Abstract
Flower color and plant architecture are important commercially valuable features for ornamental petunias (Petunia x hybrida Vilm.). Photoperception and light signaling are the major environmental factors controlling anthocyanin and chlorophyll biosynthesis and shade-avoidance responses in higher plants. The genetic regulators of these processes were investigated in petunia by in silico analyses and the sequence information was used to devise a reverse genetics approach to probe mutant populations. Petunia orthologs of photoreceptor, light-signaling components and anthocyanin metabolism genes were identified and investigated for functional conservation by phylogenetic and protein motif analyses. The expression profiles of photoreceptor gene families and of transcription factors regulating anthocyanin biosynthesis were obtained by bioinformatic tools. Two mutant populations, generated by an alkalyting agent and by gamma irradiation, were screened using a phenotype-independent, sequence-based method by high-throughput PCR-based assay. The strategy allowed the identification of novel mutant alleles for anthocyanin biosynthesis (CHALCONE SYNTHASE) and regulation (PH4), and for light signaling (CONSTANS) genes.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-013-0212-4) contains supplementary material, which is available to authorized users.
Keywords: Data mining, Flavonoid, Light signaling, Mutagenesis, Solanaceae, TILLING
Introduction
Reverse genetics aims to uncover gene function by investigating the mutation in a determined sequence without a priori knowledge of the mutant phenotype (Alonso and Ecker 2006). In the recent years, the development of inexpensive and high-throughput sequencing technologies has helped to increase the accumulation of sequence data from genomic and expressed sequence tag (EST) projects; thus, facilitating the use of reverse genetics to investigate gene function in several species (Pérez-de-Castro et al. 2012) . Reverse genetics is generally less time-demanding than forward genetics and circumvents functional redundancy often found in plant genomes (Alonso and Ecker 2006; Pérez-de-Castro et al. 2012). In plants, reverse genetic methods frequently employ polymerase chain reaction (PCR)-based tools to investigate saturated mutant populations and identify mutants in the sequence(s) of interest (Alonso and Ecker 2006). Targeting Induced Local Lesions In Genomes (TILLING) is a reverse genetic technique that identifies unique, induced mutations within target genes due to the formation of wild-type and mutant DNA strand heteroduplexes (McCallum et al. 2000; Pérez-de-Castro et al. 2012). Aberrant pairings caused by the mutation are recognized and cleaved by a mismatch-specific endonuclease, CELI, so fragments of size distinct from that of the wild-type gene indicate the presence a mutation in the sequence of interest. In petunia, functional gene analysis has profited from the abundance of endogenous transposable elements (Vandenbussche et al. 2008), however the transient nature of the insertion and the consequent instability of the phenotypes reduce its effectiveness (Alonso and Ecker 2006; Vandenbussche et al. 2008). Moreover, insertion transposon-tagging approaches frequently generate loss-of-function mutations by disrupting the gene or its regulatory sequences, rarely identifying genes with redundant action, common to eukaryotic genomes (Alonso and Ecker 2006).
Light is one of the most important environmental factors controlling several aspects of plant development and metabolism (Schäfer and Nagy 2006). In higher plants, it is perceived by a complex system of photoreceptor molecules: the blue (B) and ultraviolet-A (UV-A) light sensing cryptochromes (cry) (Chaves et al. 2011) and phototropins (phot) (Inoue et al. 2010) and the red (R)/far red (FR) phytochrome (phy) receptors (Nagatani 2010). More recently, a novel family of B photoreceptors has been described in Arabidopsis thaliana: the Zeitlupe (ZTL)/ Flavin-binding Kelch repeat F-box protein (FKF1)/LOV Kelch Protein (LKP2) family (Demarsy and Fankhauser 2009). Upon light perception, photoreceptor-mediated signal transduction involves a wide-range of mechanisms that include light-regulated sub-cellular localization of the photoreceptor molecules (Fankhauser and Chen 2008), a large reorganization of transcriptional programs (Casal and Yanovsky 2005) and light-regulated proteolytic degradation of photoreceptors and signaling components (Henriques et al. 2009). Studies in model species have demonstrated that photoperception, signaling and the responses elicited by light form a complex network of interconnected pathways rather than a linear cascade (Quecini and Liscum 2006; Chory 2010).
Anthocyanins are flavonoid pigments, ubiquitously found in reproductive and vegetative plant parts, involved in biotic and abiotic stress protection (Winkel-Shirley 2001; Huang et al. 2012). Anthocyanin biosynthesis occurs via a branch of the flavonoid pathway and consists of a well-known sequence of enzymatic steps, studied in Antirrhinum majus, A. thaliana, Petunia and Zea mays (Koes et al. 2005). The activity of anthocyanin biosynthetic genes is largely regulated at the transcriptional level via the function of several classes of transcriptional regulators, such as: WD40, helix-loop-helix (HLH), R2R3 and R3 MYB, WRYK, Zn finger and MADS-box (Koes et al. 2005; Ambawat et al. 2013). In several plant species, light has been demonstrated to be an important environmental factor controlling anthocyanin accumulation, directly via transcriptional control of biosynthetic genes and transcription factors or indirectly via developmental regulation (Albert et al. 2009; Lopez et al. 2012; Petroni and Tonelli 2011; Huang et al. 2012; Ambawat et al. 2013).
In this study, we have investigated the candidate genes for light perception, signal transduction and anthocyanin metabolism in petunia, employing genetic distance, phylogenetic studies, functional motif analyses and in silico expression profiling. The use of bioinformatic and comparative genomic tools allowed the establishment of the genetic framework for photoperception, light signaling and anthocyanin metabolism in petunia. The compiled sequences were used in a PCR-based reverse genetics approach to identify mutant alleles of the genes of interest in two M3 populations of Petunia x hybrida Mitchell Diploid, generated by gamma irradiation and treatment with the alkylating agent, ethyl methanesulfonate (EMS) (Berenschot et al. 2008). Novel mutant alleles were identified for genes involved in anthocyanin biosynthesis (CHALCONE SYNTHASE) and regulation (PH4), and light signaling (CONSTANS) in the absence of phenotypic screenings.
Materials and methods
Plant material
Seeds of Petunia x hybrida Vilm. Mitchell Diploid (MD), a doubled haploid derived from the complex hybrid between P. axillaris and the cultivated variety “Rose of Heaven” were kindly provided by Prof. Dr. David G. Clark from the Environmental Horticulture Department of University of Florida, United States. ‘Mitchell Diploid’ flowers are white and were deliberately chosen to investigate the efficiency of mutant identification in the absence of visual color phenotypes; thus, proving the concept of reverse genetics screening. Two mutant populations were independently generated by gamma irradiation and EMS treatment and evaluated for moderate DNA damage allowing a high saturation of mutant alleles (Berenschot et al. 2008). Seeds representing 100 progenies (M2 populations) from self-pollinations were sown and 5,000 plants per population (M3) were grown in trays under standard greenhouse conditions. Mutant individuals were grown to maturity, manually self-pollinated for three reproductive cycles and the recovered seeds were kept as individual progenies. The presence of the mutation originally identified was confirmed by gene-specific PCR.
Nucleic acid purification and PCR conditions
Young leaf material (100 mg) was harvested from individual plants in gamma and EMS populations and used for DNA extraction as described (Doyle and Doyle 1990). Isolated DNA was quantified by SYBR® Safe (Invitrogen, USA) fluorescence analysis on 0.7 % (w/v) agarose gel electrophoresis and a 5 μg aliquot of DNA from each plant was pooled into 20 composite samples consisting of DNA from 250 individuals from each mutant population. Standard TILLING PCR parameters were as follows: one cycle of 95 °C for 2 min and 94 °C for 20 s followed by 56 cycles of 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 1 min. The next step in PCR was 72 °C for 5 min, then a 99 °C step for 10 min followed by a 70 °C to 0 °C melt. Cleavage of the PCR products by CEL I and detection on agarose gels was done exactly as described (Raghavan et al. 2007). Pools exhibiting cleaved products, indicating the presence of heteroduplex mismatch due to a mutation, were opened by separating the individual DNA samples for sequencing to confirm the mutation. The gene-specific primers for the identified alleles were used for chain-terminator sequencing (ABI Prism 311 e BigDye® Terminator v3.1 Cycle Sequencing Kit, Applied Biosystem).
For expression analyses, leaf tissue was excised from five plants of each mutant line, immediately frozen in liquid nitrogen and stored at −80 °C. Three independent extractions were carried out for each genotype. RNA was extracted using the Trizol reagent (Life Technologies), combined with Ambion RiboPure™ Kit (Ambion/Life Technologies). DNA was removed by TURBO™ DNase (Life Technologies) and RNA integrity and quantity were determined by gel electrophoresis. Synthesis of cDNA was carried out using 2 μg of total RNA by reverse transcription (RT) with Superscript II (Life Technologies) and random oligo dT25 primers (Life Technologies). Steady-state mRNA levels of the genes of interest were analyzed in the mutant and control lines by semi-quantitative RT-PCR using 2 μl of cDNA, 0.2 mM of each dNTP, 2.5 mM of MgCl2, 0.25 μM of each forward and reverse primer (Table S5) and 0.2 u of Taq DNA polymerase. Amplification cycles were as follows: 2 min at 95 °C followed by the specified number of cycles for each gene at 94 °C for 30 S, annealing temperature (Table S5) for 30 s, and 1 min at 72 °C. The number of cycles were 35, 35 and 32 for CHS, PH4 and CO, respectively, and 32 for α-tubulin. Standard cDNA dilutions were used to investigate reaction efficiency and quality. Negative controls (no template cDNA) were included in all semi-quantitative RT-PCR assays, and results are representative triplicate experiments. Amplification products were separated on 1.0 % (w/v) agarose gel and stained as described earlier.
Database searches and alignments
Homologs of model species photoperception, light signaling and anthocyanin metabolism genes were identified in BLAST searches (Altschul et al. 1997) against petunia databases (Genbank at http://www.ncbi.nlm.nih.gov/BLAST/; The Institute for Genomic Research, TIGR, at http://tigrblast.tigr.org/tgi/, and the SOL Genomics Network at http://www.sgn.cornell.edu). Data validation was performed by tBLASTx and tBLASTn searches of the retrieved sequence against GenBank and P. inflata, P. axillaris transcript database (http://biosrv.cab.unina.it/454petuniadb/) (Zenoni et al. 2011). Sequences failing to retrieve the original sequence used to query the database were eliminated. The resulting alignments were filtered by a threshold e-value of 1 e−15 and the hits were further analyzed according to functional domain description. Validated sequences were translated and protein (deduced amino acid) alignments were performed using ClustalX (Thompson et al. 1997). When necessary, alignments were manually adjusted using Lasergene MegAlign (DNASTAR, Madison, WI, USA).
Motif analysis and in silico characterization
The identified genes were further investigated for the presence and sequence conservation of recognizable functional domains described in protein analysis and gene function databases (European Bioinformatics Institute-European Molecular Biology Laboratory –EMBL-EBI www.ebi.ac.uk/interpro/; Expert Protein Analysis System—ExPaSy from Swiss Institute of Bioinformatics—SIB http://www.expasy.org/prosite/; Gene Ontology database—GO, http://www.godatabase.org/cgi-bin/amigo/go.cgi; Protein Families database—Pfam http://www.sanger.ac.uk/Software/Pfam/).
Phylogenetic analysis
The functionality of recovered genes in comparison to the characterized counterparts was assessed by genetic distance and phylogenetic studies. Phylogenetic analyses were performed using parsimony methods in the software MEGA 5.2.2 (Tamura et al. 2011), using the software default parameters. The software was also used to construct resampling bootstrap trees containing 1,000 random samples. Modular functional domains were employed for genetic distance studies for genes previously characterized as having divergent regions and conserved blocks.
In silico gene expression analysis
Qualitative gene expression profiling was performed by in silico analyses of the EST databases using virtual northern blot analyses. The gene of interest was used in queries against reference sequence databases, generating an alignment of the input gene to its paralogs. The resulting alignment was then used to find sequences in the entire mRNA input that are specific to the gene family (probe). The resulting alignments were collectively used to query the EST database again using BLAST. The identity numbers of the ESTs matching the probes were recovered and the databases were used to find the names of the libraries from which those ESTs were derived. The frequency of reads of each EST contig in a given library was calculated and normalized according to the total number of reads from the investigated library and the total number of reads in all libraries. A correlation matrix between EST contigs and libraries was then generated and gene expression patterns among ESTs and libraries were obtained by hierarchical clustering based on Spearman Rank correlation matrix using the MeV 4.9 software from the TM4 Microarray Software Suite (Saeed et al. 2006). Graphic outputs are presented as color scales of the relative gene expression (Saeed et al. 2006).
A posteriori mutant phenotypic analyses
The phenotype of the identified mutant lines was screened after three cycles of selfing, using at least 20 plants from each line. The pH of petal and leaf extracts was measured by grinding the petal limbs of two corollas and two leaf mesophyll blades, from the first and second fully expanded leaves from the apice, in 6 mL of distilled water. The pH was immediately measured with a pH electrode to avoid the possibility that atmospheric CO2 would alter the pH of the extract, as described by (Quattrocchio et al. 2006). Flower longevity was considered as the number of days of fully opened flowers until senescence and days to flowering were counted from seedling emergency to flower bud appearance. Internode length was measured with a ruler in horizontally positioned plants. Leaf number was counted from seedling emergency to senescence and leaf area was estimated from digitalized images of five leaves from plant apex. Total chlorophyll contents were estimated by spectrophotometric readings of dimethyl sulfoxide (DMSO) extracts at 649 and 665 nm (Weller et al. 2001) and vegetative anthocyanin contents were determined under white and monochromatic blue light by acidic methanol extractions and spectrophotometric absorbance at 530 and 657 (Weller et al. 2001).
Results
Data mining for photoperception, light-signaling and anthocyanin metabolism
Initially, we have compiled a set of functionally characterized genes, known to be involved in light-sensing and signaling and in anthocyanin metabolism, to be searched for in petunia sequence databases (Fig. 1, Tables S1, S2, S3 and S4). In general, the relative composition of sample Petunia transcriptome and Arabidopsis proteome over the functional categories involved in light perception, signaling, anthocyanin biosynthesis and regulation was very similar (Fig. 1). The putative functionality of the retrieved sequences was further investigated by phylogenetic analysis and in silico expression profiling, allowing us to rank the candidate genes and use the sequence information to screen mutant populations.
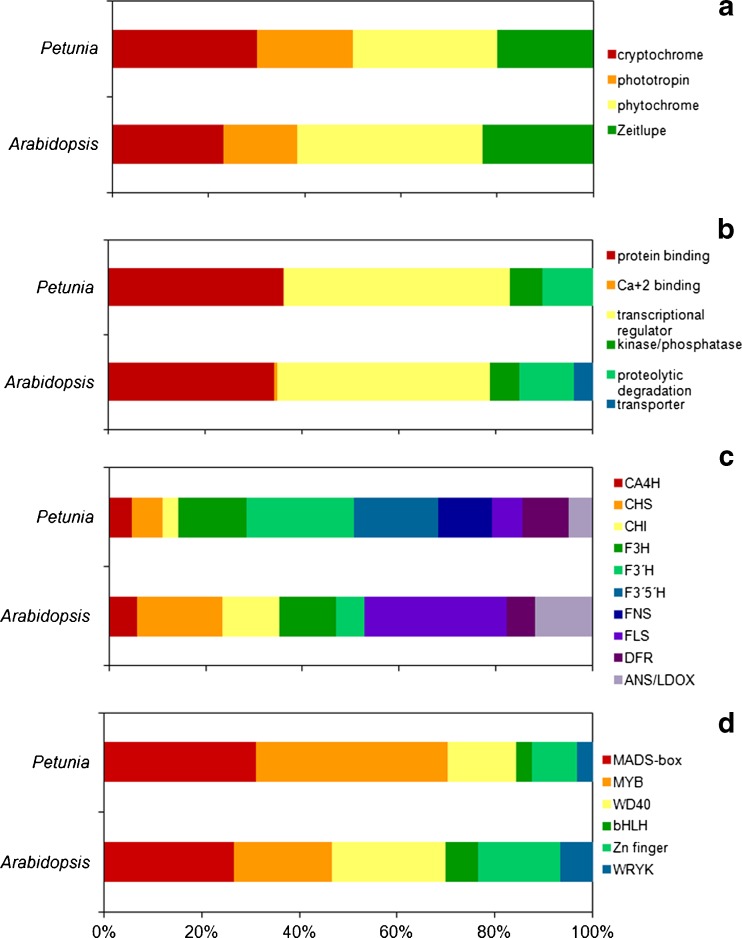
Fig. 1.
Functional classification of Petunia transcripts associated to light and anthocyanin in comparison to Arabidopsis thaliana proteome; photoreceptor families (a), light-signal transduction pathways classified into gene ontology molecular function categories (b), anthocyanin-biosynthesis structural (c) and regulatory genes classified in transcription factor families (d). Assignments are based on GO Slim terms for genes were obtained from the Arabidopsis database site (ftp://ftp.arabidopsis.org/home/tair/Ontologies)
Sequences displaying significant conservation to members of the phytochrome, cryptochrome phototropin and ZTL/FKF1/LKP2 families of plant photoreceptors were identified in the investigated petunia databases (Fig. 1a, Table S1). Homologs of the vast majority of the photoreceptor signaling partners identified in model-species were identified in petunia, providing an initial genetic framework for light signaling (Fig. 1b, Table S2). The highest levels of deduced amino acid sequence identity, in comparison to the Arabidopsis counterparts, were observed for protein kinase/phosphatase families (Table S2). In petunia, sequences corresponding to Arabidopsis transcription factors involved in light signaling are more divergent (Table S2). Currently, transcripts exhibiting significant sequence conservation to the phy-A specific signaling partner FHY1 (Desnos et al. 2001), the bZIB transcription factor HY5/HYH family (Chattopadhyay et al. 1998), the kinase substrate PKS1 family (Fankhauser et al. 1999), the EXS and SPX domain blue-light signaling partner SHB1 (Kang and Ni 2006) and the Ca+2-binding protein SUB1 (Guo et al. 2001) are absent from Petunia x hybrida databases (Fig. 1b). The absent sequences represent 27.7 % (5/18) of the light-signal transduction partners searched in petunia.
Anthocyanin metabolism has been extensively studied in several plant species, leading to the discovery of a consensus pathway shared among higher plants, for which the structural genes have been isolated. In P. x hybrida databases, we have identified 62 orthologs of the structural genes involved in anthocyanin biosynthetic pathways relevant to color, including 11 F3′5′H- and seven FNS-like sequences that are absent from Arabidopsis genome (Fig. 1c, Table S3). Sequence identity of the deduced amino-acids of petunia putative proteins ranged from 84 % to 8 % in comparison to the functionally characterized sequence used to query the databases (Table S3). Highest sequence conservation was observed for the CHS family (84.1 % to 71.7 %), where the totality of the sequences identified in petunia was more closely related to TRANSPARENT TESTA 4 (AT5G13930). In contrast, the identified CHI-like sequences exhibited lower levels of sequence conservation (60.5 to 56.4 %) and sequences similar to F3′H, FLS and ANS were generally less conserved in each group and in comparison to the bait sequence (Table S3). Thirty-nine petunia sequences exhibiting significant sequence conservation to the transcription factors regulating anthocyanin biosynthesis were identified (Fig. 1d, Table S4). All classes of transcription factor families demonstrated to be involved in the regulation of the pathway in Arabidopsis and maize were found in petunia, namely; WRYK, WD-40, MADS, MYB, HLH and Zinc finger (Koes et al. 2005; Ambawat et al. 2013) (Fig. 1d, Table S4). Petunia MYB sequences sharing significant sequence homology to Arabidopsis transcription factors involved in several developmental and physiological processes other light responses and anthocyanin metabolism were identified (Fig. S1). Petunia orthologs of anthocyanin biosynthesis regulators exhibited more divergent sequences in comparison to their Arabidopsis counterparts (deduced amino-acid identity ranging from 78.3 to 25.8 %) than the structural genes, although sequence conservation at the DNA-binding domains remained high, thus accounting for the higher identity average at the lower end of the interval (Table S4). The MADS /K-box family was the most abundant group of transcription factors affecting anthocyanin biosynthesis present in petunia databases, corresponding to approximately 51 % (20/39) of the retrieved sequences, followed by the MYB family (33 %, 13/39) (Fig. 1d, Fig. S1).
In silico transcriptional profiling of photoreceptor and anthocyanin-biosynthesis regulator genes
In order to help establishing a functional association between the sequences identified in petunia databases and their characterized orthologs from model species, the transcriptional profile of the anthocyanin regulatory and photoreceptor families was investigated by in silico analysis. The expression profile of the queried genes was determined using alignments of the searched gene, and its paralogs were collectively used to query the EST database using BLAST. This heuristic allowed us to avoid false positives or ESTs from a paralog of the input gene rather than the gene itself; thus, providing a more precise expression pattern.
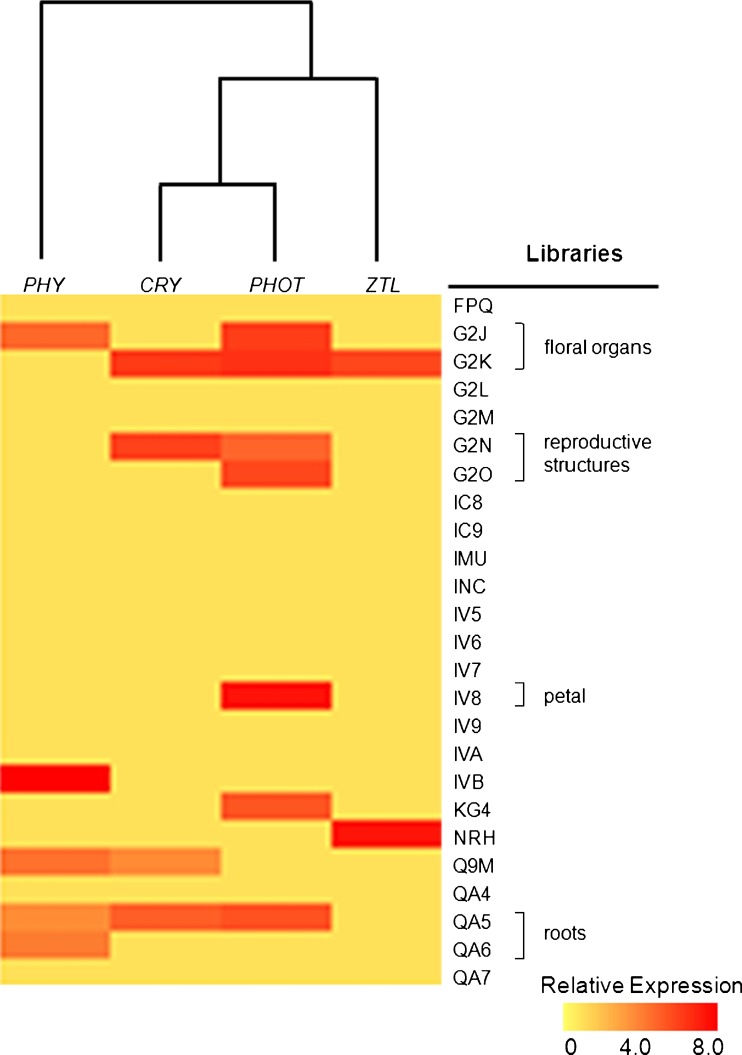
Hierarchical clustering analysis indicated that the expression profile of blue-light photoreceptor families CRY, PHOT and ZTL was similar, whereas transcripts of the red/far-red sensing PHY family exhibit a distinct pattern (Fig. 2). In spite of the observed differential expression patterns, transcripts corresponding to members of all investigated photoreceptor families were present in flower and reproductive organs libraries (Fig. 2). In root tissues, only PHY- and CRY-like transcripts were detected by in silico analysis (Fig. 2).
Fig. 2.
In silico expression analysis of photoreceptor gene-families in Petunia transcriptome. The probe set of photoreceptor genes was clustered using EST library datasets at The Institute of Genome Research (TIGR) (http://tigrblast.tigr.org/tgi/). Gene families and library data were log transformed, normalized and analyzed by Spearman’s Rank Correlation. Library descriptions are available at TIGR. Expression level heatmaps are represented as color scale
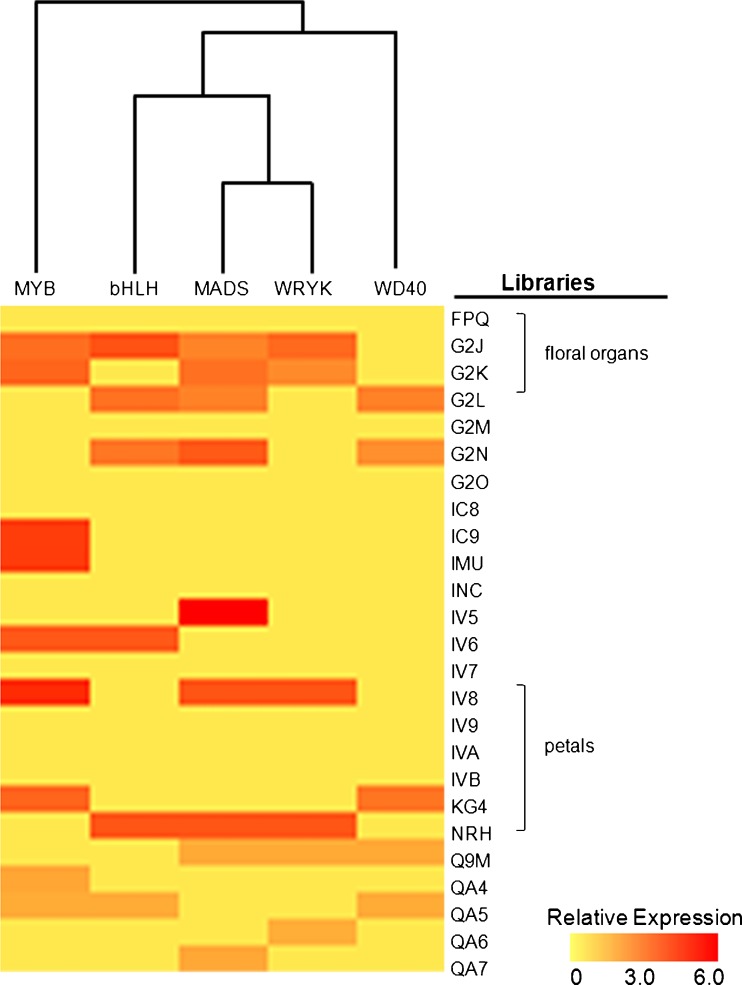
Transcripts corresponding to MADS-box and WRYK families of transcription factors were highly abundant in petunia reproductive organs (floral parts, embryo sac, ovary and inflorescences) and apex libraries (Fig. 3). In contrast, MYB transcripts were also present at high levels in developing seedlings, where transcripts corresponding to MADS-box proteins were less frequent (Fig. 3). The expression pattern of the WD40 class of transcripts was the most divergent among the transcriptional regulators investigated, whereas MADS-box, MYB and bHLH transcription factors exhibit similar expression profiles, as demonstrated by hierarchical clustering analysis (Fig. 3).
Fig. 3.
In silico expression analysis of transcription factor classes involved in anthocyanin metabolism regulation in Petunia transcriptome. The probe set of photoreceptor genes was clustered using EST library data sets at TIGR (http://tigrblast.tigr.org/tgi/). Gene families and library data were log transformed, normalized and analyzed by Spearman’s Rank Correlation. Library descriptions are available at TIGR. Expression level heatmaps are represented as color scale
Mutant identification by reverse genetics
In a previous study, two saturated mutant populations of petunia were generated by chemical (EMS, ethyl-methane sulfonate) and physical (gamma irradiation) mutagenesis and crossed to generate M2 families (Berenschot et al. 2008). The sequences of anthocyanin metabolism and light signaling identified in this study were used to design gene-specific primers to screen the populations for mutations in the genes of interest by a high-throughput PCR-based method, in the absence of direct phenotypic screenings. Gamma irradiation preferentially generates large DNA rearrangements, allowing the detection of mutations by size distinction between wild-type and mutant PCR-amplified fragments. In contrast, EMS mutagenesis induces point mutations that were detected by a modified agarose gel-based TILLING procedure (Raghavan et al. 2007).
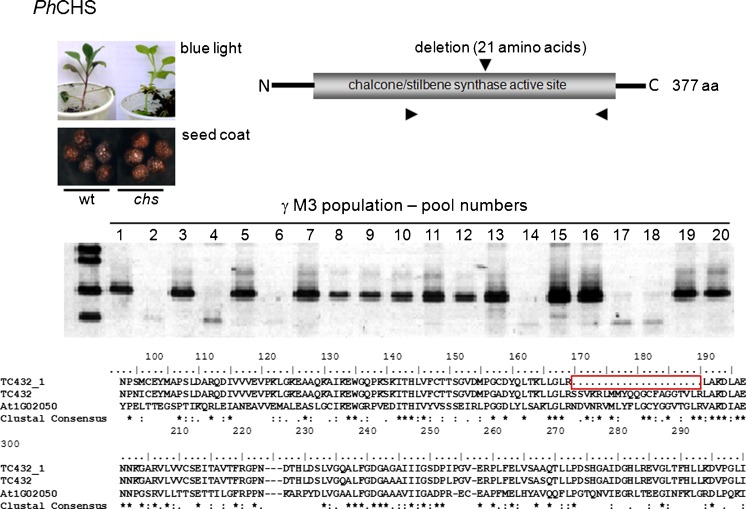
We have analyzed 10,000 plants (20 pools of 250 plants, representing 100 progenies, per M3 population) employing specific primers for petunia sequences similar to five anthocyanin metabolism genes (CHS, DFR4, AN1, AN2 and PH4) and five light-perception and signaling sequences (PHYC, CRY-DASH, CO and PFT1) (Table 5S). In non-mutagenized wild-type plants, the amplification reactions using primer-pairs designed based on bioinformatic analyses of petunia sequences resulted in fragments of the expected sizes, indicating that the identified sequences accurately represent the genes of interest (Table 5S). In the gamma population, we have identified six pools containing putative mutations in CHS sequence (Fig. 4). The plants from the pools exhibiting distinct amplification pattern were individually analyzed and one novel mutant allele of CHS was found (Fig. 4). Mutant chs lines exhibit a conditional reduction of vegetative anthocyanin production under constant blue light (Fig. 4) and no visible alteration in seed testa (Fig. 4) and corolla pigmentation.
Fig. 4.
Detection of CHS mutation in gamma-induced M3 populations of P. x hybrida MD. The numbers represent DNA pools from 25 M3 plants. The lack of amplification in pools 2, 4, 6, 14, 17 and 18 indicate mutated sequence. Plants from pool 2 were individually analyzed and the mutant CHS gene was sequenced; schematic representation of the mutation in the protein and deduced amino-acid sequence of the mutated allele are represented. Representative seed coat and vegetative anthocyanin pigmentation under monochromatic blue light are represented for wild type and mutant plants
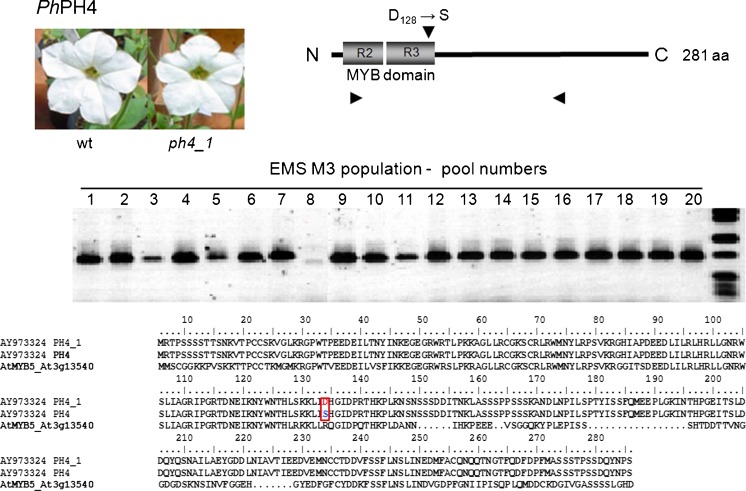
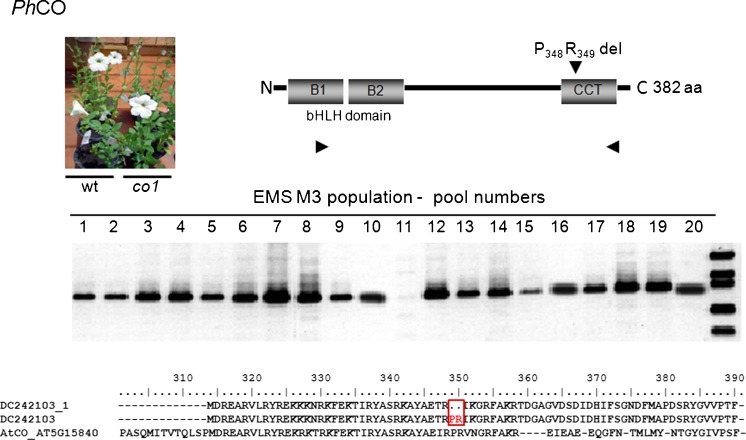
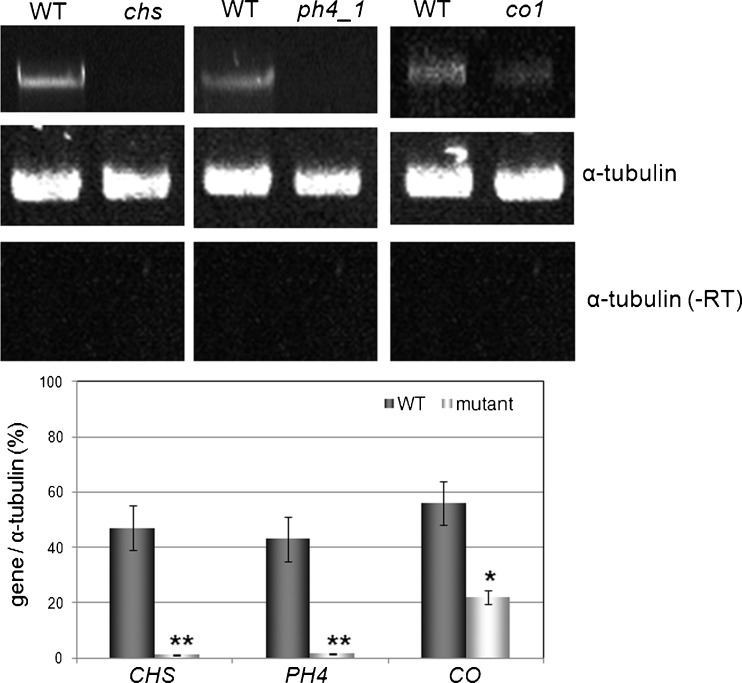
The reverse-genetics TILLING approach allows the detection of point-mutations by strand separation and re-annealing of PCR products corresponding to the gene of interest. The presence of a point mutation will lead to mismatched heteroduplexes resulting from the annealing of a wild-type strand with a mutated one, and the aberrant pairing is targeted to the mismatch-specific endonuclease activity of CELI; producing amplification products of distinct sizes in comparison to the products of the wild type. In the investigated M3 EMS population, we have identified one pool containing a putative mutation in anthocyanin-biosynthesis regulator PH4 (AtMYB5) (Fig. 5) and in a petunia sequence similar to the CONSTANS gene (Fig. 6). The presence of the mutant alleles was confirmed by PCR analyses of the individual plants and sequencing of the genes (Figs. 5 and 6). The identified plants were selfed and homozygous mutants ph4-1 and co-1 are shown in Figs. 5 and 6, respectively. The point mutation identified in PH4 caused no effect on flower color (Fig. 5) and vacuolar pH (Table 1) in the homozygous progeny plants. The presence of a two amino acid deletion in the CCT domain of CO slightly delayed flowering in petunia mutants under 16-h natural photoperiod (Fig. 6, Table 1). After three cycles of self-pollination, the identified lines exhibit reduced steady-state levels of transcripts of the mutated genes (Fig. 7).
Fig. 5.
Detection of PH4 mutation by modified TILLING in EMS-induced M3 populations of P. x hybrida MD. The numbers represent DNA pools from 25 M3 plants. The mismatch-specific cleavage of the amplification product of pool 8 indicates mutated sequence. Plants from pool 8 were individually analyzed and the mutant PH4 gene was sequenced; schematic representation of the mutation in the protein, deduced amino-acid sequence of the mutated allele and mutant plant flower phenotype are represented
Fig. 6.
Detection of CO mutation by modified TILLING in EMS-induced M3 populations of P. x hybrida MD. The numbers represent DNA pools from 25 M3 plants. The mismatch-specific cleavage of the amplification product of pool 11 indicates mutated sequence. Plants from pool 11 were individually analyzed and the mutant CO gene was sequenced; schematic representation of the mutation in the protein and deduced amino-acid sequence of the mutated allele are represented
Table 1.
Summary of the phenotypes of the petunia mutants identified by reverse genetics. The data correspond to in vivo analyses from the M3 isolated individual plants, grown in the greenhouse under 25 °C ± 5 °C, 12 h photoperiod provided by natural light and fluorescent supplementation (200 μmol.m−2.s−1). The numbers represent mean values ± standard errors from two independent evaluations and statistical significance is represented by Tukey test at P > 0.05
| Mutant | ||||
|---|---|---|---|---|
| Stage | chs1(Δ63) | ph4-1(D128→S) | co1 (ΔP348, R349) | Wild type |
| Vegetative | ||||
| Total chlorophyll (μg.mg−1) | 25.32 ± 2.24 | 25.56 ± 3.17 | 21.56 ± 3.01 | 25.41 ± 1.78 |
| Total anthocyanin (A535.mg−1) | 17.11 ± 3.25 | 18.02 ± 4.66 | 18.39 ± 4.49 | 18.54 ± 4.34 |
| Leaf pH | 6.20 ± 1.05 | 6.50 ± 0.94 | 6.20 ± 1.83 | 6.23 ± 0.93 |
| Leaf area (mm2) | 6.09 ± 2.91a | 6.21 ± 3.34a | 5.67 ± 3.76b | 5.98 ± 3.87ab |
| Internode length (cm) | 11.91 ± 3.11a | 11.82 ± 2.54a | 16.91 ± 3.23b | 11.85 ± 2.67a |
| Days to flowering | 18 ± 2.0 | 17 ± 1.7 | 25 ± 3.5 | 21 ± 2.2 |
| Reproductive | ||||
| Number of flowers per 30 days | 14 ± 1.3 | 14 ± 1.5 | 12 ± 1.8 | 14 ± 1.6 |
| Flower longevity (days) | 8.33 ± 1.12 | 7.00 ± 0.89 | 7.03 ± 1.76 | 7.55 ± 2.05 |
| Corolla pH | 5.70 ± 1.12a | 6.30 ± 0.87b | 5.60 ± 1.02a | 5.56 ± 0.99a |
Fig. 7.
Steady state mRNA levels of the genes CHS, PH4 and CO in leaves of wild type and mutant examined by RT-PCR. WT: wild type Mitchell Diploid plants. chs, ph4_1, co_1: mutant plants. Positive reference control corresponds to transcripts of α-tubulin gene. A reaction mixture without template cDNA was used to confirm the absence of amplification from genomic DNA contamination of the RNA samples (RT-)
Discussion
In P. x hybrida databases, we have identified sequences similar to the PHY, CRY, PHOT and ZTL photoreceptor family genes (Table S1). Arabidopsis is the sole plant species for which the full set of photoreceptor genes has been characterized (Demarsy and Fankhauser 2009; Inoue et al. 2010; Nagatani 2010; Chaves et al. 2011). In Solanaceae, photoperception and light signaling have been mainly investigated in tomato, tobacco and potato (Perrotta et al. 2000; Weller et al. 2001; Fernández et al. 2005; Rutitzky et al. 2009). High-throughput transcriptomic analyses have demonstrated extensive sequence and functional conservation among phenotypically distinct Solanaceae species potato, tomato, pepper, petunia, tobacco and Nicotiana benthamiana, although species-specific transcripts were also indentified (Rensink et al. 2005; Rutitzky et al. 2009). Similarly, global gene-expression profiling in Nicotiana and Solanum species has demonstrated that light perception and signaling employ shared and exclusive pathways to control several photomorphogenic and photoperiodic responses (Rensink et al. 2005; Rutitzky et al. 2009). Petunia transcriptome exhibits high sequence and functional conservation to tomato transcript profiles (Rensink et al. 2005) and, in this species de-etiolation and anthocyanin biosynthesis are mainly controlled by phyA- and cry1-mediated signaling (Weller et al. 2001). However, the overexpression of CRY2 has also led to enhanced anthocyanin accumulation in tomato fruits (Giliberto et al. 2005; Lopez et al. 2012). Petunia PHY and CRY transcripts share extensive sequence conservation to the functional domains of the photoreceptors; thus, being likely to be functionally equivalent to their Arabidopsis and tomato counterparts (Table S1). Photoreceptor mutants were absent from the investigated M3 populations (Figs. 5 and 6); however, the number of progenies screened (50 per population) is considered small for reverse genetic approaches (Alonso and Ecker 2006). The identification of photoreceptor mutants by forward genetic approaches requires monochromatic light conditions and intensive experimental care (Fankhauser and Casal 2004). The identified sequences may provide important reverse genetic tools for further in-depth investigation of available petunia mutant populations (Berenschot et al. 2008; Vandenbussche et al. 2008).
Light signal transduction is virtually unknown in petunia, in spite of its role in commercially important traits such as plant architecture and flower pigmentation. By reverse genetics, we have identified candidate genes to regulators of shade-avoidance responses; ATHB2 orthologs (Ciarbelli et al. 2008), photomorphogenesis and seedling establishment; FHY3/FAR1, NDPK2, PIF/PIL family, PP7, RAP1/ATMYC2, RFI2 (Shen et al. 2005; Yadav et al. 2005; Chen and Ni 2006; Lin et al. 2007; Genoud et al. 2008; Leivar et al. 2008), light-regulated proteolysis; COP1, EID1, LAF1, SPA1 (Yi and Deng 2005; Marrocco et al. 2006; Yang et al. 2009; Chen et al. 2010; Fankhauser and Ulm 2011) and developmental transitions; CO, PFT1 (Suárez-López et al. 2001; Cerdán and Chory 2003) (Table S2). Using the obtained sequence information in a reverse genetics approach, we have identified a novel petunia mutant carrying a mutation in CCT (CO, CO-like, TOC1) domain (Strayer et al. 2000; Valverde 2011). In Arabidopsis, the CONSTANS family of zinc finger proteins acts between the circadian clock and genes controlling meristem identity in the photoperiodic flowering pathway (Turck et al. 2008). Arabidopsis is a facultative long-day plant that flowers earlier under long days (LDs) than under short days (SDs), whereas in rice, flowering is induced by SDs (Thomas and Vince-Prue 1996). Photoperiodic-flowering induction in model plants occurs via internal and external coincidence model (Song et al. 2010), where the coincidence between the levels of CO mRNA and the illuminated or dark period of the day activates the transcription of FLOWERING LOCUS T (FT) or its orthologs, which, in turn, are involved in the control of meristem-identity genes, shifting from vegetative to reproductive development (Turck et al. 2008; Song et al. 2010). In day-neutral tomato, the FT ortholog, SINGLE FLOWER TRUSS (SFT), has been demonstrated to trigger the systemic signal responsible for pleiotropic regulation of growth and flowering (Lifschitz et al. 2006). In potato, the ectopic expression of a rice FT gene elucidated the role of CO in controlling the photoperiodic processes flowering and tuberization (Navarro et al. 2011). The mutation identified in petunia CO ortholog is similar to the 4-bp deletion in the CCT domain of the Arabidopsis late-flowering co-5 and co-7 mutant alleles (Koornneef et al. 1991), generating a peptide lacking the P and R conserved residues (Fig. 6). Homozygous petunia plants carrying the two amino acid deletion exhibit a slightly delayed flower induction under natural LD (Fig. 6). However, the phenotype under distinct photoperiod conditions remains to be investigated in functional characterization studies. Further physiological and molecular investigations of the identified mutant allele will help to elucidate the control of developmental transitions in facultative long-day petunia.
Anthocyanin biosynthesis and regulation have been intensively studied in petunia (Winkel-Shirley 2001; Koes et al. 2005; Ambawat et al. 2013). Genetic analyses of flower color mutants have been instrumental to the identification of several structural and regulatory genes in the species (Koes et al. 2005; Ambawat et al. 2013). In a large number of higher plant species, the activity of chalcone synthase displays gene redundancy and it is encoded by at least nine genes in Medicago truncatula, three in grapevine, two in maize and only one in Antirrhinum (Desnos et al. 2001; Koes et al. 2005; Abe and Morita 2010). The CHS gene family in Ipomoea, a genus of the Solanales order, consists of five genes: CHSA until E (Abe and Morita 2010); whereas in Petunia, twelve CHS genes have been described, although their functionality has not been accessed (Koes et al. 2005). Our bioinformatic analyses indicate that at least four distinct transcripts of CHS are present in petunia transcriptome (Table S3) and a novel mutant allele has been identified (Fig. 4). Petunia plants carrying a loss-of-function mutated chs allele are defective in functional pollen tube formation, although the pollen remains viable, and produce shorter and scarcer root hairs in comparison to the wild type (Taylor and Grotewold 2005). No visible phenotype was observed in the identified chs mutant; except for a conditional reduction in blue-light induced vegetative anthocyanin biosynthesis (Fig. 4). The fertility of homozygous lines is reduced in comparison to wild-type plants (Table 1). Extensive gene redundancy in the CHS family in petunia (Koes et al. 2005; Morita et al. 2012) may be responsible for the lack of visible phenotype in the identified line. A novel mutant allele of the gene coding for the R2R3 MYB protein PH4 in white-flowered plants was also identified (Fig. 5). PH4 has been hypothesized to activate vacuolar acidification by interacting with other basic Helix-Loop-Helix transcription factors (Quattrocchio et al. 2006). In the absence of functional PH4 protein, the expression levels of a petal-specific cysteine (cys) proteinase–like protein, CAC16.5, are significantly reduced; however, cys proteinases have several regulatory functions, including defense and programmed cell death responses (Quattrocchio et al. 2006). Flowers from homozygous ph4-1 plants are smaller (Fig. 6) and shorter-lived than those from wild-type flowers (Table 1). The presence of unaccounted secondary mutations may affect the phenotype in M3. Thus, advanced lines carrying the novel ph4 mutant allele may help to elucidate its function in anthocyanin metabolism in petunia.
Electronic supplementary material
(DOC 175 kb)
Acknowledgments
The authors would like to acknowledge Rauly Moretti for the excellent technical assistance. Petunia research is supported by a FAPESP grant (2006/06306-0) to VQ. VQ is the recipient of a CNPq productivity research fellowship (307031/2010-1). The present work is part of the MSc. dissertation of ASB in Tropical and Subtropical Agriculture at the Instituto Agronômico (IAC), Campinas, Brazil (Genetics, Plant Breeding and Biotechnology).
References
- Abe I, Morita H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat Prod Rep. 2010;27:809–838. doi: 10.1039/b909988n. [DOI] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Irvin LJ, Jameson PE, Davies KM. Light-induced vegetative anthocyanin pigmentation in Petunia. J Exp Bot. 2009;60:2191–2202. doi: 10.1093/jxb/erp097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Ecker JR. Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Gen. 2006;7:524–536. doi: 10.1038/nrg1893. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 2013 doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenschot AS, Zucchi MI, Tulmann-Neto A, Quecini V. Mutagenesis in Petunia x hybrida Vilm and isolation of a novel morphological mutant. Braz J Plant Physiol. 2008;20:95–103. doi: 10.1590/S1677-04202008000200002. [DOI] [Google Scholar]
- Casal JJ, Yanovsky MJ. Regulation of gene expression by light. Int J Dev Biol. 2005;49:501–511. doi: 10.1387/ijdb.051973jc. [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J. Regulation of flowering time by light quality. Nature. 2003;423:881–885. doi: 10.1038/nature01636. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- Chen M, Ni M. RED AND FAR-RED INSENSITIVE 2, a RING-domain zinc finger protein, mediates phytochrome-controlled seedling deetiolation responses. Plant Physiol. 2006;140:457–465. doi: 10.1104/pp.105.073163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D, Deng XW. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J. Light signal transduction: an infinite spectrum of possibilities. Plant J. 2010;61:982–991. doi: 10.1111/j.1365-313X.2009.04105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarbelli AR, Ciolfi A, Salvucci S, Ruzza V, Possenti M, Carabelli M, Fruscalzo A, Sessa G, Morelli G, Ruberti I. The Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol Biol. 2008;68:465–478. doi: 10.1007/s11103-008-9383-8. [DOI] [PubMed] [Google Scholar]
- Demarsy E, Fankhauser C. Higher plants use LOV to perceive blue light. Curr Op Plant Biol. 2009;12:69–74. doi: 10.1016/j.pbi.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP. FHY1: a phytochrome A-specific signal transducer. Genes Dev. 2001;15:2980–2990. doi: 10.1101/gad.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid total DNA preparation procedure for fresh plant tissue. Focus. 1990;12:13–15. [Google Scholar]
- Fankhauser C, Casal JJ. Phenotypic characterization of a photomorphogenic mutant. Plant J. 2004;39:747–760. doi: 10.1111/j.1365-313X.2004.02148.x. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. Transposing phytochrome into the nucleus. Trends Plant Sci. 2008;13:596–601. doi: 10.1016/j.tplants.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Ulm R. Light-regulated interactions with SPA proteins underlie cryptochrome-mediated gene expression. Genes Dev. 2011;25:1004–1009. doi: 10.1101/gad.2053911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Fernández AP, Gil P, Valkai I, Nagy F, Schäfer E. Analysis of the function of the photoreceptors phytochrome B and phytochrome D in Nicotiana plumbaginifolia and Arabidopsis thaliana. Plant Cell Physiol. 2005;46:790–796. doi: 10.1093/pcp/pci073. [DOI] [PubMed] [Google Scholar]
- Genoud T, Santa-Cruz MT, Kulisic T, Sparla F, Fankhauser C, Métraux JP. The protein phosphatase 7 regulates phytochrome signaling in Arabidopsis. PLoS One. 2008;3(7):e2699. doi: 10.1371/journal.pone.0002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giliberto L, Perrotta G, Pallara P, Weller JL, Fraser PD, Bramley PM, Fiore A, Tavazza M, Giuliano G (2005) Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol 137(1):199–208. doi: 10.1104/pp.104.051987 [DOI] [PMC free article] [PubMed]
- Guo H, Mockler T, Duong H, Lin C. SUB1, an Arabidopsis Ca2+−binding protein involved in cryptochrome and phytochrome coactions. Science. 2001;291:487–490. doi: 10.1126/science.291.5503.487. [DOI] [PubMed] [Google Scholar]
- Henriques R, Jang IC, Chua NH. Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol. 2009;12:49–56. doi: 10.1016/j.pbi.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Huang Z-A, Zhao T, Fan H-J Wang N, Zheng S-S, Ling H-Q. The upregulation of NtAN2 expression at low temperature is required for anthocyanin accumulation in juvenile leaves of Lc-transgenic tobacco (Nicotiana tabacum L) J Genet Genomics. 2012;39:149–156. doi: 10.1016/j.jgg.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Inoue S, Takemiya A, Shimazaki K. Phototropin signaling and stomatal opening as a model case. Curr Opin Plant Biol. 2010;13:587–593. doi: 10.1016/j.pbi.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Kang X, Ni M. Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell. 2006;18:921–934. doi: 10.1105/tpc.105.037879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes RE, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci U S A. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318:1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez L, Carbone F, Bianco L, Giuliano G, Facella P, Perrotta G. Tomato plants overexpressing cryptochrome 2 reveal altered expression of energy and stress-related gene products in response to diurnal cues. Plant Cell Environ. 2012;35(5):994–1012. doi: 10.1111/j.1365-3040.2011.02467.x. [DOI] [PubMed] [Google Scholar]
- Marrocco K, Zhou Y, Bury E, Dieterle M, Funk M, Genschik P, Krenz M, Stolpe T, Kretsch T. Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J. 2006;45:423–438. doi: 10.1111/j.1365-313X.2005.02635.x. [DOI] [PubMed] [Google Scholar]
- McCallum MC, Comai L, Greene EA, Henikoff S. Targeting Induced Local Lesions In Genomes (TILLING) for plant functional genomics. Plant Physiol. 2000;123:439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Saito R, Ban Y, Tanikawa N, Kuchitsu K, Ando T, Yoshikawa M, Habu Y, Ozeki Y, Nakayama M. Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant J. 2012;70:739–749. doi: 10.1111/j.1365-313X.2012.04908.x. [DOI] [PubMed] [Google Scholar]
- Nagatani A. Phytochrome: structural basis for its functions. Curr Opin Plant Biol. 2010;13:565–570. doi: 10.1016/j.pbi.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Navarro C, Abelenda JA, Cruz-Oró E, Cuéllar CA, Tamaki S, Silva J, Shimamoto K, Prat S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478:119–122. doi: 10.1038/nature10431. [DOI] [PubMed] [Google Scholar]
- Pérez-de-Castro AM, Vilanova S, Cañizares J, Pascual L, Blanca JM, Díez MJ, Prohens J, Picó B. Application of genomic tools in plant breeding. Curr Genomics. 2012;13(3):179–195. doi: 10.2174/138920212800543084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta G, Ninu L, Flamma F, Weller JL, Kendrick RE, Nebuloso E, Giuliano G. Tomato contains homologues of Arabidopsis cryptochromes 1 and 2. Plant Mol Biol. 2000;42:765–773. doi: 10.1023/A:1006371130043. [DOI] [PubMed] [Google Scholar]
- Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181:219–229. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell. 2006;18:1274–1291. doi: 10.1105/tpc.105.034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quecini V, Liscum E. Signal transduction in blue light-mediated responses. In: Schäfer E, Nagy F, editors. Photomorphogenesis in Plants and Bacteria: function and signal transduction mechanisms. Dordrecht: Kluwer Academic Publishers; 2006. pp. 305–327. [Google Scholar]
- Raghavan C, Naredo MEB, Wang H, Atienza G, Liu B, Qiu F, McNally KL, Leung H. Rapid method for detecting SNPs on agarose gels and its application in candidate gene mapping. Mol Breed. 2007;19:87–101. doi: 10.1007/s11032-006-9046-x. [DOI] [Google Scholar]
- Rensink WA, Lee Y, Liu J, Iobst S, Ouyang S, Buell CR. Comparative analyses of six solanaceous transcriptomes reveal a high degree of sequence conservation and species-specific transcripts. BMC Genomics. 2005;6:124. doi: 10.1186/1471-2164-6-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutitzky M, Ghiglione HO, Curá JA, Casal JJ, Yanovsky MJ. Comparative genomic analysis of light-regulated transcripts in the Solanaceae. BMC Genomics. 2009;10:60. doi: 10.1186/1471-2164-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Schäfer E, Nagy F. Photomorphogenesis in plants and bacteria: Function and signal transduction mechanisms. Dordrecht: Springer; 2006. [Google Scholar]
- Shen Y, Kim JI, Song PS. NDPK2 as a signal transducer in the phytochrome-mediated light signaling. J Biol Chem. 2005;280:5740–5749. doi: 10.1074/jbc.M408965200. [DOI] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T. Similarities in the circadian clock and photoperiodism in plants. Curr Opin Plant Biol. 2010;13:594–603. doi: 10.1016/j.pbi.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. Flavonoids as developmental regulators. Curr Opin Plant Biol. 2005;8:317–323. doi: 10.1016/j.pbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in plants. San Diego: Academic; 1996. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Physiol Plant Mol Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Valverde F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J Exp Bot. 2011;62:2453–2463. doi: 10.1093/jxb/erq449. [DOI] [PubMed] [Google Scholar]
- Vandenbussche M, Janssen A, Zethof J, van Orsouw N, Peters J, van Eijk MJ, Rijpkema AS, Schneiders H, Santhanam P, de Been M, van Tunen A, Gerats T. Generation of a 3D indexed Petunia insertion database for reverse genetics. Plant J. 2008;54:1105–1114. doi: 10.1111/j.1365-313X.2008.03482.x. [DOI] [PubMed] [Google Scholar]
- Weller JL, Perrotta G, Schreuder ME, van Tuinen A, Koornneef M, Giuliano G, Kendrick RE. Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochromes A, B1 and B2. Plant J. 2001;25:427–440. doi: 10.1046/j.1365-313x.2001.00978.x. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 2005;17:1953–1966. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Jang IC, Henriques R, Chua NH. FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell. 2009;21:1341–1359. doi: 10.1105/tpc.109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15:618–625. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Zenoni S, D’Agostino N, Tornielli GB, Quattrocchio F, Chiusano ML, Koes R, Zethof J, Guzzo F, Delledonne M, Frusciante L, Gerats T, Pezzotti M. Revealing impaired pathways in the an11 mutant by high-throughput characterization of Petunia axillaris and Petunia inflata transcriptomes. Plant J. 2011;68(1):11–27. doi: 10.1111/j.1365-313X.2011.04661.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 175 kb)