Abstract
Antigen-presenting cells (APCs) induce T cell activation as well as T cell tolerance. The molecular basis of the regulation of this critical ‘decision’ is not well understood. Here we show that HDAC11, a member of the HDAC histone deacetylase family with no prior defined physiological function, negatively regulated expression of the gene encoding interleukin 10 (IL-10) in APCs. Overexpression of HDAC11 inhibited IL-10 expression and induced inflammatory APCs that were able to prime naive T cells and restore the responsiveness of tolerant CD4+ T cells. Conversely, disruption of HDAC11 in APCs led to upregulation of expression of the gene encoding IL-10 and impairment of antigen-specific T cell responses. Thus, HDAC11 represents a molecular target that influences immune activation versus immune tolerance, a critical ‘decision’ with substantial implications in autoimmunity, transplantation and cancer immunotherapy.
Bone marrow–derived antigen-presenting cells (APCs) are important in the initiation of productive antigen-specific T cell responses1,2 and in the induction of T cell tolerance3–5. This apparently dual function was initially explained by the existence of specific APC subpopulations that ‘preferentially’ trigger T cell priming, whereas other subpopulations were identified as inducers of T cell anergy6–8. The demonstration that a single APC subpopulation can elicit both T cell outcomes9, however, led to the alternative explanation that the functional status of the APC at the time of antigen presentation, rather than its phenotypic characteristics, might be the critical determinant of antigen-specific T cell responses10.
Several factors have been linked to influencing the functional status of the APC. Among them, the production of pro- and anti-inflammatory mediators at the site of antigen encounter have been shown to shape the magnitude and duration of the immune response initiated by the APC11. Interleukin 12 (IL-12; A002864 and A002865) and IL-10 (A001243), cytokines with divergent inflammatory properties, are at the center of this delicate balance. IL-12 is required for resistance to infection, but persistently increased concentrations can result in autoimmunity12. Conversely, IL-10 can serve a key function in tolerance induction by keeping immune responses in check and preventing self tissue damage13–15. A better understanding of the molecular mechanisms that regulate the production of these mediators would probably lead to the identification of new targets for influencing T cell activation versus T cell tolerance.
In the past, special attention has been given to chromatin modification by acetylation or deacetylation of histone tails and its involvement in regulating gene transcription, including that of genes involved in the inflammatory response16. For example, cytokine production by APCs can be influenced by changes in the acetylation status of the gene promoter17,18. Here we show that histone deacetylase 11 (HDAC11), by interacting with the distal segment of the promoter of the gene encoding IL-10 (Il10), negatively regulated the expression of this cytokine in mouse and human APCs. Such an effect not only determined the inflammatory status of these cells but also influenced priming versus tolerance of antigen-specific CD4+ T cells.
RESULTS
Histone deacetylases and Il10 expression
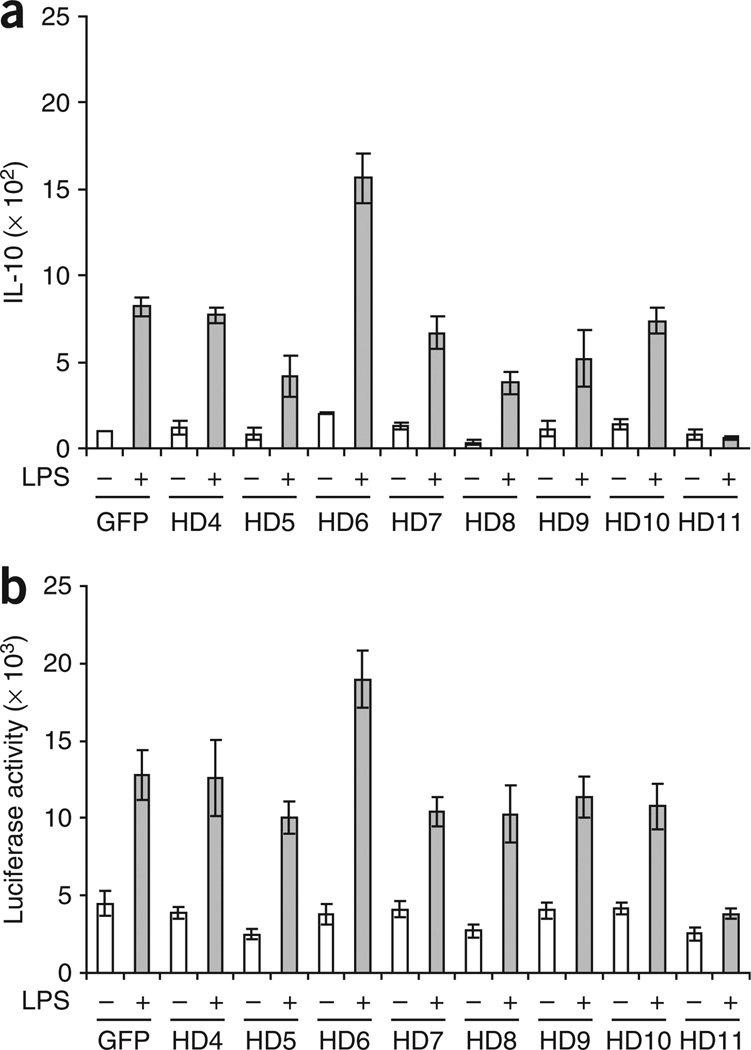
Chromatin accessibility in genes involved in inflammatory responses is influenced by the acetylation status of their promoters. In general, whereas histone acetylation results in transcriptionally active chromatin, histone deacetylation mediated by HDAC proteins is associated with an inactive chromatin. Although the involvement of HDAC proteins in regulation of gene transcription in nonimmune cells is well established, little is known about the function of specific HDAC proteins in influencing the inflammatory status of APCs. Given the dominant function of IL-10 in tolerance induction and regulation of inflammation14,19, we sought to determine whether overexpression of specific HDAC proteins might influence the transcriptional activity of Il10 in APCs. We therefore infected the mouse macrophage cell line RAW264.7 with adenovirus encoding Flag- and GFP-tagged versions of several known HDAC proteins20–22. In initial experiments, we evaluated HDAC1 and HDAC2, but given their nonspecific effects as repressors of several cytokine promoters, we decided to focus our attention on the remaining HDAC proteins. Unstimulated RAW264.7 cells infected with adenovirus vector expressing green fluorescent protein (GFP) had minimal expression of IL-10 mRNA (Fig. 1a). After in vitro stimulation with lipopolysaccharide (LPS), these macrophages had higher expression of IL-10 mRNA (Fig. 1a). Infection of macrophages with adenovirus encoding HDAC4, HDAC5, HDAC7, HDAC8, HDAC9 or HDAC10 did not affect the ability of these cells to express IL-10 mRNA in response to LPS stimulation. Overexpression of HDAC6 (A001723) in RAW264.7 cells, however, was associated with enhanced IL-10 mRNA expression in response to LPS (Fig. 1a). Overexpression of HDAC11 resulted in blunted expression of IL-10 mRNA in LPS-treated RAW264.7 cells (Fig. 1a).
Figure 1.
Overexpression of HDAC11 abrogates the expression of IL-10 mRNA in LPS-treated macrophages. (a) Quantitative real-time RT-PCR analysis of IL-10 mRNA among total RNA from RAW264.7 cells infected in vitro with adenovirus encoding HDAC4–HDAC11 (HD4–HD11) or GFP and, 48 h later, left unstimulated (−) or stimulated with LPS (1 µg/ml) for an additional 3 h (+). Results are normalized to GAPDH expression and are presented relative to that of control cells infected with adenovirus encoding GFP alone. Data are representative of three independent experiments with similar results (error bars, s.d. of triplicates). (b) Luciferase activity of lysates of RAW264.7 cells transfected by electroporation with plasmid containing a luciferase reporter gene plus the Il10 promoter, then subjected to adenoviral infection and left unstimulated or stimulated with LPS (1 µg/ml) for an additional 3 h, then analyzed after 24 h. Results are normalized to protein concentrations. Data are from one experiment representative of three independent experiments with similar results (error bars, s.d.).
HDAC11 is a newly identified member of the HDAC family and thus far no known physiological function for HDAC11 has been demonstrated23. A putative function for HDAC11 as a negative regulator of Il10 transcriptional activity was unexpected. To confirm our observation, we transfected RAW264.7 cells with a reporter gene containing the Il10 promoter fused to a luciferase gene. Again, unlike overexpression of other HDAC proteins, only HDAC11 overexpression resulted in the inhibition of luciferase activity in response to LPS stimulation (Fig. 1b).
Effects of HDAC11 overexpression in mouse and human APCs
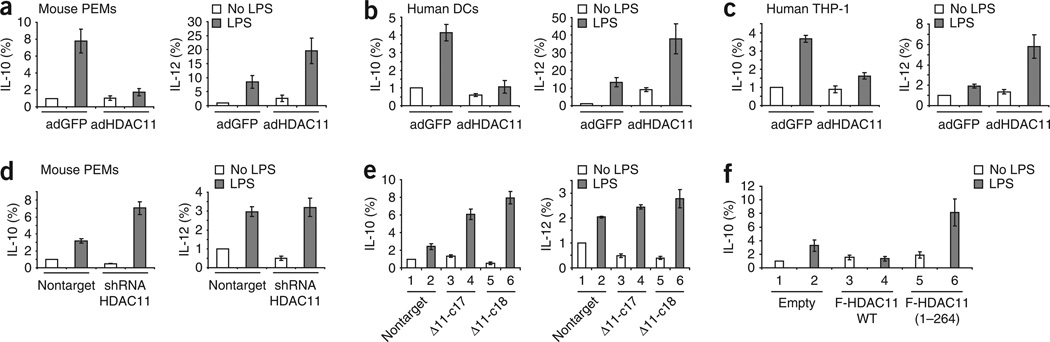
To expand our studies beyond a mouse macrophage cell line, we determined the effects of HDAC11 overexpression in primary mouse macrophages (PEMs) as well as in human APCs. LPS stimulation of PEMs infected with adenovirus vector encoding GFP resulted in higher expression of IL-10 mRNA than that of unstimulated PEMs (Fig. 2a). Reminiscent of our studies of RAW264.7 cells (Fig. 1), PEMs infected with adenovirus encoding HDAC11 did not increase IL-10 mRNA expression in response to LPS (Fig. 2a). Of note, overexpression of HDAC11 was associated with higher IL-12 mRNA expression in PEMs in response to LPS than that of PEMs infected with adenovirus vector encoding GFP (Fig. 2a). Similarly, we also noted less IL-10 mRNA and more IL-12 mRNA in primary human dendritic cells (Fig. 2b) and THP-1 human monocytic cells (Fig. 2c) overexpressing HDAC11.
Figure 2.
Expression of IL-10 and IL-12 mRNA in APCs overexpressing or lacking HDAC11. (a–c) Real-time RT-PCR analysis of the expression of IL-10 and IL-12 mRNA by PEMs from BALB/c mice (a), primary human dendritic cells (b) or human THP-1 monocytic cells (c) infected with adenovirus encoding HDAC11 (adHDAC11) or GFP (adGFP) and then, 48 h later, left unstimulated or stimulated with LPS (1 µg/ml) for an additional 3 h. Results are normalized to GAPDH expression and are presented relative to that of control cells infected with adenovirus encoding GFP in the absence of LPS stimulation. (d) Real-time RT-PCR analysis of IL-10 and IL-12 mRNA in PEMs transiently transduced with lentivirus particles containing HDAC11-specific shRNA (shRNA HDAC11) or a nonspecific nontargeting control (Nontarget) and then stimulated and assessed as described in a–c. (e) Real-time RT-PCR analysis of IL-10 and IL-12 mRNA in RAW264.7 clones 17 and 18 (which lack HDAC11 expression; Δ11-c17 and Δ11-c18, respectively), and in RAW264.7 cells transduced with a nontargeting shRNA, stimulated and assessed as described in a–c. (f) Real-time RT-PCR analysis of IL-10 mRNA in RAW264.7 cells transfected by electroporation with plasmid encoding wild-type HDAC11 (F-HDCA11 WT) or an HDAC11 construct lacking enzymatic activity (F-HDAC11 (1–264)) or with empty vector (Empty), then stimulated and assessed as described in a–c. Data are representative of three independent experiments with similar results (a–c; error bars, s.d.) or are from one experiment representative of three (d) or two (e,f) independent experiments with similar results (d–f; error bars, s.d.).
Given that overexpression of HDAC11 was associated with inhibition of expression of the gene encoding IL-10, we sought to determine whether ‘knocking down’ HDAC11 in APCs would lead to the opposite effect. Transduction of PEMs with short hairpin RNA (shRNA) specific for mouse HDAC11 resulted in higher expression of IL-10 mRNA in response to LPS stimulation relative to that of PEMs transduced with nontargeting shRNA (Fig. 2d). The observed effect was indeed mediated by inhibition of HDAC11, as there was less HDAC11 protein in cells transduced with a lentivirus encoding HDAC11-specific shRNA (Supplementary Fig. 1a online). Inhibition of HDAC11 expression in PEMs did not affect IL-12 mRNA expression in response to LPS relative to that of control cells (Fig. 2d). To further confirm those results, we generated two stable cell lines derived from RAW264.7 cells lacking HDAC11 expression because of transduction with lentiviral particles encoding HDAC11-specific shRNA (clones 17 and 18; Supplementary Fig. 1b). Similar to the results obtained with PEMs (Fig. 2d), stimulation of these two clones with LPS resulted in more IL-10 mRNA expression than that of cells transduced with nontargeting shRNA (Fig. 2e). The absence of HDAC11 was associated with lower baseline expression of IL-12 mRNA in both clones relative to that of control cells (Fig. 2e). In response to LPS stimulation, there was a trend toward more IL-12 mRNA expression in RAW264.7 clones (Fig. 2e).
Next we determined whether the enzymatic deacetylase activity of HDAC11 was required for the regulation of IL-10 mRNA expression in APCs. We generated a HDAC11 construct that lacked enzymatic activity because of deletion of its deacetyltransferase domain but was still able to form dimers (Supplementary Fig. 2 online). Overexpression of wild-type HDAC11 inhibited IL-10 mRNA expression in LPS-treated RAW264.7 cells relative to that of cells transfected with empty vector (Fig. 2f). There was no such inhibition, however, in RAW264.7 cells transfected with the enzymatically inactive mutant HDAC11. Instead, these cells had more IL-10 mRNA expression in response to LPS (Fig. 2f), which suggested that the HDAC11 mutant might be acting as a dominant negative variant. Thus, whereas overexpression of HDAC11 in APCs resulted in inhibition of the transcriptional activity of Il10, targeting HDAC11 by RNA-mediated interference inhibition led to the opposite outcome. Furthermore, intact enzymatic deacetylase activity was required for HDAC11-mediated inhibition of Il10 expression in APCs.
HDAC 11 interacts with the Il10 promoter
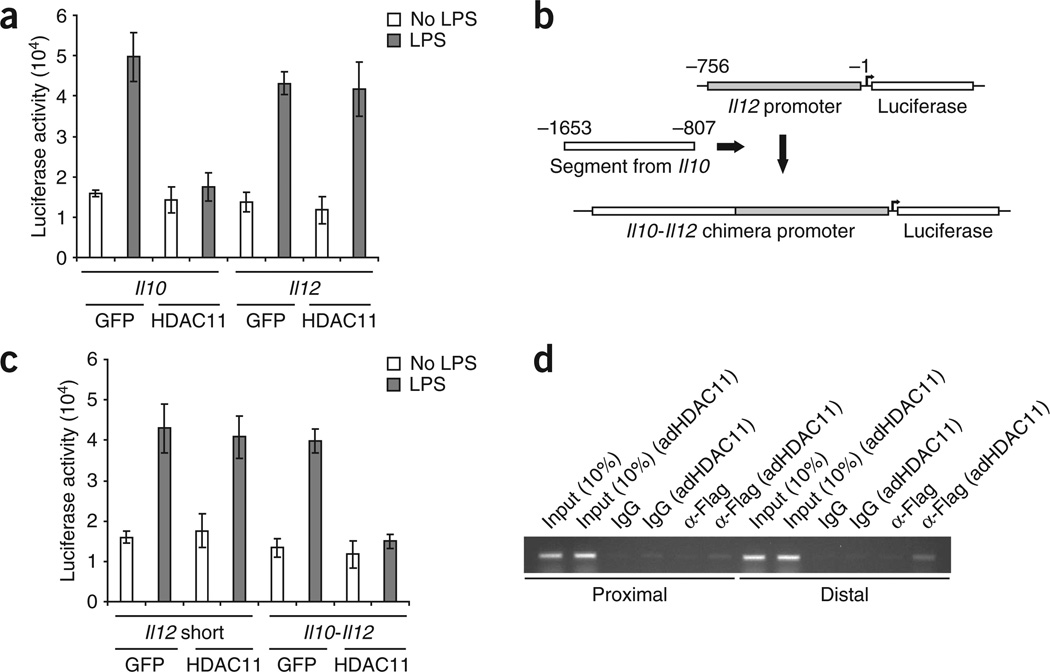
The results presented above demonstrated that HDAC11 repressed mainly Il10 expression. However, there were also changes in IL-12 mRNA expression in cells overexpressing HDAC11 (Fig. 2a–c) or lacking HDAC11 (Fig. 2e). To address whether HDAC 11 interacts at the Il10 and/or Il12 promoter, we transfected RAW264.7 cells with a reporter gene containing either the Il12 or Il10 promoter fused to a luciferase gene. We then infected these cells with adenovirus encoding HDAC11 and left them unstimulated or stimulated them with LPS. There was again strong inhibition of lucifease activity in LPS-stimulated cells transfected with the Il10 reporter gene and overexpressing HDAC11 (Fig. 3a). In contrast, in cells transfected with the Il12 reporter gene, overexpression of HDAC11 did not inhibit luciferase activity in response to LPS (Fig. 3a).
Figure 3.
The distal region of the Il10 promoter is required for HDAC11-mediated gene repression. (a) Luciferase activity of lysates of RAW264.7 cells transfected by electroporation with plasmid containing a luciferase reporter gene plus the Il10 or Il12 promoter, then subjected to infection with adenovirus encoding GFP or HDAC11 and left unstimulated or stimulated with LPS (1 µg/ml) for an additional 3 h, then analyzed after 24 h. Results are normalized to protein concentrations. Data are from three independent experiments with similar results (error bars, s.d. of triplicates). (b) Generation of a chimeric reporter gene with the distal region of the Il10 promoter fused to the ‘short region’ of the Il12 promoter (Il10-Il12 chimera). (c) Luciferase activity of RAW264.7 cells transfected with plasmid containing a luciferase reporter gene plus the Il12 promoter ‘short region’ or the Il10-Il12 chimera promoter construct, then infected with adenovirus encoding GFP or HDAC11 and left unstimulated or stimulated with LPS (1 µg/ml) for an additional 3 h. Data are from one experiment representative of three independent experiments with similar results (error bars, s.d.). (d) ChIP analysis of RAW264.7 cells left uninfected or infected for 48 h with adenovirus encoding HDAC11 (adHDAC11), assessed with anti-Flag (α-Flag) to evaluate HDAC11 binding or with normal mouse IgG as a control. Input (10%), analysis of 10% of the input DNA before immunoprecipitation. Data are representative of three experiments.
Additional confirmation that the Il10 promoter represents the target for HDAC11 was provided by studies with an Il10-Il12 chimeric promoter construct. We transfected RAW264.7 cells with the proximal region of the Il12 promoter (positions −1 to −756 relative to the transcription start site; ‘short region’) fused to a luciferase gene (Fig. 3b) or with a chimeric construct composed of the same Il12 promoter (‘short region’) fused to the distal segment of the Il10 promoter (positions −807 to −1653; Fig. 3b). Overexpression of HDAC11 or GFP in RAW264.7 cells transfected with the ‘short region’ of the Il12 promoter resulted in similar luciferase activity in response to LPS stimulation (Fig. 3c). In contrast, overexpression of HDAC11 in RAW264.7 cells transfected with the Il10-Il12 chimeric promoter completely abrogated the increase in luciferase activity in response to LPS stimulation (Fig. 3c). Therefore, simply the addition of the distal segment of the Il10 promoter to the ‘short region’ of the Il12 promoter resulted in strong inhibition of gene expression in RAW264.7 infected with adenovirus encoding HDAC11. To further confirm to which segment of the Il10 promoter HDAC11 was being recruited, we infected RAW264.7 cells with adenovirus encoding HDAC11. We then analyzed the presence of HDAC11 on either the proximal region (positions −1 to −807) or the distal region (positions −807 to −1653) of the Il10 promoter. We detected HDAC11 mainly on the distal region of the Il10 promoter (Fig. 3d). Using a similar approach, we did not detect HDAC11 in the Il12 promoter region (Supplementary Fig. 3a online). Our results collectively provide evidence that HDAC11 exerts its negative regulatory effect at the distal segment of the Il10 promoter.
Changes in the Il10 promoter induced by HDAC11
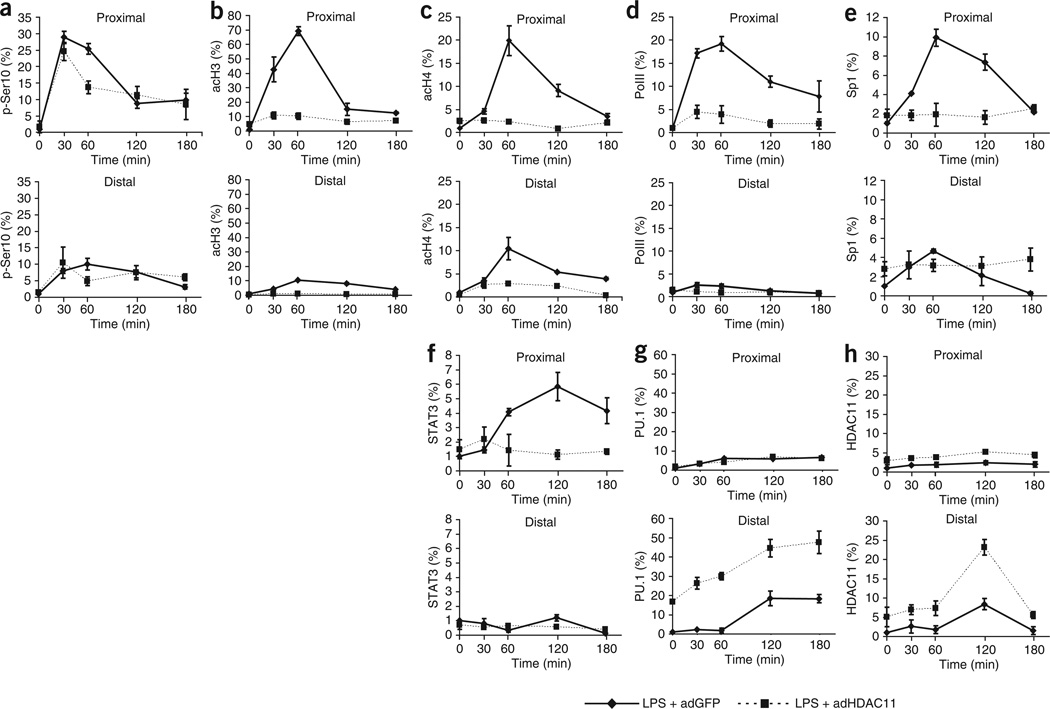
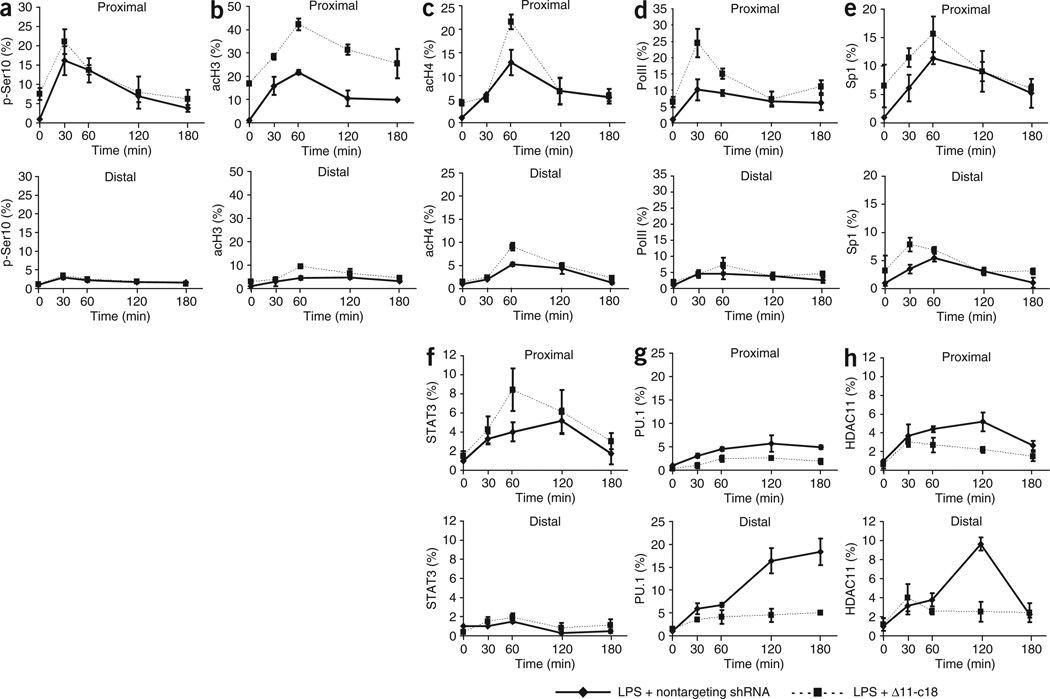
To gain insight into the potential chromatin modifications induced by HDAC11, we used chromatin immunoprecipitation (ChIP) analysis to evaluate histone changes in the proximal and distal regions of the Il10 promoter after LPS stimulation of macrophages overexpressing HDAC11. Published studies have shown that phosphorylation of histone H3 at the serine residue at position 10 (Ser10) is needed for transcriptional activation of the Il10 promoter24. Indeed, in control cells stimulated with LPS, there was such phosphorylation in the proximal region of the Il10 promoter, which reached its peak by 30 min and was followed by a progressive decrease (Fig. 4a). In macrophages overexpressing HDAC11, we found a similar pattern of phosphorylation; the only difference was that after the peak at 30 min, there was a more rapid decrease in this phosphorylation (Fig. 4a). Unlike the proximal region, the distal region of the Il10 promoter had only minimal changes in such phosphorylation in response to LPS in either control cells or cells overexpressing HDAC11 (Fig. 4a).
Figure 4.
Chromatin modifications induced by HDAC11 overexpression. ChIP analysis of RAW264.7 cells infected for 24 h with adenovirus encoding GFP or HDAC11, then stimulated with LPS (1.0 µg/ml) and collected at baseline (time 0) or at 30, 60, 120 or 180 min after stimulation, assessed with antibody to phosphorylated Ser10 of histone H3 (p-Ser10; a), hyperacetylated histone H3 (acH3; b), hyperacetylated histone H4 (acH4; c), RNA polymerase II (PolII; d), Sp1 (e), STAT3 (f), PU.1 (g) or HDAC11 (h), followed by real-time RT-PCR analysis of the proximal region (positions −10 to −367) and distal region (positions −922 to −1324) of the Il10 promoter. Values were obtained with the Pfaffl method and are presented relative to input before immunoprecipitation. Data are from one experiment representative of two independent experiments with similar results (error bars, s.d. of triplicates)
ChIP analysis of the acetylation status of histones H3 and H4 in LPS-stimulated control cells (infected with adenovirus expressing GFP) showed peak acetylation in the proximal region of the Il10 promoter by 60 min, followed by a rapid decrease (Fig. 4b,c). In contrast, in macrophages overexpressing HDAC11, there was no acetylation of histones H3 and H4 in the proximal promoter at any time points evaluated (Fig. 4b,c). In the distal region of the Il10 promoter, there were only minimal changes in histone H3 acetylation in control cells or HDAC11-overexpressing cells (Fig. 4b). The greater H4 acetylation in the distal region was of a lesser magnitude than that in the proximal promoter region. It reached a peak by 60 min, followed by a progressive decrease (Fig. 4c). Again, we found no such changes in cells overexpressing HDAC11 (Fig. 4c). Of note, ChIP analysis of the proximal and distal segments of the Il12 promoter showed no inhibition in the acetylation of histones H3 and H4 in macrophages overexpressing HDAC11 (Supplementary Fig. 3b,c).
We next evaluated the effect of HDAC11 overexpression on the transcriptional activity of the Il10 promoter by determining the binding of RNA polymerase II to the Il10 promoter. Macrophages infected with adenovirus expressing GFP and stimulated with LPS had a rapid increase in the binding of RNA polymerase II, followed by a progressive decrease in the proximal but not in the distal region of the Il10 promoter (Fig. 4d). In contrast, there was only a minimal and transient increase in the binding of RNA polymerase II in macrophages infected with adenovirus expressing HDAC11 (Fig. 4d). Next we determined the kinetics of the expression of Sp1 and STAT3, transcription factors known to interact with the Il10 promoter25,26. First, ChIP analysis of the proximal region of the Il10 promoter showed that unlike LPS-stimulated control cells, in which binding of Sp1 peaked at 1 h and was then followed by a rapid decrease at 3 h (Fig. 4e), macrophages overexpressing HDAC11 did not show such changes in response to LPS stimulation (Fig. 4e). Similarly, whereas binding of STAT3 to the proximal region of the Il10 promoter was evident within 60 min of LPS stimulation and reached its peak within 2 h in control macrophages (Fig. 4f), there was no binding of STAT3 to the proximal promoter in macrophages overexpressing HDAC11 (Fig. 4f). There were no substantial alterations in the abundance of Sp1 or STAT3 in the distal Il10 promoter region for either group of macrophages (Fig. 4e,f). Notably, in control macrophages, the changes in histone acetylation and binding of transcription factors to the proximal region of the Il10 promoter coincided temporally with maximum expression of IL-10 mRNA (60–120 min; Supplementary Fig. 4 online).
PU.1 is a transcription repressor that interacts with the Il10 promoter27,28. Unlike STAT3 and Sp1, which bind to the proximal region of the Il10 promoter, PU.1 showed no changes to its binding in either control macrophages or in cells overexpressing HDAC11, by ChIP analysis of this region (Fig. 4g). In contrast, ChIP analysis of the distal promoter region of control macrophages showed that PU.1 binding reached a peak within 2 h and remained increased for the duration of the analysis (Fig. 4g). In macrophages overexpressing HDAC11, binding of PU.1 to the distal promoter was already increased at time 0 and it increased further in response to LPS (Fig. 4g).
Given the finding that the distal segment of the Il10 promoter represented the common region in which we detected both HDAC11 (Fig. 3d) and PU.1 (Fig. 4g), we next evaluated the kinetics of the binding of HDAC11 to the distal Il10 promoter. In control macrophages, changes in HDAC11 were detectable 2 h after LPS stimulation, were modest in magnitude and returned to baseline within 3 h (Fig. 4h). In macrophages overexpressing HDAC11, there was more binding of this molecule at time 0; it reached its peak within 2 h of LPS stimulation and was followed by a rapid return to baseline within 3 h (Fig. 4h). There were no such changes in the proximal Il10 promoter region (Fig. 4h).
Changes in the Il10 promoter in cells lacking HDAC11
Although the results reported above demonstrated the chromatin modifications induced by overexpression of HDAC11 in APCs, the physiological function of endogenous HDAC11 in these cells remained to be defined. To determine this, we did a ChIP analysis similar to that reported above but used cells in which HDAC11 expression was diminished by RNA-mediated interference. We simulated control RAW 264.7 cells infected with nontargeting shRNA or one of clones reported above lacking HDAC11 (clone 18) with LPS. First, we found no differences in the kinetics of phosphorylation of Ser10 of histone H3 in the proximal Il10 promoter of cells with or without expression of HDAC11 (Fig. 5a). Unlike cells overexpressing HDAC11, in which we found no acetylation of histone H3 or H4 in the proximal Il10 promoter (Fig. 4b,c), cells lacking HDAC11 had more acetylation of histones H3 and H4 than did control cells in response to LPS (Fig. 5b,c). Of note, the transcriptional activity of the Il10 promoter, as determined by binding of RNA polymerase II to the proximal promoter, was of greater magnitude in cells lacking HDAC11 than in control cells stimulated with LPS (Fig. 5d). There were again no substantial chromatin changes in the distal Il10 promoter of cells lacking HDAC11 (Fig. 5a–d).
Figure 5.
Chromatin modification in APCs lacking HDAC11. ChIP analysis of RAW264.7 clone 18 (lacking HDAC11) or RAW264.7 cells transduced with a nontargeting control, then stimulated with LPS (1.0 µg/ml) and collected at baseline (time 0) or at 30, 60, 120 or 180 min after stimulation, assessed with antibody to phosphorylated Ser10 of histone H3 (a), hyperacetylated histone H3 (b), hyperacetylated histone H4 (c), RNA polymerase II (d), Sp1 (e), STAT3 (f), PU.1 (g) or HDAC11 (h), followed by real-time RT-PCR analysis of the proximal region (positions −10 to −367) and distal region (positions −922 to −1324) of the Il10 promoter. Values were obtained with the Pfaffl Method and are presented relative to input before immunoprecipitation. Data are from one experiment representative of two independent experiments with similar results (error bars, s.d. of triplicates).
Kinetic analysis of the Il10-transcription activators Sp1 and STAT3 in cells lacking HDAC11 showed earlier and greater detection of both transcription factors in the proximal Il10 promoter relative to that of control cells (Fig. 5e,f). In contrast, the transcription repressor PU.1 was minimally detected in the distal Il10 promoter of cells lacking HDAC11 relative to that of control cells (Fig. 5g). As expected, HDAC11 was barely detected in the distal Il10 promoter of cells in which HDAC11 expression had been inhibited (Fig. 5h).
Functional changes in APCs overexpressing or lacking HDAC11
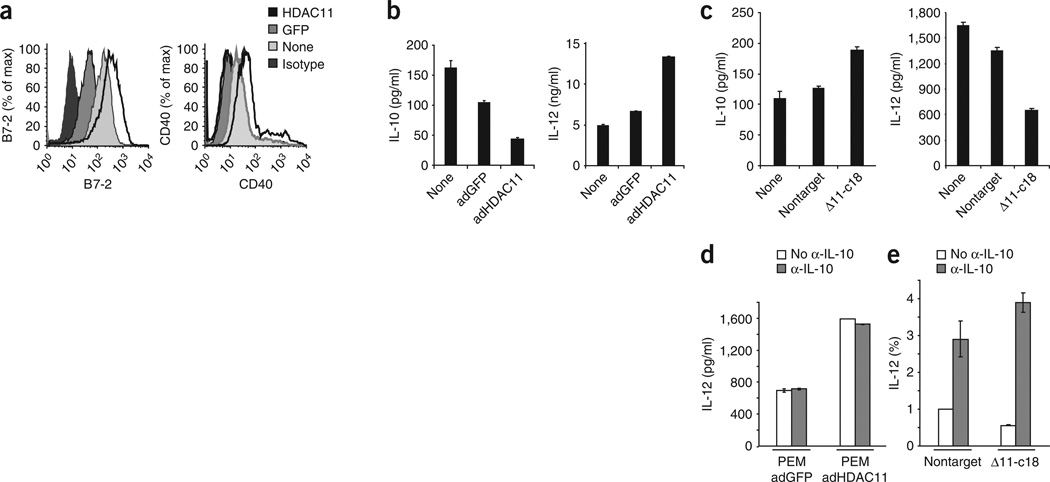
Next we determined the functional consequences of overexpressing or inhibiting HDAC11 in APCs. For this, first we infected PEMs with adenovirus encoding GFP or HDAC11 or left the cells uninfected. Macrophages overexpressing HDAC11 had more expression of the costimulatory molecules CD86 and CD40 (Fig. 6a) and produced less IL-10 than did uninfected PEMs or PEMs infected with adenovirus expressing GFP alone (Fig. 6b). Conversely, IL-12 production was higher in HDAC11-overexpressing PEMs than in control PEMs (Fig. 6b). Next we did a similar analysis of RAW264.7 clone 18, which lacks HDAC11, RAW264.7 cells infected with nontargeting shRNA and wild-type RAW264.7 cells. We did not detect IL-10 or IL-12 by enzyme-linked immunosorbent assay (ELISA) in the supernatants of these cells in the absence of LPS stimulation (data not shown). After treatment with LPS, we detected more production of IL-10 and less production of IL-12 in cells lacking HDAC11 than in control wild-type cells or cells transfected with nontargeting shRNA (Fig. 6c).
Figure 6.
Phenotypic and functional analysis of PEMs overexpressing or lacking HDAC11. (a,b) Flow cytometry of the expression of B7-2 and CD40 (a) and ELISA of the production of IL-12 and IL-10 (b) by BALB/c PEMs left uninfected (None) or infected for 24 h in vitro with adenovirus encoding HDAC11 or GFP alone. Isotype, isotype-matched control antibody (for anti-B7-2 and anti-CD40). (c) ELISA of IL-12 and IL-10 in supernatants of RAW264.7 clone 18 (lacking HDAC11), RAW264.7 cells transduced with a nontargeting control and wild-type RAW264.7 cells (None) stimulated for 2 h with LPS (2 µg/ml). (d) ELISA of IL-12 in supernatants of PEMs treated as described in b, except in the presence or absence of IL-10-neutralizing antibody (α-IL-10; 10 µg/ml); cells were assessed at 24 h. (e) Real-time RT-PCR analysis of IL-12 mRNA in RAW264.7 clone 18 and RAW264.7 cells transduced with nontargeting shRNA, stimulated for 24 h with LPS (1 µg/ml) in the presence or absence of IL-10-neutralizing antibody (10 µg/ml) and assessed as described in Figure 2e. Data are from one experiment representative of three (a,b) or two (c–e) independent experiments with similar results (error bars, s.d.).
One notable observation from the experiments reported above was the reciprocal changes in the production of IL-10 and IL-12 after overexpression and inhibition of HDAC11 in APCs. Given that IL-10 negatively regulates IL-12 expression, the presence of IL-10 in our experimental system might have masked a potential direct regulatory effect of HDAC11 on Il12 expression. To address this, we assessed IL-12 expression in APCs either overexpressing or lacking HDAC11 when IL-10-neutralizing antibodies were added to the in vitro culture system. First we found that PEMs overexpressing HDAC11 produced more IL-12 than did control PEMs (Fig. 6d).When IL-10-neutralizing antibodies were added, there were no changes in the production of IL-12 in either PEMs overexpressing HDAC11 or control PEMs (Fig. 6d). In RAW264.7 cells lacking HDAC11 (clone 18), we noted more production of IL-10 and less production of IL-12 (Fig. 6c). After the addition of IL-10-neutralizing antibodies, RAW264.7 clone 18 cells had higher IL-12 mRNA expression than did clone 18 cells cultured without antibody to IL10 (anti-IL-10; Fig. 6e). This higher IL-12 expression, however, was not different from the enhancement in IL-12 in control cells treated with IL-10-neutralizing antibodies (Fig. 6e). These results, together with our failure to detect HDAC11 in the Il12 promoter, suggest that HDAC11 did not directly influence Il12 expression in APCs. Instead, it is likely that the changes in IL-12 expression were secondary to the effect of this HDAC on Il10 expression.
HDAC11 influences antigen-specific CD4+ T cell responses
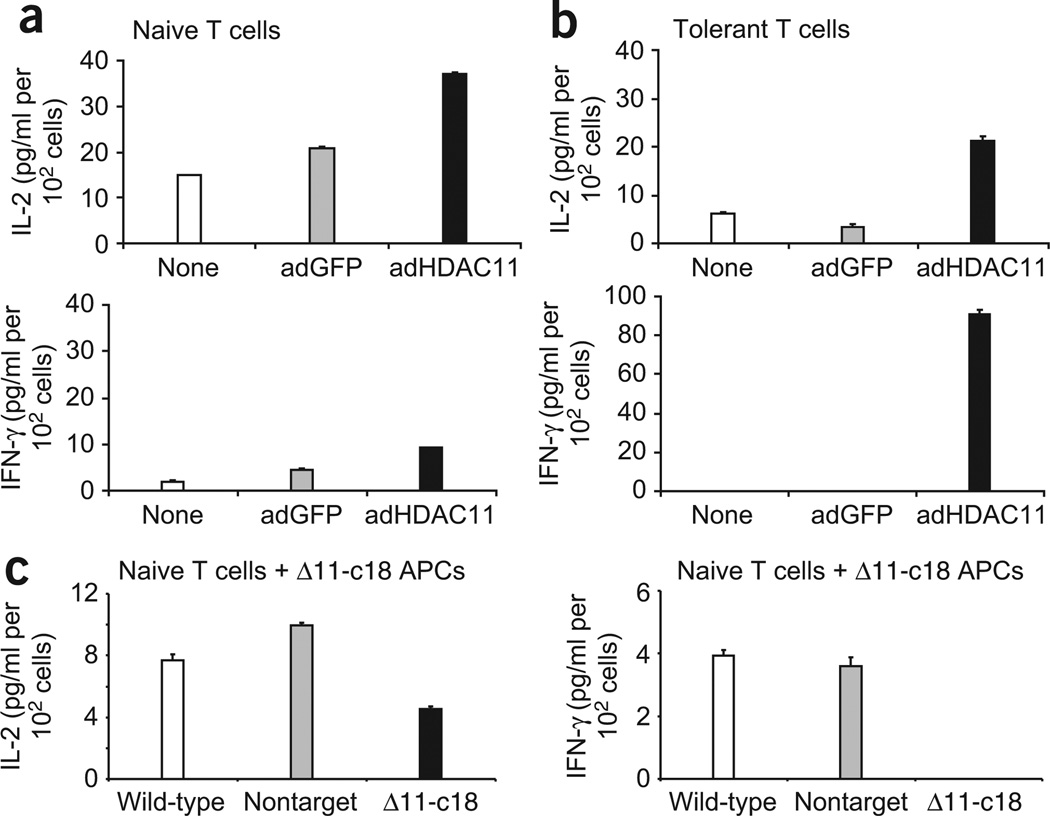
Next we assessed the antigen-presenting ability of APCs overexpressing or lacking HDAC11. First we cultured PEMs overexpressing HDAC11 as well as control cells (uninfected PEMs or PEMS infected with adenovirus expressing GFP) together with naive CD4+ T cells specific for a major histocompatibility complex class II–restricted epitope of influenza hemagglutinin in the presence or absence of cognate hemagglutinin peptide. Clonotypic T cells encountering hemagglutinin peptide on PEMs infected with HDAC11 were better activated, as they produced more IL-2 and interferon-γ (IFN-γ; Fig. 7a) than did clonotypic T cells recognizing antigen on uninfected PEMs or PEMs infected with adenovirus encoding GFP. More notably, PEMs overexpressing HDAC11 were able to restore the responsiveness of tolerant CD4+ T cells. Studies with a T cell receptor–transgenic model have demonstrated that anti-hemagglutinin CD4+ transgenic T cells are rendered tolerant after in vivo exposure to high doses of hemagglutinin peptide29. Indeed, anti-hemagglutinin CD4+ T cells reisolated from these mice had minimal production of IL-2 and lacked IFN-γ production in response to restimulation with hemagglutinin peptide presented by uninfected control PEMs (Fig. 7b). Similarly, T cells remained unresponsive to hemagglutinin peptide presented by PEMs infected with adenovirus encoding GFP (Fig. 7b). In contrast, tolerant T cells encountering antigen on PEMs overexpressing HDAC11 regained their ability to produce IL-2 and IFN-γ (Fig. 7b).
Figure 7.
CD4+ T cell responses to cognate antigen presented by PEMs overexpressing or lacking HDAC11. (a,b) ELISA of IL-2 and IFN-γ in supernatants of BALB/c PEMs left uninfected (None) or infected with adenovirus encoding HDAC11 or GFP, then washed, counted and plated at a density of 1 × 105 cells per well and cultured for 48 h with 5 × 104 purified naive (a) or tolerant (b) antigen-specific CD4+ T cells in the presence of cognate hemagglutinin peptide. Data are from one experiment representative of three independent experiments with similar results (error bars, s.d.). (c) ELISA of IL-2 and IFN-γ in supernatants of RAW264.7 clone 18, RAW264.7 cells transduced with nontargeting control shRNA or wild-type RAW264.7 cells (1 × 105 cells per well) treated with LPS (2 µg/ml) and cultured for 48 h with 5 × 104 purified naive antigen-specific CD4+ T cells in the presence of hemagglutinin peptide. Data are from one experiment representative of two independent experiments with similar results (error bars, s.d.).
Next we assessed the antigen-presenting ability of RAW264.7 cells lacking HDAC11 (clone 18), RAW264.7 cells transduced with nontargeting shRNA and wild-type RAW264.7 cells cultured in vitro with naive anti-hemagglutinin CD4+ T cells in the presence or absence of hemagglutinin peptide. In contrast to naive CD4+ T cells activated by APCs overexpressing HDAC11 (Fig. 7a), clonotypic CD4+ T cells encountering antigen in APCs lacking HDAC11 were functionally impaired, as they produced less IL-2 and lacked IFN-γ production (Fig. 7c) relative to the clonotypic T cells recognizing cognate antigen on wild-type RAW264.7 cells or RAW264.7 cells transduced with nontargeting shRNA. These data collectively indicate that whereas overexpression of HDAC11 in APCs effectively activated naive antigen-specific CD4+ T cells and restored the responsiveness of tolerant T cells, APCs devoid of HDAC11 induced the opposite effect, resulting in impairment of antigen-specific CD4+ T cell responses.
DISCUSSION
The most recently identified member of the family of HDAC proteins, HDAC11, is a 39-kilodalton protein encoded on chromosome 3. Although there is much information about its structure, enzymatic activity and tissue distribution23,30, little is known about the function of this HDAC in normal and/or transformed cells. Here we have unambiguously identified HDAC11 as a negative transcriptional regulator of Il10 expression in mouse and human APCs.
Several lines of evidence indicated that the distal Il10 promoter region is the target for HDAC11. First, we detected HDAC11 only in the distal region of the Il10 promoter. Second, ChIP analysis of the proximal and distal Il10 promoter regions showed changes in HDAC11 mainly in the distal promoter region. Perhaps the most convincing proof of the required interaction between HDAC11 and the distal Il10 promoter region was provided by experiments in which we found strong inhibition of gene expression by HDAC11 after adding only the distal segment of the Il10 promoter to the ‘short region’ of the Il12 promoter. Notably, although we detected HDAC11 mainly in the distal promoter region, most inhibitory effects on histone acetylation and recruitment of transcription factors occurred in the proximal region of the Il10 promoter. One plausible explanation for the repressor function of the distal Il10 promoter region is that perhaps HDAC11 induces changes in the three-dimensional chromatin structure that result in ‘scaffolding’ of the distal region toward the proximal region. This physical interaction might allow putative regulatory factors bound to the distal region to regulate the proximal promoter, including changes in histone acetylation and binding of key transcription factors needed for Il10 activation. In support of that possibility, studies have shown that conformational chromatin changes can regulate the transcription of several genes31.
Dynamic changes in the chromatin structure of the Il10 promoter in T cells differentiated into the TH1 or TH2 phenotype closely regulate IL-10 expression32. Similarly, in macrophages, more acetylation of the Il10 promoter has been associated with enhanced transcription activity17. However, the molecules involved and the sequence of events mediating these chromatin modifications are not fully understood. In our studies, an early event after stimulation of macrophages with LPS was phosphorylation of Ser10 of histone H3 in the proximal Il10 promoter. This was followed by more acetylation of histones H3 and H4 and subsequent recruitment to the proximal promoter of the transcription factors Sp1 and STAT3. That sequence of events ultimately led to transcriptional activation of Il10 that reached its peak by 2 h after LPS treatment. Of note, detection of the transcription repressors PU.1 and HDAC11 in the distal promoter region was a late event that peaked at 2 h, perhaps as a counter-regulatory mechanism to diminish Il10 transcriptional activation. It is plausible, therefore, that this specific order of events and the highly coordinated binding of transcription activators and repressors to the Il10 promoter region might determine not only the initiation but also the intensity and duration of IL-10 production by an APC in response to inflammatory stimuli. Conversely, disruption of this sequence of molecular events would negatively affect IL-10 production by the APC. This seemed to be the scenario in macrophages overexpressing HDAC11. In these cells, with early phosphorylation of histone H3 at Ser10 as the only exception, all of the subsequent events were substantially altered, which resulted in less binding of necessary transcription activators such as STAT3 or Sp1 to the proximal Il10 promoter region. The lack of detection of these transcription factors in the proximal Il10 promoter region could reflect the diminished accessibility of a less acetylated and, thus, more compact chromatin. The enzymatic activity of HDAC11 seemed to be required for the process described above, as overexpression of a HDAC11 mutant with a deleted deacetyltransferase domain failed to inhibit Il10 expression. Instead, there was more IL-10 mRNA expression, which was perhaps a result of the HDAC11 mutant’s acting as a dominant negative variant and therefore competing with endogenous HDAC11. There was similar enhancement in Il10 expression in APCs lacking HDAC11. In these cells, the greater acetylation of histones H3 and H4 that led to less compacted chromatin might have allowed access of the transcription activators Sp1 and STAT3 to the proximal region of the Il10 promoter. Such changes, together with the absence of negative regulation in the distal promoter mediated by HDAC11, provide a plausible explanation for the enhanced Il10 expression in APCs lacking HDAC11. In summary, here we have identified a previously unknown function for HDAC11 as a transcriptional repressor of Il10 expression in APCs. The additional demonstration that genetic manipulation of HDAC11 influenced the inflammatory status of APCs and their ability to determine the functional response of antigen-specific CD4+ T cells indicates that this molecule is a likely target for influencing APC-mediated immune activation versus immune tolerance.
METHODS
Mice
Male BALB/c mice 6–8 weeks of age were from the National Institutes of Health. Transgenic mice expressing an αβ T cell antigen receptor specific for amino acids 110–120 of influenza hemagglutinin presented by I-Ed were from H. von Boehmer33. All animal experiments were in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of South Florida College of Medicine.
Cell lines
The mouse macrophage cell line RAW264.7 has been described34. The human monocytic cell line THP-1 was provided by A. List. Cells were cultured in vitro in RPMI-1640 media supplemented with 10% (vol/vol) FCS, penicillin and streptomycin (50 U/ml), l-glutamine (2 mM) and β-mercaptoethanol (50 mM; complete media) and were grown as a suspension culture at 37 °C in 5% CO2.
Isolation of PEMs and human dendritic cells
BALB/c mice were injected intraperitoneally with 1 ml thioglycollate (Difco Laboratories). Then, 4 d later, PEMs were isolated by peritoneal lavage as described29. Dendritic cells were isolated from buffy coats of the peripheral blood of volunteer blood donors (samples without identification) obtained from the Florida Blood Bank (exempt from the Institutional Review Board). Monocyte-enriched PBMC fractions were isolated from total PBMCs with a plastic adherence technique. Adherent cells were cultured further in RPMI-1640 medium supplemented with 1–5% (vol/vol) autologous plasma, recombinant human IL-4 (1,000 U/ml; R&D Systems) and recombinant human granulocyte-macrophage colony-stimulating factor (1,000 U/ml; Berlex). On days 2 and 4 of incubation, half the medium was replaced with fresh culture medium supplemented with IL-4 and granulocyte-macrophage colony-stimulating factor and the culture was continued. On day 6, half the medium was replaced with culture medium supplemented with IL-4, granulocyte-macrophage colony-stimulating factor, tumor necrosis factor (10 ng/ml), IL-1β (400 IU), IL-6 (1,000 IU; all from R&D Systems) and prostaglandin E2 (1 mg/ml; Sigma-Aldrich). On day 9, cells were collected and were used as monocyte-derived dendritic cells.
Real-time RT-PCR
Cell lines and primary APCs were plated at a density of 2 × 106 cells per 35-mm well and were cultured in the conditions described for each experiment. Total RNA was extracted with TRIzol reagent (Qiagen) and cDNA was obtained with the iScript cDNA synthesis kit (Bio-Rad) with procedures that have been described35. Target mRNA was quantified with the MyiQ single-color real-time PCR detection system and iQ SYBR Green Supermix (Bio-Rad; primers, Supplementary Table 1 online). Single-product amplification was confirmed by melting-curve analysis, and primer efficiency was near 100% in all experiments. Quantification is expressed in arbitrary units, and target mRNA abundance was normalized to the expression of GAPDH (glyceraldehyde phosphate dehydrogenase) with the Pfaffl method36. All real-time RT-PCR experiments were repeated at least three times with similar results.
Antibodies and immunoblot analysis
Total cell lysates were prepared in lysis buffer II (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% (vol/vol) Nonidet P-40 and protease inhibitors). For immunoblot analysis, samples were resolved by SDS-PAGE and were transferred onto nitrocellulose membranes. Membranes were probed with the appropriate antibodies and proteins were visualized with a Chemiluminescent Detection kit (Pierce). Anti-GAPDH (sc-25778), anti–RNA polymerase II (sc-9001) and anti-PU.1 (sc-352) were from Santa Cruz Biotechnology; antibodies to hyperacetylated histones H3 (06–599) and H4 (06–598), as well as anti-Sp1 (07–645) and anti-STAT3 (06–596), were from Millipore. Two different antibodies to HDAC11 were used: ab47036 (Abcam), for immunoblot analysis, and H4539 (Sigma), for ChIP analysis.
Adenovirus and lentivirus infection, transient transfection and luciferase reporter assays
Adenovirus was used as the vector for overexpression of HDAC4–HDAC11 as described37. All HDAC constructs were tagged with Flag and GFP. The lentivirus transduction particles containing shRNA specific for mouse HDAC11 (SHVRS-NM_144919) or nontargeting shRNA (SHC002V) were from Sigma. Plasmid details are in the Supplementary Methods online
Cells were infected in conditions and with titrations to minimize cellular death and to obtain at least 70–90% of the cell population expressing the protein of interest. All cells were transfected by electroporation with a Gene Pulser II according to the manufacturer’s instructions (Bio-Rad). Cells were grown in 100-mm dishes and then were scraped from the plates and washed twice with 1× PBS. Cells (1 × 107) were resuspended in 300 ml media and were mixed with 20 mg plasmid DNA; cell suspensions were subjected to electroporation with 0.2 kV and 1,070 mF. All adenoviruses were purified by the CsCl gradient method. Cells were infected so that over 80% of cells expressed GFP-tagged protein (different multiplicities of infection were used). For transduction of lentivirus particles encoding HDAC11-specific shRNA, the protocol provided by the manufacturer was strictly followed, with a final multiplicity of infection of 75. The shRNA specific for HDAC11 was a combination of five sequences targeting different segments of HDAC11 mRNA. The nontargeting control shRNA was a single random sequence not present in the human or mouse genome. For reporter-gene analysis, all protein concentrations were determined with the Bradford reagent (Bio-Rad), and relative light units were measured in a luminometer with the Luciferase kit (Promega). All assays were done in triplicate and protein expression was evaluated by immunoblot analysis.
ChIP
These studies were done as described38 with some modifications (Supplementary Methods).
Phenotypic and functional analysis of APCs
CD86 expression in PEMs was determined by staining with biotin-conjugated anti-CD86 (GL1; BD Pharmingen) followed by streptavidin-phycoerythrin (Caltag). CD40 expression was determined with monoclonal anti-CD40 (3/23; BD Pharmingen). Gated events (1 × 103) were collected on a FACScan (Becton Dickinson) and were analyzed with FlowJo software. In a parallel plate, PEMs were left unstimulated or were stimulated with LPS and supernatants were collected after 12 h. The production of IL-12 and IL-10 was then measured by ELISA.
Tolerance model
For the in vivo generation of tolerized antigen-specific CD4+ T cells, a well established experimental model of intravenous injection of high-dose peptide-induced tolerance was used (details, Supplementary Methods)29,39. For antigen-presentation studies, PEMs (1 × 105 cells per well) from the various experimental groups were cultured with 5 × 104 purified naive antigen-specific CD4+ T cells (isolated from the spleens of hemagglutinin TCR–transgenic mice) or with a similar number of tolerized antigen-specific CD4+ T cells in the presence or absence of cognate peptide (hemagglutinin peptide of amino acids 110–120: SFERFEIFPKE). After 48 h, supernatants were collected and were stored at −70 °C until ELISA of the production of IL-2 and IFN-γ (R&D Systems). Values for T cells cultured in media alone were usually less than 10% of the values for antigen-stimulated T cells. The amount of cytokine production is expressed as pg/ml per 1 × 102 clonotype-positive CD4+ T cells.
Supplementary Material
ACKNOWLEDGMENTS
Mice transgenic for a T cell antigen receptor specific for hemagglutinin peptide were from H. von Boehmer (Harvard University); the THP-1 cell line was provided by A. List (H. Lee Moffitt Cancer Center). Supported by the US Public Health Service (CA78656 and CA87583 to E.M.S.).
Footnotes
Accession codes. UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org): A002864, A002865, A001243 and A001723.
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
A.V. did ChIP, quantitative real-time RT-PCR, cloning of reporter genes and reporter gene assays, overexpression of HDAC proteins and generation of stable cell lines lacking HDAC11, designed the overall project and prepared part of the manuscript; F.C. isolated B cells and T cells and did ELISAs, tolerance experiments and flow cytometry; H.-W.W. provided technical and experimental support for B cell and T cell isolation, ELISA and tolerance experiments; I.S. provided technical and experimental support for the overexpression of HDAC proteins and quantitative real-time RT-PCR; M.G. cloned mutant HDAC11; M.M. purified adenovirus encoding GFP and HDAC11; D.N. provided technical and experimental assistance for the cloning of reporter genes and reporter gene assays; K.L.W., P.W.A., K.B. and J.P.-I. helped design experiments, provided reagents and discussed the project throughout; and E.M.S. directed the project, designed the overall project, oversaw all experiments, secured funding and was mainly responsible for manuscript writing.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/natureimmunology/.
References
- 1.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Guermonprez P, Valladeau J, Zitvogel L, Thery C, Amigorena S. Antigen presentation and T cell stimulation by dendritic cells. Annu. Rev. Immunol. 2002;20:621–667. doi: 10.1146/annurev.immunol.20.100301.064828. [DOI] [PubMed] [Google Scholar]
- 3.Kurts C, Kosaka H, Carbone FR, Miller JF, Heath WR. Class I-restricted cross-presentation of exogenous self-antigens leads to deletion of autoreactive CD8+ T cells. J. Exp. Med. 1997;186:239–245. doi: 10.1084/jem.186.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adler AJ, et al. CD4+ T cell tolerance to parenchymal self-antigens requires presentation by bone marrow-derived antigen-presenting cells. J. Exp. Med. 1998;187:1555–1564. doi: 10.1084/jem.187.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinman RM, Hawiger D, Nussenzweig MC. Tolerogenic dendritic cells. Annu. Rev. Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- 6.Huang FP, et al. A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J. Exp. Med. 2000;191:435–444. doi: 10.1084/jem.191.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munn DH, et al. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science. 2002;297:1867–1870. doi: 10.1126/science.1073514. [DOI] [PubMed] [Google Scholar]
- 8.Scheinecker C, McHugh R, Shevach EM, Germain RN. Constitutive presentation of a natural tissue autoantigen exclusively by dendritic cells in the draining lymph node. J. Exp. Med. 2002;196:1079–1090. doi: 10.1084/jem.20020991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belz GT, et al. The CD8α+ dendritic cell is responsible for inducing peripheral self-tolerance to tissue-associated antigens. J. Exp. Med. 2002;196:1099–1104. doi: 10.1084/jem.20020861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rabinovich GA, Gabrilovich D, Sotomayor EM. Immunosuppressive strategies that are mediated by tumor cells. Annu. Rev. Immunol. 2007;25:267–296. doi: 10.1146/annurev.immunol.25.022106.141609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Napolitani G, Rinaldi A, Bertoni F, Sallusto F, Lanzavecchia A. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 2005;6:769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 13.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 14.Li MO, Flavell RA. Contextual regulation of inflammation: a duet by transforming growth factor-β and interleukin-10. Immunity. 2008;28:468–476. doi: 10.1016/j.immuni.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 16.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Edwards JP, Mosser DM. Dynamic and transient remodeling of the macrophage IL-10 promoter during transcription. J. Immunol. 2006;177:1282–1288. doi: 10.4049/jimmunol.177.2.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Y, Li W, Kaplan MH, Chang CH. Interleukin (IL)-4 inhibits IL-10 to promote IL-12 production by dendritic cells. J. Exp. Med. 2005;201:1899–1903. doi: 10.1084/jem.20050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akbari O, DeKruyff RH, Umetsu DT. Pulmonary dendritic cells producing IL-10 mediate tolerance induced by respiratory exposure to antigen. Nat. Immunol. 2001;2:725–731. doi: 10.1038/90667. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, et al. Activation of the growth-differentiation factor 11 gene by the histone deacetylase (HDAC) inhibitor trichostatin A and repression by HDAC3. Mol. Cell. Biol. 2004;24:5106–5118. doi: 10.1128/MCB.24.12.5106-5118.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol. Cell. Biol. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rezai-Zadeh N, et al. Histone deacetylases: purification of the enzymes, substrates, and assay conditions. Methods Enzymol. 2004;377:167–179. doi: 10.1016/S0076-6879(03)77009-8. [DOI] [PubMed] [Google Scholar]
- 23.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 24.Lucas M, Zhang X, Prasanna V, Mosser DM. ERK activation following macrophage FcγR ligation leads to chromatin modifications at the IL-10 locus. J. Immunol. 2005;175:469–477. doi: 10.4049/jimmunol.175.1.469. [DOI] [PubMed] [Google Scholar]
- 25.Benkhart EM, Siedlar M, Wedel A, Werner T, Ziegler-Heitbrock HW. Role of Stat3 in lipopolysaccharide-induced IL-10 gene expression. J. Immunol. 2000;165:1612–1617. doi: 10.4049/jimmunol.165.3.1612. [DOI] [PubMed] [Google Scholar]
- 26.Tone M, Powell MJ, Tone Y, Thompson SAJ, Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J. Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 27.Reuss E, et al. Differential regulation of interleukin-10 production by genetic and environmental factors - a twin study. Genes Immun. 2002;3:407–413. doi: 10.1038/sj.gene.6363920. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Oikawa T. Direct association between PU.1 and MeCP2 that recruits mSin3A-HDAC complex for PU.1-mediated transcriptional repression. Oncogene. 2003;22:8688–8698. doi: 10.1038/sj.onc.1207182. [DOI] [PubMed] [Google Scholar]
- 29.Cheng F, et al. A critical role for Stat3 signaling in immune tolerance. Immunity. 2003;19:425–436. doi: 10.1016/s1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- 30.Bradbury CA, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19:1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 31.West AG, Fraser P. Remote control of gene transcription. Hum. Mol. Genet. 2005;14:R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 32.Im S-H, Hueber A, Monticelli S, Kang K-H, Rao A. Chromatin-level regulation of the IL10 gene in T cells. J. Biol. Chem. 2004;279:46818–46825. doi: 10.1074/jbc.M401722200. [DOI] [PubMed] [Google Scholar]
- 33.Kirberg J, et al. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex-restricted receptor. J. Exp. Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicente-Suarez I, et al. Identification of a novel negative role of flagellin in regulating IL-10 production. Eur. J. Immunol. 2007;37:3164–3175. doi: 10.1002/eji.200737306. [DOI] [PubMed] [Google Scholar]
- 35.Villagra A, et al. Histone deacetylase 3 down-regulates cholesterol synthesis through repression of lanosterol synthase gene expression. J. Biol. Chem. 2007;282:35457–35470. doi: 10.1074/jbc.M701719200. [DOI] [PubMed] [Google Scholar]
- 36.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X-J, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villagra A, et al. Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein β-dependent recruitment of SWI/SNF activity. J. Biol. Chem. 2006;281:22695–22706. doi: 10.1074/jbc.M511640200. [DOI] [PubMed] [Google Scholar]
- 39.Kearney ER, Pape KA, Loh DY, Jenkins MK. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity. 1994;1:327–339. doi: 10.1016/1074-7613(94)90084-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.