Abstract
Escherichia coli ST131 is an important cause of multidrug-resistant infections. Thus, the aim of this study was to evaluate the concomitant presence of resistance plasmids and pathogenicity islands (PAIs) in ST131 E. coli. From 97 extraintestinal E. coli characterized for antimicrobial susceptibility and extended-spectrum β-lactamase production, 16% of isolates were identified as CTX-M-15 producers. These strains were studied by PFGE, MLST, and phylogroups, plasmid groups, PAIs, and plasmid-mediated quinolone-resistance determinants. MLST identified one ST10 strain from phylogroup A and the remaining isolates were ST131, from group B2. Despite the genetic variability, 64% of ST131 strains presented a profile composed by PAI IV536, PAI ICFT073, and PAI IICFT073, IncF plasmid, blaCTX-M-15, and aac(6’)-lb-cr genes. The prevalent virulence and resistance profile detected among the strains may constitute an optimal combination of factors, which allow E. coli ST131 to maintain both features becoming concomitantly virulent and extremely resistant.
Keywords: Escherichia coli ST131, pathogenicity island, plasmid replicon typing, CTX-M-15, PMQR, phylogeny
Introduction
The high levels of resistance observed in Escherichia coli, a pathogen responsible for several infections in the human host, to several important antibiotics groups, such as cephalosporins and quinolones, have become a main concern. One of the most successful E. coli clone, ST131, has emerged as very important cause of multidrug-resistant infections worldwide.1
The E. coli ST131 clone has been usually associated with resistance to β-lactams, mostly due to the production of extended-spectrum β-lactamase (ESBL) CTX-M-15, which confers resistance to penicillins, cephalosporins, and monobactams but not to cephamycins.2 Resistance to fluoroquinolones is also frequently found in E. coli ST131 clones, probably potentiated by the presence of plasmid-mediated quinolone resistance (PMQR), like qnr encoding genes or aminoglycoside modifying enzyme AAC(6’)-Ib-cr, usually present in ST131 strains.2 This mobile resistance determinants (ESBLs and PMQRs), responsible for antimicrobial resistance, can be carried and disseminated through plasmids. In Enterobacteriaceae family, plasmids are gathered into major replicon families HI2, HI1, I1-ϒ, X, L/M, N, FIA, FIB, FIC, W, Y, P, A/C, T, K, and B/O.3
Additionally to the resistance capacities, E. coli ST131 strains have been described to harbor several virulence factors, contributing to the pathogenicity and invasion of the hosts. These determinants are generally involved in escaping the host immune system, adhesion, collection of nutrients under limited conditions, and induction of inflammation, instigating the development of extraintestinal pathology. Virulence factors in this clone include toxins, adhesins, and siderophore systems among others.2 Furthermore, the virulence factors can be carried and disseminated by horizontal gene transfer through pathogenicity islands.4 Although several virulence factors have been identified in E. coli ST131 strains,5 the lack of pathogenicity islands could make it less virulent than other uropathogenic bacteria.
Regardless of the general assumption that the acquisition of resistance may have a fitness cost which leads to decreased virulence, E. coli ST131 has proven to be able to maintain both features.6 Thus, the aim of this work was to evaluate the concomitant presence of plasmids as resistance determinants transporters and pathogenicity islands carrying several virulence factors in multidrug-resistant ST131 E. coli clinical isolates.
Results and Discussion
ST131 clone is a highly adapted E. coli with a genetic structure with several virulence and resistance genes which contribute to the efficient and global spread of this pathogen. The virtual omnipresence of ST131 E. coli strains, normally associated with CTX-M-15 ESBL production, impelled us to investigate the relationship between the presence of plasmids and PAIs within the cell. Thus, the present study aimed the investigation of the optimal relationship between these two mobile elements, within multidrug-resistant ST131 E. coli strains presenting PMQRs and ESBL.
From the total of 97 isolates, the co-analysis of the antibiogram performed by Vitek2Advanced Expert System and the double disk synergy results indicated the expression of ESBL in 15 strains, and suggested the production of CTX-M enzymes. This was further confirmed using PCR and sequencing techniques which identified the blaCTX-M-15 gene, whose expression was responsible for the ESBL phenotype in all strains.
In order to establish a phylogenetic relationship between the isolates, MLST was first executed, which allowed the identification of ST131 clone in 93% of the strains, all belonging to phylogroup B2. The remaining strain was integrated in the phylogenetic group A and presented a MLST corresponding to the ST10. ST131 has been mainly known as a worldwide extraintestinal pathogenic E. coli from phylogroup B2 with the capacity to acquire simultaneously virulence and resistance, thus having the capacity to cause severe antimicrobial-resistant infections.2 This clone contradicts the hypothesis that bacteria displaying high levels of resistance have a high fitness cost which results in decreased pathogenic capacities.6
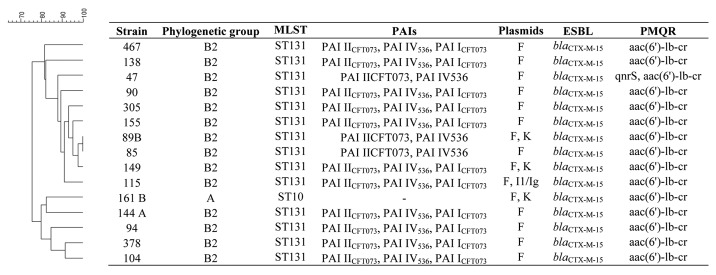
In this study, despite of the genetic variability observed in the PFGE profiles for the ST131 isolates, it was possible to observe that 64% of the strains presented the same characteristics in terms of virulence and resistance determinants (Fig. 1). These strains presented one IncF plasmid, containing a blaCTX-M-15 and aac(6’)-lb-cr genes and three PAIs (PAI IV536, PAI ICFT073, and PAI IICFT073). This may constitute a favorable and equilibrated combination of resistance and virulence factors for E. coli ST131 clones which may enhance its fitness and adaptation capacity, contributing to the dissemination of this clone.
Figure 1. Clonal relatedness of strains and characterization of phylogeny, virulence, and resistance determinants, and plasmid incompatibility group.
The resistance determinants blaCTX-M-15, and the aac(6’)-lb-cr enhance the survival of E. coli ST131 strains regarding the action of the main antibiotics used in clinical practice such as cephalosporins and quinolones. In fact, CTX-M-15 enzyme is now worldwide disseminated mostly by E. coli strains, and it has been previously detected among Portuguese clinical strains.7 The association between blaCTX-M-15 and aac(6’)-lb-cr genes is common, because they are usually found in the same plasmid, namely in IncF and in IncI1 plasmids, since they share the same genetic platform.3 Actually, in all the strains we have detected one plasmid from the group F. Moreover, in one ST131 strain an IncI1/Iγ was also detected, and in two ST131 strains and one ST10 isolate the IncK was also identified. After plasmid extraction we detected one plasmid carrying blaCTX-M-15, aac(6’)-lb-cr as well as PAI IV536 which was assigned by replicon typing to IncF group. Regardless of the high dissemination of blaCTX-M-15 associated to aac(6’)-lb-cr, the acquisition of the mobile element containing these determinants may have an implicated fitness cost. In fact Sandegren and colleagues have studied the fitness cost of the acquisition of a plasmid containing the multiresistance gene cassette associated with ST131 clone, composed namely by these two resistance elements, and found that it originated a reduction of 3–4% of exponential growth rate.8
The virulence factors encoded in the prevalent combination of PAIs, PAI IV536, PAI ICFT073, and PAI IICFT073 include P-fimbriae, siderophore systems, and hemolysin and may be responsible for the enhancement of the virulence potential of E. coli strains, and thus the development of more invasive disease.4,9 In fact, 80% of the strains were isolated from urinary tract, where P-fimbriae mediates bacterial adherence to human epithelial cells through di-galactoside-specific binding to the P-blood group antigens, which are present all over the urinary tract and enable ascending infection of the ureter and kidney.9 Furthermore, siderophore systems, as efficient methods to acquire iron, have been described to be essential to bacterial growth especially under limiting conditions such as the ones found in the urinary tract.9 These virulence factors present in the detected islands increase the E. coli ST131 capacity to subsist in the host and ascend in the urinary tract and may therefore contribute to the survival of this clone and to the evolution of its pathogenesis.
Regardless of the presence of PAIs carrying important virulent factors, we have not detected other islands which are present in more virulent uropathogenic strains. The results are supported by a genomic mapping performed by Lavigne and colleagues in which they have not detected typical extraintestinal PAIs such as PAI I536, PAI II536, and PAI III536.5
In conclusion, the E. coli ST131 clone despite not being an extremely virulent clone presents an efficient organization of virulence and resistance determinants. The predominant arrangement of virulence and resistance determinants found in the ST131 isolates may therefore constitute an optimal and balanced combination, providing important resistance and virulence capacities without having an excessive fitness cost to bacteria. This may allow the dissemination and survival of these clonal strains among the clinical and communitarian sets.
Materials and Methods
From a total group of 97 non-duplicate isolates of E. coli collected between November and December 2007 from different clinical samples and wards of the University Hospitals of Coimbra (HUC), located in the Centro region of Portugal, presumable ESBL producers were selected based in the antimicrobial susceptibility tests. ESBL production was further confirmed with the double disk synergy test using the antimicrobials disks of amoxicillin/clavulanic acid, cefotaxime, cefatzidime, cefepime, and aztreonam (OXOID). Results interpretation was done according Clinical and Laboratory Standards Institute guidelines.10 E. coli ATCC 25922 was used as a quality control strain.
Initially, multi-locus sequence typing (MLST) was performed based in the University of College Cork scheme for E. coli.11 To strengthen the phylogenetic relationship of the strains, pulsed-field gel electrophoresis (PFGE) was performed as previously described7 and PFGE images were analyzed with Bionumerics 6.6. Clustering was performed using the Dice band-based similarity coefficient, with a band position tolerance of 1.0% and an optimization of 1.8. A cutoff value of 80% similarity was determined by the cluster cutoff method according to Bionumerics software. Isolates with a Dice band-based similarity coefficient value >80% were assigned to the same cluster.
In order to evaluate the presence of resistance determinants, PMQR and ESBL encoding genes (qnrA, B, and S, aac[6’]-Ib-variant, qepA, and blaCTX-M) were screened by PCR.12-14 E. coli 12HUC carrying the blaCTX-M-15 and aac(6’)-lb-cr determinants, Klebsiella pneumoniae5, carrying qnrA and qnrB, and the plasmid pSTV that carries qepA, were used as positive controls in PCR. Resulting PCR products were submitted to purification using the QIAquick PCR Purification kit (QIAGEN), according to the producer’s instructions and were sequenced at Macrogen.
The detection of plasmid-mediated resistance mechanisms implied the plasmid replicon typing. For this, a PCR-based replicon typing scheme described by Carattoli et al. detecting the principal incompatibility groups in enterobacteria: HI2, HI1, I1-ϒ, X, L/M, N, FIA, FIB, FIC,W, Y, P, A/C, T, K, and B/O was performed.15 When needed, plasmid extraction was performed using QIAGEN Plasmid Midi Kit.
Finally, to evaluate the pathogenicity potential of these E. coli strains, pathogenicity island (PAI) markers were detected using the method of Sabaté et al. consisting in three multiplex-PCR which allow the detection of eight PAIs,4 reinforced by the determination of the major E. coli phylogenetic groups (A, B1, B2, and D), according to the Clermont method.16
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
All authors contributed equally to this work. N Mendonça was supported by a grant SFRH/BPD/45815/2008 from Fundação para a Ciência e a Tecnologia, Lisbon, Portugal. This work was supported financially by the European Society of Clinical Microbiology and Infectious Diseases 2010 Research Grant and by the Center for Pharmaceutical Studies, University of Coimbra. The authors would like to thank all the members of the Bacteriology Laboratory of the Clinical Pathology Service of University Hospitals of Coimbra for the collaboration in isolation and identification of bacteria. K. pneumoniae5 and the plasmid pSTV were kindly provided by Sónia Mendo, University of Aveiro, and Kunikazu Yamane, National Institute of Infectious Diseases, respectively. Positive controls used in plasmid replicon typing were kindly provided by Alessandra Carattoli, Instituto Superiore di Sanità.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/26552
References
- 1.Totsika M, Beatson SA, Sarkar S, Phan MD, Petty NK, Bachmann N, Szubert M, Sidjabat HE, Paterson DL, Upton M, et al. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PLoS One. 2011;6:e26578. doi: 10.1371/journal.pone.0026578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 3.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–38. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabaté M, Moreno E, Pérez T, Andreu A, Prats G. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin Microbiol Infect. 2006;12:880–6. doi: 10.1111/j.1469-0691.2006.01461.x. [DOI] [PubMed] [Google Scholar]
- 5.Lavigne JP, Vergunst AC, Goret L, Sotto A, Combescure C, Blanco J, O’Callaghan D, Nicolas-Chanoine MH. Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One. 2012;7:e34294. doi: 10.1371/journal.pone.0034294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark G, Paszkiewicz K, Hale J, Weston V, Constantinidou C, Penn C, Achtman M, McNally A. Genomic analysis uncovers a phenotypically diverse but genetically homogeneous Escherichia coli ST131 clone circulating in unrelated urinary tract infections. J Antimicrob Chemother. 2012;67:868–77. doi: 10.1093/jac/dkr585. [DOI] [PubMed] [Google Scholar]
- 7.Mendonça N, Leitão J, Manageiro V, Ferreira E, Caniça M. Spread of extended-spectrum beta-lactamase CTX-M-producing escherichia coli clinical isolates in community and nosocomial environments in Portugal. Antimicrob Agents Chemother. 2007;51:1946–55. doi: 10.1128/AAC.01412-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandegren L, Linkevicius M, Lytsy B, Melhus A, Andersson DI. Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J Antimicrob Chemother. 2012;67:74–83. doi: 10.1093/jac/dkr405. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. CLSI document M100-S20. Clinical and Laboratory Standards InstituteWayne, PA. 2010. [Google Scholar]
- 11.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007;60:394–7. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- 13.Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, Lü D, Huang L, Zhang Y, Liu J, et al. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother. 2009;53:519–24. doi: 10.1128/AAC.00886-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006;50:3953–5. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]