Abstract
22q11.2 deletion syndrome (22q11DS) results from a hemizygous microdeletion on chromosome 22 and is characterized by extensive phenotypic variability. Penetrance of signs, including congenital heart, craniofacial, and neurobehavioral abnormalities, varies widely and is not well correlated with genotype. The three-dimensional structure of the genome may help explain some of this variability. The physical interaction profile of a given gene locus with other genetic elements, such as enhancers and co-regulated genes, contributes to its regulation. Thus, it is possible that regulatory interactions with elements outside the deletion region are disrupted in the disease state and modulate the resulting spectrum of symptoms. COMT, a gene within the commonly deleted ~3 Mb region has been implicated as a contributor to the neurological features frequently found in 22q11DS patients. We used this locus as bait in a 4C-seq experiment to investigate genome-wide interaction profiles in B lymphocyte and fibroblast cell lines derived from both 22q11DS and unaffected individuals. All normal B lymphocyte lines displayed local, conserved chromatin looping interactions with regions that are lost in atypical and distal deletions, which may mediate similarities between typical, atypical, and distal 22q11 deletion phenotypes. There are also distinct clusterings of cis interactions based on disease state. We identified regions of differential trans interactions present in normal, and lost in deletion-carrying, B lymphocyte cell lines. This data suggests that hemizygous chromosomal deletions such as 22q11DS can have widespread effects on chromatin organization, and may contribute to the inherent phenotypic variability.
Keywords: DiGeorge syndrome, long-range interactions, chromosome conformation capture, genome organization, schizophrenia
Introduction
Chromosomal disorders include deletions, duplications and inversions, and may be mediated by low-copy repeats (LCRs).1 Nonallelic homologous recombination involving LCRs on chromosome 22 results in hemizygous deletions of approximately 3 Mb, leading to the 22q11.2 deletion syndrome (22q11DS).2 22q11DS is estimated to occur in 1 in 4000 live births, making it the most common microdeletion syndrome.3,4 Phenotypic variation is frequently observed in chromosomal deletion disorders,5,6 and this is especially true for 22q11DS, even though approximately 90% of individuals with 22q11DS share the typical 3 Mb deletion.7-9 The remaining 10% of deletions are composed of a nested 1.5 Mb deletion and of variable sized atypical deletions. Deletions occurring downstream of the 3 Mb deletion are considered part of a separate syndrome.10 There are over 180 distinct clinical features with variable expressivity associated with 22q11DS.11 The most common features include DiGeorge syndrome, congenital heart disease and craniofacial abnormalities as part of velocardiofacial syndrome and conotruncal anomaly face syndrome, and neurobehavioral phenotypes including learning difficulties. This deletion is also the leading known genetic cause of schizophrenia.12
Attempts to correlate phenotype with the 22q11DS patient genotype have largely failed. Patients with identical deletions, including monozygotic twins, have presented with discordant phenotypes.13-15 Moreover, patients with non-overlapping deletions may share many of the same features.16,17 Varying penetrance and the large number of clinical features have led to consideration of the dosage effect of candidate genes located in the commonly deleted region, of haplotype and single nucleotide polymorphisms (SNP) of the 22q11.2 locus, and of the contribution of genetic modifiers from elsewhere in the genome. The T-box transcription factor (Tbx1), located in the deleted region, is implicated in craniofacial and congenital heart malformations. Manipulating Tbx1 levels in mice resulted in a nonlinear phenotypic response,18 suggesting that moderate perturbations in the expression of developmental genes on the remaining 22q11.2 allele may contribute to the observed non-uniform symptoms. Thus far, SNP analyses of TBX1 in humans have failed to identify positive associations with the 22q11DS cardiac or cleft palate phenotypes.19,20 Catechol-O-methyltransferase (COMT), also deleted in 22q11DS, functions in degrading catecholamines and thus is a candidate for many neurological 22q11DS phenotypes. A valine-to-methionine substitution at codon 158 (COMT Val158Met) results in a form of COMT with decreased enzymatic activity; the presence of this allele in the non-deleted chromosome may further enhance the effects of COMT haploinsufficiency caused by the deletion.
Epigenetic mechanisms that result in altered expression may also contribute to the observed phenotypic variability. In agreement with a position effect, phenotypes commonly attributed to TBX1 and COMT haploinsufficiency have been reported in shorter atypical and distal deletions not encompassing either gene.17,21 It is becoming clear that nuclear architecture, including global structure and local chromosome conformation, plays an important role in gene regulation.22 For example, point mutations in the Shh enhancer located 1 Mb away from Shh interfere with its developmental regulation, resulting in preaxial polydactyly.23 The pluripotency factor Nanog was recently found to participate in pluripotent and differentiation-specific long-range interactions in both cis and trans, many of which preceded changes in expression.24
One possibility that may account for phenotypic variability at a locus is the status of its physical interaction profile with other genetic elements. As a first step to assess the role of long-range DNA interactions in the clinical manifestations of the 22q11.2 deletion syndrome, we used 4C-seq to investigate the local cis and genome-wide interaction profiles of COMT in four normal B lymphocyte cell lines and four B lymphocyte lines with the typical large 22q11.2 deletion. We also compare these profiles to a normal and 22q11.2 deletion-containing fibroblast cell line. We hypothesized that some COMT local or long-range physical interactions, which may play crucial roles in normal development, might occur much less frequently with the haploinsufficency seen in the deletion syndrome.
Results
COMT expression profiles in normal and 22q11DS cell lines
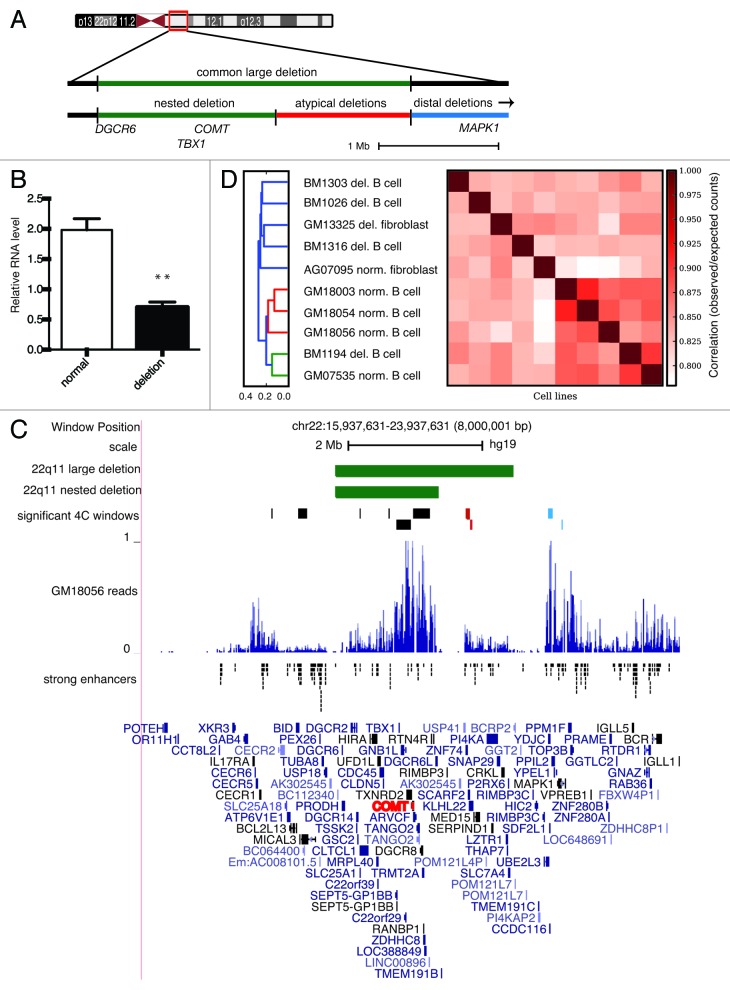
Haploinsufficiency of COMT is believed to contribute to neurobehavioral phenotypes in 22q11DS.25 COMT is located approximately at the center of the common, large deletion on chr22 and is also encompassed by the smaller nested deletion (Fig. 1A). To verify haploinsufficiency of COMT expression in our 22q11DS cell lines, we compared COMT mRNA levels in 4 normal and 4 large 3 Mb deletion-carrying B lymphocyte cell lines. As expected, COMT mRNA expression was significantly lower in the deletion-carrying cell lines (t test, P < 0.01) (Fig. 1B).
Figure 1. (A) Diagram of deletions on chromosome 22q11.2. The common large deletion (green) encompasses the shorter nested deletion (green) and the region of atypical deletions (red). The region containing distal deletions is highlighted green. Locations of DGCR6, COMT, TBX1 and MAPK1 are shown. (B) COMT expression in 4 normal and 4 deletion B lymphocyte cell lines. **, P < 0.01. (C) 4C-seq local interaction profiles observed in 4 normal B lymphocyte cell lines. The bait locus COMT (red) is located in both the large typically deleted region and the shorter nested deletion (green bars). Normalized cis reads in the normal B cell line GM18056. Image generated from UCSC genome browser GRCh37/hg19. Significant regions displayed underneath deletions were obtained using a sliding window of 20 restriction sites and a z-score cutoff of 3. Red bars indicate interactions in the region comprising atypical deletions and blue bars indicate interactions in the distal deletion region. Locations of the predicted B lymphocyte, strong enhancers were extracted from the Broad chromatin state segmentation track.26 (D) Hierarchical clustering by cis interactions profiles. Each square represents a pairwise comparison of the similarity between each cell line using Pearson correlation between the vectors of intrachromosomal z scores assigned using a sliding window of 20 restriction sites.
COMT interaction profiles in normal and 22q11DS cell lines
Next, we performed 4C-seq using the COMT promoter as bait to capture genome-wide interactions of COMT in normal and deletion-carrying cell lines. Examination of local interactions occurring within ± 4 Mb from COMT and conserved in all of our normal B lymphocyte cell lines reveals numerous significant peaks of interaction (Fig. 1C, Z-score > 3). Several of these peaks occur in regions that are lost in patients with atypical deletions located downstream of the nested deletion (red color) and distal deletions located downstream of the typical large deletion (blue color). This leaves open the possibility that regulatory interactions between COMT and regions outside the common or nested deletion contribute to the disease state when they are lost. Local cis interactions overlap with putative strong enhancers,26 and there is a notable absence of interactions flanking the typical large and nested deletions due to the presence of LCRs (Fig. 1C).
To assess whether interactions with the COMT bait could distinguish between disease states, we performed hierarchical clustering on cis interaction profiles of all ten cell lines. This analysis indicates a large degree of similarity among normal B lymphocyte cell lines (Fig. 1D). The majority of deletion-carrying B lymphocytes and both fibroblast cell lines are in a separate cluster with less similarity to the normal B lymphocytes and low similarity to one another. The exception was the deletion-carrying cell line BM1194, which was more closely related to the normal B lymphocyte cell lines. Interestingly, BM1194 was also grouped with the normal B lymphocyte cell lines when clustering by similarity of trans interaction profiles (Fig. S2).
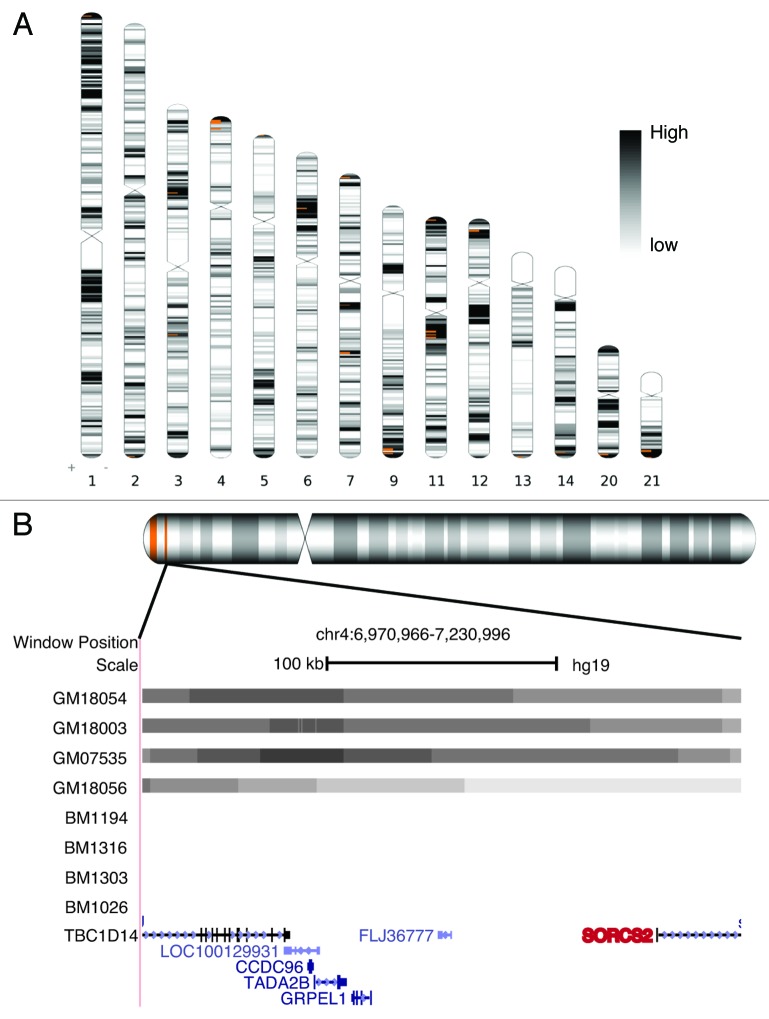
We next investigated differentially interacting regions in between normal and 22q11DS cell lines. We defined differential interactions as those interactions that are present in normal and absent in 22q11DS B lymphocyte cell lines. As cis interactions were generally conserved among both normal and deletion cell lines, we focused our analysis on interactions in trans. Figure 2A highlights COMT interacting regions that were significantly enriched in all 4 of the normal B cell lines and in either 1 or none of the deletion-carrying B cell lines. These differential interactions occurred across 14 separate chromosomes and were all located in regions of high gene density. One highlighted region of note is the 3p21.1 locus; a recent genome-wide association study meta-analysis found the strongest association shared among five psychiatric disorders occurs at this 3p21.1 locus.27
Figure 2. Differential trans interactions. (A) Ideograms of chromosomes with regions interacting with COMT in all 4 normal B lymphocyte cell lines and ≤ 1 22q11DS B lymphocyte cell lines are depicted by orange bars. Heat map represents known gene density. Region defined as 20 consecutive overlapping windows of 100 restriction sites, sliding by 1 site. (B) Magnified view of 1 selected differentially interacting region on chromosome 4. Significant COMT interacting windows (gray bars) are present in all normal and none of the 22q11DS B lymphocyte cell lines. The transcription start site of SORCS2 a bipolar disorder, and schizophrenia associated gene (red) is highlighted. Image generated from UCSC genome browser GRCh37/hg19 and Ideographica software.
To further narrow down potential functional regions, we focused on differential interactions present in all 4 normal and in none of the 22q11DS B lymphocyte cell lines. There were a total of 245 interacting genes in 18 regions with an average size of 500 kb. We chose genes with a transcription start site located within these regions and further selected those positively associated with a characteristic common to 22q11DS in the genetic association database (GAD) (Table 1). The majority of 22q11DS GAD phenotypes associated with these genes are neurobehavioral in nature, and many were directly related to schizophrenia or were in linkage disequilibrium with a schizophrenia-associated locus. One example, Sortilin-Related VPS10 Domain Containing Receptor 2 (SORCS2) on chr4, is located in a significant differentially interacting region identified using the most strict window size thresholds (Table 1; Fig. 2B). SORCS2 is highly expressed in the central nervous system, is associated with bipolar disorder and is in linkage disequilibrium with schizophrenia, making it an interesting candidate for influencing the 22q11DS phenotype.
Table 1. Disease relevant gene interactions present in all normal cells lines and no deletion cell lines.
| 10 | 20 | 50 | gene | chr | Relevant GAD record |
|---|---|---|---|---|---|
| * | * | THAP4 | 2 | brain structure | |
| * | * | BOK | 2 | brain structure | |
| * | * | AMT | 3 | LD with schizophrenia | |
| * | * | BSN | 3 | LD with schizophrenia, RDC schizoaffective disorder, bipolar | |
| * | * | DAG1 | 3 | LD with schizophrenia | |
| * | * | * | SORCS2 | 4 | bipolar, LD with schizophrenia |
| * | * | ELN | 7 | congenital heart malformation | |
| * | * | LIMK1 | 7 | neurite outgrowth | |
| * | * | NRXN2 | 11 | Autism, schizophrenia |
*, 0 out of 10, 20 or 50 consecutive, overlapping 100 restriction site windows are significant in deletion cell lines; LD, linkage disequilibrium
Discussion
Chromatin organization is a key regulator of gene expression and we hypothesized it could contribute to the unexplained phenotypic variability associated with 22q11DS. Dysregulation of developmentally important genes can result in varying phenotypes.18 The effect of COMT haploinsufficiency may be exacerbated by differential regulatory interactions either diminished or lost in 22q11DS or affecting the intact 22q11.2 allele. We identify numerous interactions conserved in normal B lymphocytes that are lost in deletion-carrying cell lines, and describe genes in these regions implicated in phenotypes consistent with 22q11DS. It is not clear why a deletion from only one chromosome would lead to complete loss of certain long-range DNA interactions. It is possible that there are inherent 22q11 allele-specific interactions. Parent of origin specific interactions are seen with several imprinted genes,28,29 and DiGeorge critical region 6 (DGCR6), a gene within the deleted region (Fig. 1A), has been reported to be more variably expressed among 22q11DS subjects, especially on the maternally inherited allele.30 Interestingly, a significant majority of 22q11 deletions are of maternal origin.31 Alternatively, some interactions that occur when both alleles are present may occur relatively infrequently, and thus the loss of one allele may reduce the interaction frequency below the level of detection.
Additionally, we established the presence of looping interactions conserved in all normal B lymphocyte cell lines assayed that reside in regions disrupted by atypical and distal deletions. Phenotypes associated with these alternate deletions share many characteristics of 22q11DS. For instance, several cases of psychiatric disorders and congenital heart disease, including tetralogy of Fallot, are observed in patients with atypical deletions not encompassing COMT or TBX1.17,32 Features overlapping with 22q11DS have also been observed with 22q11 distal deletions.10 One gene implicated in the craniofacial, neurodevelopmental and congenital heart malformations associated with 22q11 distal deletions is mitogen-Activated Protein Kinase 1 (MAPK1).33 Interestingly, a significant interaction involving COMT and MAPK1 was conserved in all normal B lymphocyte cell lines (Fig. 1A and C). Cluster analysis grouped all but one of the B lymphocyte cell lines by disease state. Because a hallmark of 22q11 deletion syndrome is phenotypic variability, this result is not completely unexpected. In this case, only by incorporating more specific neurological phenotype data from longitudinal studies in 22q11DS patients can this be further elucidated.
Chromatin organization is characterized by both ubiquitous and cell type specific interactions. We began characterizing chromatin interaction profiles in patient-derived B lymphocyte cell lines, which are the predominant cell type available. Our initial results identify the loss of long-range chromatin interactions of the 22q11.2 locus and COMT gene in 22q11DS cell lines, and local interactions that may have a regulatory function in normal B lymphocytes. Using 5C, Dekker’s group found large numbers of statistically significant promoter looping interactions, of which ~60% were observed in only one of three cell lines (K562, HeLa-S3, or GM12878).34 This illustrates that although the majority of interactions are cell type specific, a proportion of interactions are shared among distinct cell types. Blood cells have been used to model neuronal function in the past35; however, to fully characterize the molecular mechanisms that may be involved in the cognitive or psychiatric components of the 22q11DS, it is critical to study a cell type that is most relevant to the phenotype. To address this issue we have begun generating iPSCs from 22q11DS patient cells (Manuscript under preparation). Future experiments will be directed toward elucidating the role of chromatin organization in 22q11DS in neuronal cell types differentiated from these patient derived iPSCs.
Methods
Cell culture
B-Lymphocytes GM18054, GM18003, GM07535, and GM18056 (Coriell), and 22q11.2 deletion harboring B-Lymphocytes BM1194, BM1303, BM1316, BM1026, a kind gift from Bernice Morrow, (Albert Einstein College of Medicine) were cultured in RPMI 1640 GlutaMAX media supplemented with 15% fetal bovine serum, 1% penicillin-streptomycin, 1% NEAA (Gibco) at 37 °C in 5% CO2. GM13325 and AG07095 fibroblast cell lines were purchased from Coriell and cultured in Dulbecco's modified Minimal Essential Medium + GlutaMAX (DMEM), 10% fetal bovine serum, 1% penicillin-streptomycin, 1% NEAA.
Circular chromosome conformation capture (4C) sequencing assay
4C was performed as in Zeitz et al.36 with minor modifications. See supplement for detailed methodology. 4C libraries were analyzed on a MultiNA microchip electrophoresis system (Shimadzu) and mixed in equimolar amounts. Multiplex sequencing was performed on 2 lanes (5 samples per lane) of an Illumina genome analyzer IIx (Illumina).
RNA extraction and quantitative RT-PCR
RNA was extracted from 3 × 106 cells using the RNeasy Mini Kit and QIAshredder mini column (Qiagen) according to the manufacturer’s instructions. DNA was digested on a column using RNase free DNase set (Qiagen). 1μg of RNA was reverse transcribed with Superscript III first-strand synthesis supermix for qRT-PCR (Life Technologies). qRT-PCR was performed using KAPA SYBR Fast ABI PRISM qPCR mix (KAPA) on an ABI 7900HT Real-Time PCR System (Applied Biosystems). Primers for target genes and reference genes (TBP, HPRT) were purchased from RealTimePrimers.com. Reaction efficiency for each primer set was calculated using Real-time PCR Miner.37
Mapping and filtering of 4C reads
We first de-multiplexed the 76 bp single-end reads using barcodes for each cell line. We retained only those reads that contained one of the valid barcodes followed by the primer sequence and a HindIII cleavage site with at most one mismatch and truncated them to obtain the prey sequence. We mapped the truncated reads to a database of all sequences flanking HindIII sites in the human genome (UCSC hg19) using Bowtie0.12.8 with the settings `-v 2 –best –k 5`, and post-processed the alignment results to extract only the reads that satisfied the following criteria: (1) mapped uniquely to one location in the reference genome (with at most 2 mismatches), and (2) mapped to a location within 2kb of a flanking MspI site. We then identified the restriction fragments interacting (those flanking the cleavage sites with a read count of at least one) with the bait region for each of the barcoded samples.
Statistical analysis of 4C data
We first identified all the HindIII sites in the genome and eliminated the ones with no MspI site within 2kb downstream of the HindIII site, resulting in ~850k restriction fragments for downstream analysis (considering both forward and reverse fragments for HindIII site). In order to avoid PCR artifacts, we binarized the interaction counts as was done previously in other 4C analysis pipelines.38 In order to account for the difference in the number of interacting fragments between cell types and the effect of genomic distance on the intrachromosomal interaction probability, we applied a statistical significance assignment procedure similar to the one described in Splinter et al.38 Due to the limited resolution of current 4C methods, we combined multiple consecutive restriction fragments to enable us to assign statistical confidences for interactions at varying resolutions. We first counted the number of interacting fragments within a window, and then generated a background distribution by randomly shuffling the interacting and non-interacting fragments for each group and repeated this randomization 100 times. For intrachromosomal interactions, we took into account the linear distance of each region to the bait when generating the background as previously described.38 For both intrachromosomal and interchromosomal interactions, we calculated the z-score threshold at which the false discovery rate (FDR) is 0.01 to identify the windows that significantly interact with the 4C bait (4C-enriched windows/regions) as previously described.38 For determining differential trans interactions we used z-scores from a window size of 100 restriction sites. We used the z-scores gathered by a window size of 20 for determining significant local interactions occurring within ± 4 Mb from the bait.
Hierarchical clustering of cell lines according to 4C profiles
We performed hierarchical clustering using the average/UPGMA linkage method of Scipy library in Python. For cis interaction profiles we used the z-scores gathered as previously described38 by a window size of 20 consecutive restriction sites. For each pair of cell lines we measured cis interaction profile similarity as the correlation between the two z-score vectors. For trans interactions we simply used the binarized interaction counts from the bait to all interchromosomal restriction fragments.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We would like to thank Bernice Morrow for providing us with cell lines. This study was supported by an NIH R01 grant (GM090314), the Research Service of the Department of Veterans Affairs, and a Computing Research Association CIFellows award (NSF award CIF 1136996 to Ay F).
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/nucleus/article/27364
Glossary
Abbreviations:
- LCR
low-copy repeats
- 22q11DS
22q11.2 deletion syndrome
- TBX1
T-box transcription factor
- COMT
Catechol-O-methyltransferase
- SNP
single nucleotide polymorphism
- GAD
genetic association database
- SORCS2
sortilin-related VPS10 domain containing receptor 2
- DGCR6
DiGeorge critical region 6
- MAPK1
mitogen-activated protein kinase 1
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/27364
References
- 1.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/S0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 2.Edelmann L, Pandita RK, Morrow BE. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am J Hum Genet. 1999;64:1076–86. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scambler PJ. The 22q11 deletion syndromes. Hum Mol Genet. 2000;9:2421–6. doi: 10.1093/hmg/9.16.2421. [DOI] [PubMed] [Google Scholar]
- 4.Shprintzen RJ. Velo-cardio-facial syndrome: 30 Years of study. Dev Disabil Res Rev. 2008;14:3–10. doi: 10.1002/ddrr.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–99. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–67. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- 8.Shaikh TH, Kurahashi H, Saitta SC, O’Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 9.Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, Wadey R, Patanjali SR, Weissman SM, Anyane-Yeboa K, Warburton D, et al. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet. 1997;61:620–9. doi: 10.1086/515508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fagerberg CR, Graakjaer J, Heinl UD, Ousager LB, Dreyer I, Kirchhoff M, Rasmussen AA, Lautrup CK, Birkebaek N, Sorensen K. Heart defects and other features of the 22q11 distal deletion syndrome. Eur J Med Genet. 2013;56:98–107. doi: 10.1016/j.ejmg.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Robin NH, Shprintzen RJ. Defining the clinical spectrum of deletion 22q11.2. J Pediatr. 2005;147:90–6. doi: 10.1016/j.jpeds.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–5. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 13.Yamagishi H, Ishii C, Maeda J, Kojima Y, Matsuoka R, Kimura M, Takao A, Momma K, Matsuo N. Phenotypic discordance in monozygotic twins with 22q11.2 deletion. Am J Med Genet. 1998;78:319–21. doi: 10.1002/(SICI)1096-8628(19980724)78:4<319::AID-AJMG3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 14.Goodship J, Cross I, Scambler P, Burn J. Monozygotic twins with chromosome 22q11 deletion and discordant phenotype. J Med Genet. 1995;32:746–8. doi: 10.1136/jmg.32.9.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh SM, Murphy B, O’Reilly R. Monozygotic twins with chromosome 22q11 deletion and discordant phenotypes: updates with an epigenetic hypothesis. J Med Genet. 2002;39:e71. doi: 10.1136/jmg.39.11.e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amati F, Conti E, Novelli A, Bengala M, Diglio MC, Marino B, Giannotti A, Gabrielli O, Novelli G, Dallapiccola B. Atypical deletions suggest five 22q11.2 critical regions related to the DiGeorge/velo-cardio-facial syndrome. Eur J Hum Genet. 1999;7:903–9. doi: 10.1038/sj.ejhg.5200399. [DOI] [PubMed] [Google Scholar]
- 17.Verhagen JM, Diderich KE, Oudesluijs G, Mancini GM, Eggink AJ, Verkleij-Hagoort AC, Groenenberg IA, Willems PJ, du Plessis FA, de Man SA, et al. Phenotypic variability of atypical 22q11.2 deletions not including TBX1. Am J Med Genet A. 2012;158A:2412–20. doi: 10.1002/ajmg.a.35517. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Baldini A. In vivo response to high-resolution variation of Tbx1 mRNA dosage. Hum Mol Genet. 2008;17:150–7. doi: 10.1093/hmg/ddm291. [DOI] [PubMed] [Google Scholar]
- 19.Guo T, McDonald-McGinn D, Blonska A, Shanske A, Bassett AS, Chow E, Bowser M, Sheridan M, Beemer F, Devriendt K, et al. International Chromosome 22q11.2 Consortium Genotype and cardiovascular phenotype correlations with TBX1 in 1,022 velo-cardio-facial/DiGeorge/22q11.2 deletion syndrome patients. Hum Mutat. 2011;32:1278–89. doi: 10.1002/humu.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman SB, Guo T, McGinn DM, Blonska A, Shanske AL, Bassett AS, Chow EW, Bowser M, Sheridan M, Beemer F, et al. International Chromosome 22q11.2 Consortium Overt cleft palate phenotype and TBX1 genotype correlations in velo-cardio-facial/DiGeorge/22q11.2 deletion syndrome patients. Am J Med Genet A. 2012;158A:2781–7. doi: 10.1002/ajmg.a.35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, Hummel M, Amato S, Tartaglia N, Berg J, Sutton VR, et al. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008;82:214–21. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–35. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 24.Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boot E, Booij J, Zinkstok J, Abeling N, de Haan L, Baas F, Linszen D, van Amelsvoort T. Disrupted dopaminergic neurotransmission in 22q11 deletion syndrome. Neuropsychopharmacology. 2008;33:1252–8. doi: 10.1038/sj.npp.1301508. [DOI] [PubMed] [Google Scholar]
- 26.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smoller JW, Craddock N, Kendler K, Lee PH, Neale BM, et al. Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic Risk Outcome of Psychosis (GROUP) Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–72. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 29.Lonfat N, Montavon T, Jebb D, Tschopp P, Nguyen Huynh TH, Zakany J, Duboule D. Transgene- and locus-dependent imprinting reveals allele-specific chromosome conformations. Proc Natl Acad Sci U S A. 2013;110:11946–51. doi: 10.1073/pnas.1310704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das Chakraborty R, Bernal AJ, Schoch K, Howard TD, Ip EH, Hooper SR, Keshavan MS, Jirtle RL, Shashi V. Dysregulation of DGCR6 and DGCR6L: psychopathological outcomes in chromosome 22q11.2 deletion syndrome. Transl Psychiatry. 2012;2:e105. doi: 10.1038/tp.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delio M, Guo T, McDonald-McGinn DM, Zackai E, Herman S, Kaminetzky M, Higgins AM, Coleman K, Chow C, Jalbrzikowski M, et al. Enhanced maternal origin of the 22q11.2 deletion in velocardiofacial and DiGeorge syndromes. Am J Hum Genet. 2013;92:439–47. doi: 10.1016/j.ajhg.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Miñaur S, Fantes J, Murray RS, Porteous ME, Strain L, Burns JE, Stephen J, Warner JP. A novel atypical 22q11.2 distal deletion in father and son. J Med Genet. 2002;39:E62. doi: 10.1136/jmg.39.10.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuels IS, Karlo JC, Faruzzi AN, Pickering K, Herrup K, Sweatt JD, Saitta SC, Landreth GE. Deletion of ERK2 mitogen-activated protein kinase identifies its key roles in cortical neurogenesis and cognitive function. J Neurosci. 2008;28:6983–95. doi: 10.1523/JNEUROSCI.0679-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rausch JL, Johnson ME, Li J, Hutcheson J, Carr BM, Corley KM, Gowans AB, Smith J. Serotonin transport kinetics correlated between human platelets and brain synaptosomes. Psychopharmacology (Berl) 2005;180:391–8. doi: 10.1007/s00213-005-2178-6. [DOI] [PubMed] [Google Scholar]
- 36.Zeitz MJA, Ay F, Heidmann JD, Lerner PL, Noble WS, Steelman BN, Hoffman AR. Genomic interaction profiles in breast cancer reveal altered chromatin architecture. PLoS One. 2013;8:e73974. doi: 10.1371/journal.pone.0073974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–64. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Splinter E, de Wit E, van de Werken HJ, Klous P, de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 2012;58:221–30. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.