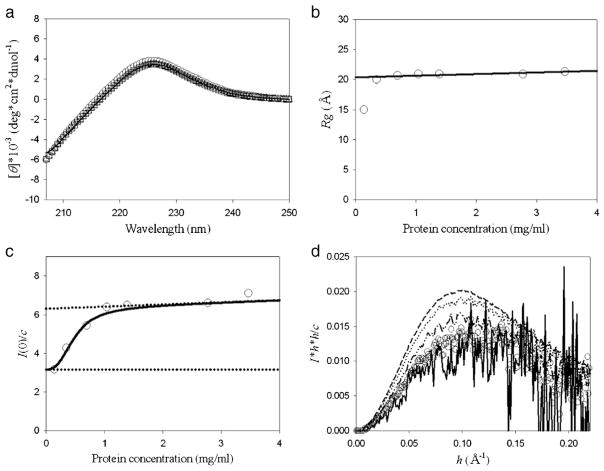
As the first step, we did far-UV CD and XSS experiments on hNck2 SH3 domain were performed at different protein concentration at pH 6. Fig. 1(a) shows far-UV CD spectra of hNck2 SH3 domain at various protein concentrations from 0.26 to 0.99 mg/ml at pH 6. All far-UV CD spectra of the protein are very similar with previous report regarding the native state at pH 6.5, reported by Liu and Song [25]. This agreement indicates that our purified hNck2 SH3 domain took the native structure at pH 6. All far-UV CD spectra at all protein concentrations were overlapped very well, indicating that the secondary structure of hNck2 SH3 domain is independent of protein concentration up to 0.99 mg/ml. Thus, protein concentration does not affect the secondary structure of hNck2 SH3 domain significantly in this concentration range.
Fig. 1.
Far-UV CD and XSS measurements of protein concentration dependence of hNck2 SH3 domain at pH 6. (a) Far-UV CD spectra. 0.26 mg/ml (circle), 0.37 mg/ml (triangle up), 0.69 mg/ml (square) and 0.99 mg/ml (solid line), respectively. (b) Rg dependence of hNck2 SH3 domain protein concentration. The solid line is the fitting line except 0.14 mg/ml. (c) I(0) / c plots against hNck2 SH3 domain protein concentration. A continuous line represents the theoretical curve based on Eq. (1) [10]. Broken lines indicate the I(0) / c(M) and I(0) / c(D) values. (d) Normalized Kratky plots of hNck2 SH3 domain at various protein concentrations. 0.14 mg/ml (solid line), 0.35 mg/ml (circle), 0.69 mg/ml (dash–dot–dot), 1.0 mg/ml (dot) and 3.5 mg/ml (dash), respectively.
Fig. 1(b) shows Rg dependence of the hNck2 SH3 domain at various protein concentrations. Rg value was 15.0 Å at the protein of 0.14 mg/ml, whereas Rg was nearly constant (20.8 Å) above 0.35 mg/ml. Rg, extrapolated to 0 mg/ml, was 20.3 Å with the assumption of linear concentration dependence above 0.35 mg/ml (Fig. 1(b)). The Rg value at 0.14 mg/ml (15.0 Å), is in good agreement with that of the native state of src SH3 domain (14.7 Å) [4,5], whereas the Rg values above 0.35 mg/ml. are significantly larger. As hNck2 SH3 domain and src SH3 domain are homologous with each other in sequence (33.3%) and in tertiary structure, we judged that hNck2 SH3 domain forms monomer at 0.14 mg/ml [2,26,27].
I(0) values were also obtained from Guinier analysis of the same experimental data, and normalized I(0) values, I(0) / c, are plotted against the protein concentration, c. The parameter I(0) / c is proportional to molecular weight [38]. As seen in Fig. 1(c), I(0) / c values above 1.0 mg/ml were found to be nearly twice as big as that at 0.14 mg/ml, suggesting that hNck2 SH3 domain structure would change from monomer to dimer. Normalized Kratky plots of hNck2 SH3 domain at various protein concentrations are presented in Fig. 1(d). It is clear that all Kratky plots have a significant peak. This indicates that hNck2 SH3 domain was compact globule state at any concentrations both at monomeric and dimeric states.
From these protein concentration-dependent experiments by far-UV CD and XSS, it is clear that hNck2 SH3 domain takes compact globule native β-structured monomeric and dimeric states depending on protein concentration.
3.2. pH dependent structural change of hNck2 SH3 domain
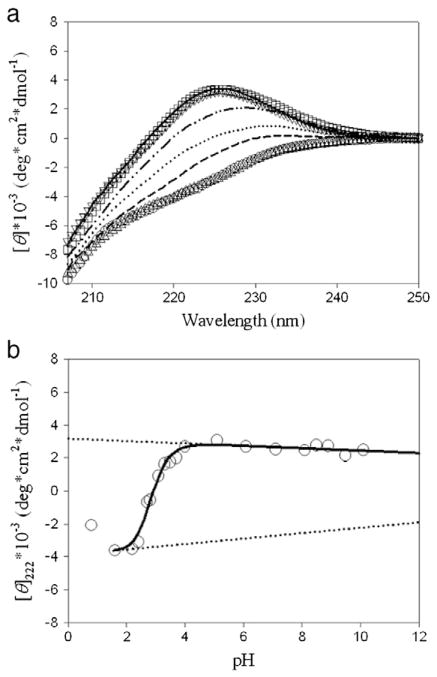
Far-UV CD experiments of hNck2 SH3 domain were performed as a function of pH. Far-UV CD spectra at various pHs of the hNck2 SH3 domain are shown in Fig. 2(a). Both far-UV CD spectra at pH 1.6 and 2.2 were overlapped very well. The shape of the far-UV CD spectra at acidic pH is also very similar to the previous report at pH 2 by Liu and Song, as is the case at pH 6 [25]. This indicates that hNck2 SH3 domain took the equilibrium helix-rich intermediate at pH 1.6 and 2.2, reproductively with data reported in ref. 25. We estimated that helix fraction of the equilibrium helix-rich intermediate at acidic pH using CONTIN program [41]. Result was that the helix fraction of the intermediate was 12%. Previously, Liu and Song also estimated the helix fraction of the equilibrium helix-rich intermediate at acidic pH by CONTINLL [25]. The estimated helix fraction of the intermediate was 12% and was consistent with our estimation. These two results are in excellent agreement with each other. In contrast, the far-UV CD spectrum at pH 0.8 was different from those at pH 1.6 and 2.2. It is considered that hNck2 SH3 domain took another denatured state phase at very low pH.
Fig. 2.
Far-UV CD experiments of hNck2 SH3 domain as a function of pH. Protein concentration was between 0.9 and 1.1 mg/ml below pH 7, and between 0.5 and 0.8 mg/ml above pH 7. (a) Far-UV CD spectra at various pH. pH 0.8 (dash), pH 1.6 (circle), pH 2.2 (triangle up), pH 2.7 (dot), pH 3.1 (dash–dot–dot), pH 4.0 (square), pH 6.1 (solid line) and pH 8.1 (triangle down), respectively. (b) [θ]222 dependence of pH-induced helix-β transition of hNck2 SH3 domain. A continuous line represents the theoretical curve based on Eq. (1) (except pH 0.8) [10]. Broken lines indicate the θH and θN values.
[θ]222 values are plotted against pH in Fig. 2(b), showing a big change of [θ]222 values between pH 2 and 4. This result is also very consistent with the previous report by Liu and Song [25]. The native [θ]222 values (ca. 3000 [deg *cm2*dmol−1]) was kept from pH 4 to 10 (Fig. 2(b)). This suggests that hNck2 SH3 domain maintained the native conformation up to alkaline pH. In fact, far-UV CD spectra at pH 4.0, 6.1 and 8.1 were overlapped very well (Fig. 2(a)).
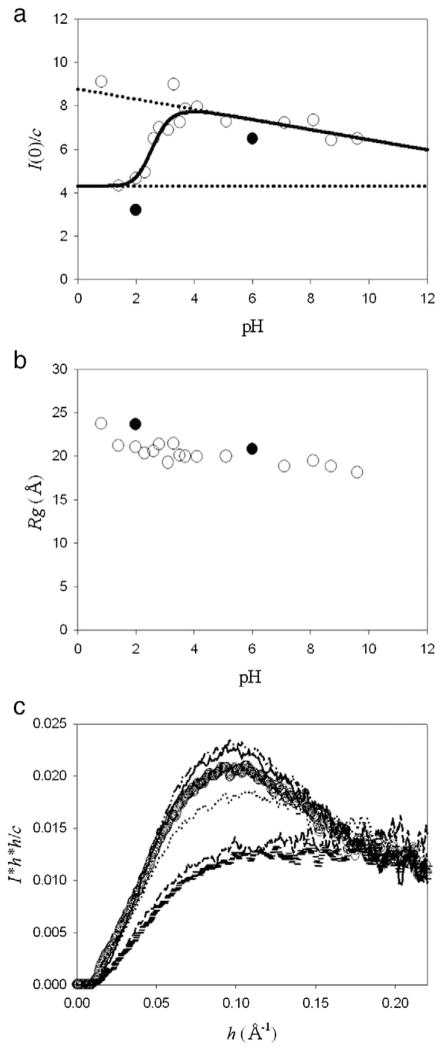
Next, XSS experiments of hNck2 SH3 domain were carried out as a function of pH. Protein concentration of 2.9 to 3.9 mg/ml were used for data collection. At those concentrations, the protein forms dimer at pH 6, as described above (Fig. 1(c)).
Fig. 3(a) represents plots of I(0) / c against pH. Curve fitting was performed based on the strategy described in Discussion (Eq. (1)). The steep increase of I(0) / c between pH 2 and 4, and the gradual decrease of I(0) / c above pH 4. As the protein forms dimer at pH 6, it took mainly monomer at pH 2. Gradual decrease of I(0) / c above pH 4 might suggest the dimer–monomer equilibrium shift to monomer at high pH. As the ionic strength becomes slightly higher with pH, such gradual decrease of I(0) / c may be due to contrast decrease of protein against solvent and not indicate structural change of the protein.
Fig. 3.
XSS experiments of hNck2 SH3 domain as a function of pH. Protein concentration was kept to be 2.9 to 3.9 mg/ml. At these concentrations, the protein forms dimer at pH 6, as described in the text (Fig. 1(c)). (a) I(0) / c plots against pH. A continuous line represents the theoretical curve based on Eq. (1) (except pH 0.8 and 3.3) [10]. Broken lines indicate the I(0) / c(M′) and I(0) / c(D′) values. The data were taken at Photon Factory (open circle) and SSRL (filled circle). About the data at SSRL (filled circle), the I(0) / c value at pH 2 was the average value of that at 1.2 and 1.4 mg/ml and that at pH 6 was also the average value from 1.0 to 2.8 mg/ml. At these concentrations also, the protein forms dimer at pH 6. (b) Rg dependence of pH. The data were taken at Photon Factory (open circle) and SSRL (filled circle). Rg at pH 2 and 6 at SSRL (filled circle) are the average value at 1.2 and 1.4 mg/ml and above 0.35 mg/ml, respectively. (c) Normalized Kratky plots of hNck2 SH3 domain at various pH. pH 1.4 (horizontal mark), pH 2.0 (dash), pH 2.6 (dot), pH 3.5 (circle), pH 4.1 (dash–dot–dot) and pH 7.1 (solid line), respectively.
The I(0) / c at pH 0.8 is bigger. Actually, Guinier plot at pH 0.8 did not show good linear region, suggesting various aggregates are coexisting. Together with far-UV CD experimental results, it is suggested that hNck2 SH3 domain was another denatured state at pH 0.8.
Rg dependence of pH is shown in Fig. 3(b). Contrary to I(0) / c, Rg values seem to show no significant changes against pH above pH 1.4. We will discuss this apparent contradiction between I(0) / c and Rg later in Discussion section.
Fig. 3(c) represents the normalized Kratky plots of hNck2 SH3 domain at various pHs. As clearly seen in the figure, there appeared no peaks at acidic pH 1.4 and 2.0, while a clear peak above pH 2.6. This indicates that the protein was not compact globule state at pH 1.4 and 2.0, whereas the protein was compact above pH 2.6. Rg values at acidic pH were even a little larger than those at neutral pH (except pH 0.8) (Fig. 3(b)). This also suggests that the protein takes non-compact, helix-rich, and probably more elongated structure at acidic pH. We will discuss on this problem in the Discussion part with DAMMIF-calculated structure [28]. By Rg values and Kratky plots (Fig. 3(b) and (c)), it is taken into consideration that the equilibrium helix-rich intermediate at acidic pH of hNck2 SH3 domain was not molten globule state. At pH 0.8, Kratky plot shows a big peak at low angle region, indicating large aggregates (data not shown).
4. Discussion
4.1. Protein concentration-dependent transition at pH 6
Far-UV CD and XSS experiments indicate that hNck2 SH3 domain takes compact globule state at all concentrations investigated, and that judging from Rg value of 15.0 Å, the protein remains the native β-structured monomeric state at 0.14 mg/ml. However, I(0) / c values increased with the increase of protein concentration.
Let us analyze I(0) / c vs. c based on the two-state (M and D) model (Fig. 1(c)). With the two-state model, the transition was analyzed by Eq. (1), in which the observed value (yobs) is represented as a function of (x) [10]:
| (1) |
In the present analysis, yobs means I(0) / cobs, x means protein concentration (c), xm is the protein concentration c at the midpoint of the transition, m represents the cooperativity of the transition, and y1 and y2 are I(0) / c(M) and I(0) / c(D), respectively. Here, I(0) / c(M) and I(0) / c(D) are the I(0) / c values in the pure M and D states, respectively. In general, I(0) / c(M) and I(0) / c(D) are also dependent on c, i.e. I(0) / c(M) = I(0)/c(M1) + I(0)/c(M2) * c and I(0)/c(D) = I(0)/c(D1) +I(0) / c(D2) * c.
The obtained thermodynamic parameters for Fig. 1(c) were that m is 2.7 ± 0.5 (mg/ml)−1 and xm is 0.47 ± 0.04 mg/ml.
4.2. Helix-β transition and M′–D′ transition upon pH change
Far-UV CD experiments revealed a pH-dependent conformational change of hNck2 SH3 domain from the equilibrium helix-rich (pH 2) intermediate to the native β-structured state (pH 6). We, then, analyzed helix-β transition of the hNck2 SH3 domain as the two-state model (Fig. 2(b)), like Fig. 1(c). In the case of Fig. 2(b), the helix-β transition can also be analyzed by Eq. (1) [10]. In this case, yobs is θobs, x is pH, xm is the pH at the midpoint of helix-β transition, m represents the cooperativity of the transition, and y1 and y2 are θH and θN, respectively. θH and θN are [θ]222 values in the pure equilribrium helix-rich intermdiate and the native state, respectively. In general, θH and θN are also dependent on pH, i.e. θH = θ1 + θ2*pH and θN = θ3 + θ4*pH. The obtained thermodynamic parameters were that m is 9.7 ± 1.0 pH−1 and xm is 2.88 ± 0.04.
In the same way, the observed transition of I(0) / c, shown in Fig. 3(a), was analyzed by the two-state (M′ and D′) model with Eq. (1) [10] (except pH 0.8 and 3.3). In this case, yobs, y1, y2 and m are the same of Fig. 1(c) and x and xm are the same with those of pH dependent experiments by far-UV CD (Fig. 2(b)), respectively. That is, yobs means I(0) / cobs, x means pH and xm denotes the pH at the midpoint of the M′–D′ transition. m represents the cooperativity of the transition, and y1 and y2 are I(0) / c(M′) and I(0) / c(D′), respectively, where I(0) / c(M′) and I(0) / c(D′) are the I(0) / c values in the pure M′ and D′ states, respectively. In general, I(0) / c(M′) and I(0) / c(D′) are also dependent on pH, i.e. I(0) / c(M′) = I(0) / c(M′1) + I(0) / c(M′2)*pH and I(0) / c(D′) = I(0) / c(D′1) + I(0) / c(D′2)*pH. The obtained thermodynamic parameters were that m is 8.2 ± 2.0 pH−1 and xm is 2.63 ± 0.08.
When the obtained parameters of helix-β transition by pH (Fig. 2(b)) are compared with those of the M′–D′ transition by pH (Fig. 3(a)), both cooperativities and midpoints of the transitions are in excellent agreement within the experimantal error. This suggests that helical conformation to the native β conformation change and M′ to D′ formation are simultaneously occurred by pH.
4.3. Comparison of I(0) / c, Rg, volumes obtained by DAMMIF and molecular weight calculated by SAXSMow
As described above, M state (0.14 mg/ml at pH 6) resulted in the monomer, judging from Rg of 15.0 Å. To discuss D, M′ and D′ mentioned above in further depth, we calculated volume by DAMMIF [28] and molecular weight by SAXSMow program [42]. Results are summarized in Table 1 as well as I(0) / c and Rg.
Table 1.
Comparison of I(0) / c, volume and molecular weight obtained by various methods.
| Protein
|
hNck2 SH3 domain
|
Bovine β-lactoglobulin
|
||||||
|---|---|---|---|---|---|---|---|---|
| pH | 1.4 | 2 | 8 | 2 | 6 | 6 | 2 | |
| Protein concentration (mg/ml) | 3.23 | 3.18 | 3.28 | 2.38 | 3.47 | 0.14 | 1.45 | |
| Measured facility | PF | PF | PF | SSRL | SSRL | SSRL | SSRL | |
| Figure obtained by DAMMIF | 4(b) | 4(a) | 4(d) | |||||
| I(0) / c | 4.33 | 4.66 | 7.34 | 3.18 | 7.07 | 3.14 | ||
| Rg (Å) | 21.2 | 21.1 | 19.5 | 23.8 | 21.3 | 15.0 | 19.3 | |
| Volume (Å3) obtained by DAMMIF | 21,300 | 34,800 | 28,500 | |||||
| Mw calculated by SAXSMow (kDa) | hMAX (Å −1) | |||||||
| 0.20 | 16.5 | 16.6 | 19.2 | 16.7 | 20.2 | 16.2 | 21.4 | |
| 0.25 | – | – | – | 12.7 | 18.3 | 21.2 | ||
| 0.30 | – | – | – | 10.8 | 17.9 | 19.4 | ||
| 0.40 | – | – | – | 8.4 | 17.2 | 17.2 | ||
Molecular weights calculated by SAXSMow depend on hMAX. From the value of bovine β-lactoglobulin, molecular weights calculated with hMAX up to 0.2 Å −1 are less reliable. They are also too small with hMAX = 0.4 Å −1 probably due to small signals at high angles. Thus, we use averaged value of hMAX = 0.25 Å−1 and 0.3 Å−1 hereafter.
At pH 6, averaged I(0) / c value from at 1.04 to 2.78 mg/ml is 6.46 (Figs. 1(c) and 3(a)), 2.1 times larger than that (3.14) at 0.14 mg/ml (Fig. 3(a)), which strongly suggests the protein forms dimer at these concentrations. If D is the pure dimeric state, its Rg can be estimated as (sphere) or even larger. As the averaged Rg from at 1.04 to 2.78 mg/ml was 20.9 Å (Fig. 1(b)), this result also supports D is dimer. On the other hand, molecular weight of D state at 3.47 mg/ml calculated by SAXSMow is 18 kDa (Table 1). As the molecular weight of the monomer is 11.3 kDa (94 a.a. residues), this value is significantly smaller than the pure dimer, and means D is the mixture of monomer and dimer (60%). Then we conclude D is mainly dimer, though some arguments remain with SAXSMow reliability.
At pH 2, when we compare I(0) / c, the value (3.18) is in excellent agreement with that at pH 6 with 0.14 mg/ml, suggesting the protein is monomer even at high protein concentration at pH 2. At pH 2, molecular weight calculated by SAXSMow gives 11.7 kDa, which is also in good agreement with the monomer (11.3 kDa). Thus, we can conclude that the M′ state is also monomer. Values of I(0) / c at pH 1.4 and pH 2 measured at Photon Factory (4.33 and 4.66) are a little higher. I(0) / c values were normalized by comparing I(0) / c of bovine β-lactoglobulin measured at both facilities. As seen in Fig. 3(a), the normalization seems to be modified (the values at SSRL is systematically smaller). However, the ratio of I(0) / c at pH 8 against at pH 2 is 1.6, and the volume ratio is 1.7, which leads to the conclusion that D ′ state is mainly dimer (60 to 70%).
4.4. Proposed structures of the native β-structured dimer and equilibrium helix-rich conformation by DAMMIF analysis
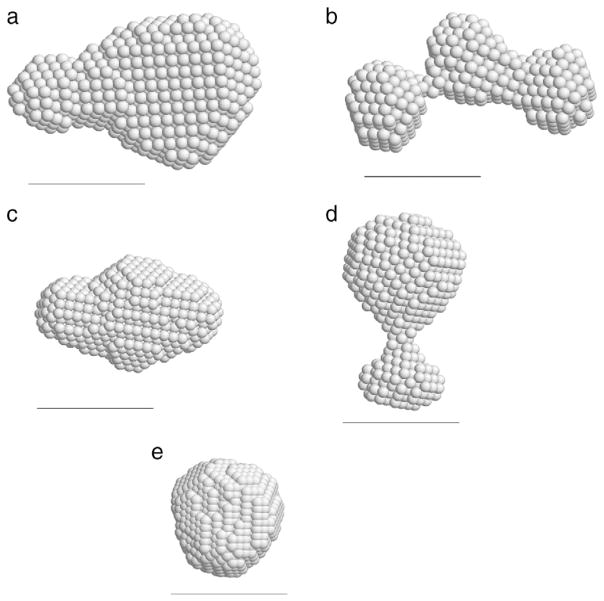
To get structural image more intuitively, we calculated structures of hNck2 SH3 domain by using DAMMIF [28] and DAMAVER [29] programs at various conditions. Structures in Fig. 4(a), (b) and (d) were calculated with the experimental data, and structures in Fig. 4(c) and (e) were calculated from atomic models (scattering curves were obtained from PDB coordinates, and then structure was calculated by DAMMIF and DAMAVER programs) [27–29,43].
Fig. 4.
Calculated structures by DAMMIF [28] and DAMAVER [29] programs. The scale bar is 30 Å. Structures (a), (b) and (d) were calculated from experimental data and structures (c) and (e) were calculated from atomic models: scattering profiles were obtained from PDB coordinates and then DAMMIF calculations were done with this scattering curve. (a) hNck2 SH3 domain at pH 8 (3.3 mg/ml). Rmax was set at 70 Å. (b) hNck2 SH3 domain at pH 2 (3.2 mg/ml). Rmax was set at 65 Å. (c) dimer of Esp8 SH3 domain at pH 7. PDB code is 1I07 [41], (64 × 2 residues). (d) hNck2 SH3 domain at pH 6 (0.14 mg/ml). Rmax was set at 55 Å. (e) hNck2 SH3 domain. PDB code is 2B86 (59 residues) [27].
4.4.1. Structure of hNck2 SH3 domain protein at pH 8
As discussed above, hNck2 SH3 domain protein was mainly dimer at pH 8 at 3.3 mg/ml. When it is compared with Esp8 SH3 domain at pH 7, two structures are partly in agreement with each other. As Esp8 SH3 domain forms domain-swapped dimer at pH 7 [43], hNck2 SH3 domain may also partly form the domain-swapped dimer like Esp8 SH3 domain.
4.4.2. Structure of hNck2 SH3 domain at pH 2
As described above, hNck2 SH3 domain at acidic pH formed monomer even at higher protein concentrations, different from pH 6 (Figs. 1(c) and 3(a)). This form is not compact, judging from Kratky plot (Fig. 3(c)). However, far-UV CD measurements showed that this state is rich in helix, ca. 12% of residues in helix region, suggesting that the form is not in the randomly coiled state. Fig. 4(b) demonstrates the protein is elongated, of which Rg value is not so different from that of the dimer at pH 8. Of course, if Kratky plot does not show any peak, the protein does not take a rigid structure. It is thus indicated that DAMMIF structure in Fig. 4(b) is not the real structure but the ensemble-averaged structure fluctuated.
We have to give one more remark on the conformation of hNck2 SH3 domain structure at pH 2. Though we assign this state as helix-rich state, we have no direct evidences to assign as α-helix. It could be 310 helix either. This assignment remains to be solved.
Yamada et al. reported that equine β-lactoglobulin forms α-helix-rich conformation at acidic pH with 2 M urea at low temperatures [23]. This state is not compact, judging from Kratky plot [23]. They called this state, as C-state (cold denatured state); at C-state, Kratky plot does not show any peak, [θ]222 shows much lower values than the native or molten globule states, and Rg or Rs (Stokes radius of gyration) are nearly the same with that of the unfolded state [23].
Judging from the experimental findings in the present report, the state of hNck2 SH3 domain at pH 2 is in accordance with the criterion of C-state, and not with molten globule state.
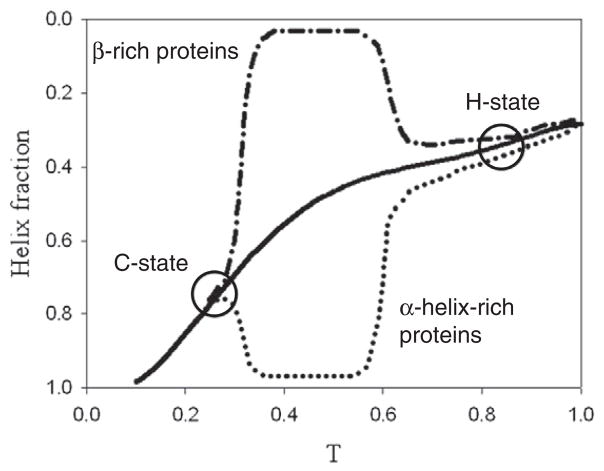
Kano reported that protein forms its native structure with the increase of hydrophobic interaction (Fig. 5) [44]. It means the native structure, when it is rich in α-helix at the native state (dot line in the figure), should have more helical fraction than the value expected by helix–coil transition (solid line in the figure), while helical fraction is lower in the case of β-rich protein (dash–dot–dash line in the figure). At H-state, heat denaturation occurs, and, at C-state, cold denaturation occurs, and below C-state, C-state is accumulated. As seen clearly, C-state has more helical fraction in the case of β-rich proteins (Fig. 5).
Fig. 5.
Model sketch of folding/unfolding diagram of helix-rich proteins, β-rich proteins, cold denatured state (C-state) and heat denatured state (H-state) reported by Kano [44]. Solid line shows helix–coil transition without any tertiary structure interaction. Dot and dash–dot–dash lines show helix–coil transition with hydrophobic interaction, which presents helix fraction of both helix-rich proteins and β-rich proteins, respectively.
We have reported that bovine and equine β-lactoglobulins take α-helix-rich intermediates in the excess amount of ethylene glycol [13,17]. These intermediates did not have any peak in Kratky plots, and Rg values were ca. 50% larger than the native state [45]. It is known that trifluoroethanol induces transition from the native β-rich state to the α-helix-rich intermediate in the case of bovine β-lactoglobulin [11,12]. Here we have also investigated such α-helix-rich intermediate by XSS method and found that the α-helix-rich intermediate of bovine β-lactoglobulin showed a significant peak in Kratky plot, indicating this intermediate is at molten globule state [11,12]. In contrast, src SH3 domain takes an α-helix-rich intermediate in 15% trifluoroethanol, of which Kratky plot did not show any peak, demonstrating the intermediate of src SH3 domain is not molten globule state but at C-state [46].
In summary, it is not clear that acid-induced transition and alcohol-induced transition are more heat denatured state-like or cold denatured state-like. However, at least we can say that denatured states do not automatically mean helices should disappear.
HIGHLIGHTS.
hNck2 SH3 domain at neutral pH forms monomer and dimer depending on concentrations.
The secondary structure is independent of the concentrations.
At acidic pH, the protein forms helix-rich monomer even at high concentration.
The helix-rich monomer is changed to β-sheeted dimer by pH increase.
The helix-rich monomer at acidic pH is not compact.
Acknowledgments
The authors are grateful to Dr. Masayuki Morita of Kansai Medical University for kindly helping them prepare E. coli expression.
We are grateful to SSRL for giving the beam time in order to do XSS experiments. XSS experiments at SSRL were done with Photon Factory proposal number of 2011G188.
XSS measurements were also performed under proposal number 2011G188 of the Photon Factory.
This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (no. 20540400).
Abbreviations
- far-UV
far-ultraviolet
- CD
circular dichroism
- XSS
X-ray solution scattering
- NMR
nuclear magnetic resonance
- C-state
cold denatured state
- Rg
radius of gyration
References
- 1.Musacchio A, Gibson T, Lehto VP, Saraste M. SH3 — an abundant protein domain in search of a function. FEBS Letters. 1992;307:55–61. doi: 10.1016/0014-5793(92)80901-r. [DOI] [PubMed] [Google Scholar]
- 2.Yu H, Rosen MK, Schreiber SL. 1H and 15N assignments and secondary structure of the Src SH3 domain. FEBS Letters. 1993;324:87–92. doi: 10.1016/0014-5793(93)81538-b. [DOI] [PubMed] [Google Scholar]
- 3.Noble ME, Musacchio A, Saraste M, Courtneidge SA, Wierenga RK. Crystal structure of the SH3 domain in human Fyn; comparison of the three-dimensional structures of SH3 domains in tyrosine kinases and spectrin. EMBO Journal. 1993;12:2617–2624. doi: 10.2210/pdb1shf/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Shinjo M, Matsumura Y, Morita M, Baker D, Ikeguchi M, Kihara H. An alpha-helical burst in the src SH3 folding pathway. Biochemistry. 2007;46:5072–5082. doi: 10.1021/bi0618262. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Matsumura Y, Shinjo M, Kojima M, Kihara H. A stable α-helix-rich intermediate is formed by a single mutation of the β-sheet protein, src SH3, at pH 3. Journal of Molecular Biology. 2007;372:747–755. doi: 10.1016/j.jmb.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Qin ZJ, Vyas S, Fink AL, Li JS, Kihara H. Transient α-helical structure during folding of src SH3 domain at subzero temperatures. Journal of Kansai Medical University. 2006;58:163–169. [Google Scholar]
- 7.Kimura T, Akiyama S, Uzawa T, Ishimori K, Morishima I, Fujisawa T, Takahashi S. Specifically collapsed intermediate in the early stage of the folding of ribonuclease A. Journal of Molecular Biology. 2005;350:349–362. doi: 10.1016/j.jmb.2005.04.074. [DOI] [PubMed] [Google Scholar]
- 8.Chaffotte AF, Guillou Y, Goldberg ME. Kinetic resolution of peptide bond and side chain far-UV circular dichroism during the folding of hen egg white lysozyme. Biochemistry. 1992;31:9694–9702. doi: 10.1021/bi00155a024. [DOI] [PubMed] [Google Scholar]
- 9.Kuwajima K, Yamaya H, Miwa S, Sugai S, Nagamura T. Rapid formation of secondary structure framework in protein folding studied by stopped-flow circular dichroism. FEBS Letters. 1987;221:115–118. doi: 10.1016/0014-5793(87)80363-0. [DOI] [PubMed] [Google Scholar]
- 10.Kuwajima K, Yamaya H, Sugai S. The burst-phase intermediate in the refolding of β-lactoblobulin studied by stopped-flow circular dichroism and absorption spectroscopy. Journal of Molecular Biology. 1996;264:806–822. doi: 10.1006/jmbi.1996.0678. [DOI] [PubMed] [Google Scholar]
- 11.Hamada D, Segawa S, Goto Y. Non-native alpha-helical intermediate in the refolding of beta-lactoglobulin, a predominantly beta-sheet protein. Natural Structural Biology. 1996;3:868–873. doi: 10.1038/nsb1096-868. [DOI] [PubMed] [Google Scholar]
- 12.Hamada D, Goto Y. The equilibrium intermediate of beta-lactoglobulin with non-native alpha-helical structure. Journal of Molecular Biology. 1997;269:479–487. doi: 10.1006/jmbi.1997.1055. [DOI] [PubMed] [Google Scholar]
- 13.Qin ZJ, Hu DM, Shimada L, Nakagawa T, Arai M, Zhou JM, Kihara H. Refolding of β-lactoglobulin studied by stopped-flow circular dichroism at subzero temperatures. FEBS Letters. 2001;507:299–302. doi: 10.1016/s0014-5793(01)02886-1. [DOI] [PubMed] [Google Scholar]
- 14.Ikeguchi M, Kato S, Shimizu A, Sugai S. Molten globule state of equine β-lactoglobulin. Proteins. 1997;27:567–575. doi: 10.1002/(sici)1097-0134(199704)27:4<567::aid-prot9>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 15.Fujiwara K, Arai M, Shimizu A, Ikeguchi M, Kuwajima K, Sugai S. Folding–unfolding equilibrium and kinetics of equine β-lactoglobulin: equivalence between the equilibrium molten globule state and a burst-phase folding intermediate. Biochemistry. 1999;38:4455–4463. doi: 10.1021/bi982683p. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T, Ikeguchi M, Sugai S. Molten globule structure of equine β-lactoglobulin probed by hydrogen exchange. Journal of Molecular Biology. 2000;299:757–770. doi: 10.1006/jmbi.2000.3761. [DOI] [PubMed] [Google Scholar]
- 17.Matsumura Y, Li J, Ikeguchi M, Kihara H. Helix-rich transient and equilibrium intermediates of equine β-lactoglobulin in alkaline buffer. Biophysical Chemistry. 2008;134:84–92. doi: 10.1016/j.bpc.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Qin ZJ, Ervin J, Larios E, Gruebele M, Kihara H. Formation of a compact structured ensemble without fluorescence signature early during ubiquitin folding. The Journal of Physical Chemistry B. 2002;106:13040–13046. [Google Scholar]
- 19.Larios E, Li JS, Schulten K, Kihara H, Gruebele M. Multiple probes reveal a native-like intermediate during low-temperature refolding of ubiqutin. Journal of Molecular Biology. 2004;340:115–125. doi: 10.1016/j.jmb.2004.04.048. [DOI] [PubMed] [Google Scholar]
- 20.Kim SJ, Matsumura Y, Dumont C, Kihara H, Gruebele M. Slow down downhill folding: a three-probe study. Biophysical Journal. 2009;97:295–302. doi: 10.1016/j.bpj.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumont C, Matsumura Y, Kim SJ, Li J, Kondrashkina E, Kihara H, Gruebele M. Solvent-tuning the collapse and helix formation time scales of λ6–85*. Protein Science. 2006;15:2596–2604. doi: 10.1110/ps.062257406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumura Y, Shinjo M, Mahajan A, Tsai MD, Kihara H. α-Helical burst on the folding pathway of FHA domains from Rad53 and Ki67. Biochimie. 2010;92:1031–1039. doi: 10.1016/j.biochi.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Yamada Y, Yajima T, Fujiwara K, Arai M, Ito K, Shimizu A, Kihara H, Kuwajima K, Amemiya Y, Ikeguchi M. Helical and expanded conformation of equine β-lactoglobulin in the cold-denatured state. Journal of Molecular Biology. 2005;350:338–348. doi: 10.1016/j.jmb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Yamada Y, Nakagawa K, Yajima T, Saito K, Tokushima A, Fujiwara K, Ikeguchi M. Structural and thermodynamic consequences of removal of a conserved disulfide bond from equine β-lactoglobulin. Proteins. 2006;63:595–602. doi: 10.1002/prot.20905. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Song J. NMR evidence for forming highly populated helical conformations in the partially folded hNck2 SH3 domain. Biophysical Journal. 2008;95:4803–4812. doi: 10.1529/biophysj.107.125641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi K, Yang H, Ng E, Park S, Sun ZJ, Reinherz EL, Wagner G. Structural and functional evidence that Nck interaction with CD3ε regulates T-cell receptor activity. Journal of Molecular Biology. 2008;380:704–716. doi: 10.1016/j.jmb.2008.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park S, Takeuchi K, Wagner G. Solution structure of the firs Src homology 3 domain of human Nck2. Journal of Biomolecular NMR. 2006;34:203–208. doi: 10.1007/s10858-006-0019-5. [DOI] [PubMed] [Google Scholar]
- 28.Franke D, Svergun D. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volkov VV, Svergun DI. Uniqueness of ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patton WF, Chung-Welch N, Lopez MF, Cambria RP, Utterback BL, Skea WM. Tris–tricine and Tris–borate buffer systems provide better estimates of human mesothelial cell intermediate filament protein molecular weights than the standard Tris–glycine system. Analytical Biochemistry. 1991;197:25–33. doi: 10.1016/0003-2697(91)90350-3. [DOI] [PubMed] [Google Scholar]
- 31.Gill SC, von Hipple PH. Calculation of extinction coefficients from amino acid data. Analytical Biochemistry. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu N, Mori T, Igarashi N, Ohta H, Nagatani Y, Kosuge T, Ito K. Refurbishing of small-angle X-ray scattering beamline, BL-6A at the Photon Factory. Journal of Physics: Conference Series. (in press) [Google Scholar]
- 33.Amemiya Y, Wakabayashi K, Hamanaka T, Wakabayashi T, Hashizume H. Design of a small-angle X-ray diffractometer using synchrotron radiation at the photon factory. Nuclear Instruments and Methods. 1983;208:471–477. [Google Scholar]
- 34.Amemiya Y, Ito K, Yagi N, Asano Y, Wakabayashi K, Ueki T, Endo T. Large-aperture TV detector with a beryllium-windowed image intensifier for X-ray diffraction. The Review of Scientific Instruments. 1995;66:2290–2294. [Google Scholar]
- 35.Ito K, Kamikubo H, Arai M, Kuwajima K, Amemiya Y, Endo T. Calibration method for contrast reduction problem in the X-ray image-intensifier. Photon Factory Activity Report. 2001;18:275. [Google Scholar]
- 36.Ito K, Kamikubo H, Yagi N, Amemiya Y. Correction method and software for image distortion and nonuniform response in charge-coupled device-based X-ray detectors utilizing X-ray image intensifier. Japanese Journal of Applied Physics. 2005;44:8684–8691. [Google Scholar]
- 37.Kihara H, Nagamura T. Development of a static flow cell for the measurement of solution X-ray scattering. KEK Tsukuba High Energy Accelerator Research Organization; 1999. p. 246. [Google Scholar]
- 38.Guinier A, Fournet G. In Small Angle Scattering of X-rays. Wiley; New York: 1955. [Google Scholar]
- 39.Shinomiya H, Shinjo M, Fengzhi L, Asano Y, Kihara H. Conformational analysis of the leukocyte-specific EF-hand protein p65/L-plastin by X-ray scattering in solution. Biophysical Chemistry. 2007;131:36–42. doi: 10.1016/j.bpc.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Kogo H, Takeuchi K, Inoue H, Kihara H, Kojima M, Takahashi K. Urea-dependent unfolding of HIV-1 protease studied by circular dichroism and small-angle X-ray scattering. Biochimica et Biophysica Acta. 2009;1794:70–74. doi: 10.1016/j.bbapap.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 41.Provencher SW, Gloeckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1984;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 42.Fischer H, de Oliveira Neto M, Napolitano HB, Polikarpov I, Craievich AF. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. Journal of Applied Crystallography. 2010;43:101–109. [Google Scholar]
- 43.Kishan KV, Newcomer ME, Rhodes TH, Guilliot SD. Effect of pH and salt bridges on structural assembly: molecular structures of the monomer and intertwined dimer of the Esp8 SH3 domain. Protein Science. 2001;10:1046–1055. doi: 10.1110/ps.50401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kano F. Helix–coil transitions in nonpolar peptides. Journal of the Physical Society of Japan. 2007;76:044603. [Google Scholar]
- 45.Matsumura Y, Shinjo M, Kihara H. Alpha-helix-rich intermediates of BLG and ELG induced in 90% ethylene glycol. Photon Factory Activity Report. 2009;26:193. [Google Scholar]
- 46.Shinjo M, Matsumura Y, Kihara H. Src SH3 forms unfolded α-helix-rich conformation in 15% trifluoroethanol. Photon Factory Activity Report. 2009;26:194. [Google Scholar]