Abstract
Cyclin-dependent kinase 6 (Cdk6) is a D-Cyclin-activated kinase that is directly involved in driving the cell cycle through inactivation of pRB in G1 phase. Increasingly, evidence suggests that CDK6, while directly driving the cell cycle, may only be essential for proliferation of specialized cell types, agreeing with the notion that CDK6 also plays an important role in differentiation. Here, evidence is presented that CDK6 binds to and promotes degradation of the EYA2 protein. The EYA proteins are a family of proteins that activate genes essential for the development of multiple organs, regulate cell proliferation, and are misregulated in several types of cancer. This interaction suggests that CDK6 regulates EYA2 activity, a mechanism that could be important in development and in cancer.
Keywords: Cdk6, EYA2, degradation, proliferation, progenitor cell, cell cycle, cancer
Introduction
Cyclin-dependent kinase 6 (cdk6) is a D Cyclin-activated kinase that phosphorylates the retinoblastoma protein (pRB) in G1 phase of the cell cycle. Following phosphorylation by a series of cdks in G1 and early S-phase, pRB releases the E2F protein, freeing it to bind DNA and induce transcription of S-phase genes, allowing cell cycle progression. Perhaps due to compensation by other cdks, the cdk6-knockout mouse is viable but displays defects in hematopoiesis.1,2 The cdk6 protein is expressed ubiquitously and is downregulated in differentiating cells. Indeed, cdk6 modulates differentiation in specific cell types, including murine erythroid leukemia,3,4 primary mouse astrocytes,5 osteoblasts and osteoclasts,6,7 hematopoietic cells,8 and thymocytes1,9 (for review, see ref. 10). Recent data indicate that CDK6 is not essential for the mammalian cell cycle and may only be essential for proliferation in specialized cells.2 In fact, a new study has shown that CDK6 can constrain cell proliferation in vivo through a transcriptional mechanism that activates the p16INK4A and the VEGF promoter, and that kinase activity is not required for these functions11.
An important role for Cdk6 in transcription and in functions besides direct pRB phosphorylation has been suggested by several earlier studies. The Cdk6 oncoprotein interacts with several factors including Runx1, Fbxo7, and Hsp90/Cdc37, and it has recently been shown to phosphorylate FoxM1.8,12,13 Cdk6 inhibits the transcriptional activity of Runx2 independent of its kinase activity.6In these studies, overexpression of Cdk6 prevented the Runx2 transcription factor from loading on the osteocalcin promoter but did not exclude pRB, a Runx2 binding cofactor.6 Cdk6 was also shown to be required for thymocyte development and tumorigenesis.1,9 The kinase activity of Cdk6 is required for both T-cell development and for Notch-dependent commitment, proliferation, survival, and differentiation of T cells.1,9 Because Cdk6 plays a role in both development and cell cycle progression, it is not surprising that the gene is overexpressed or amplified in a variety of cancers, including oral squamous cell carcinomas, breast tumors, sarcomas, melanomas, and lymphomas (for review, see ref. 14).
The EYA proteins (EYA1–4) are the mammalian counterparts of the eyes absent gene product in Drosophila. Eyes absent is one member of the retinal determination network. This network was shown to function in gonadogenesis, myogenesis, limb formation, neurogenesis, thymus and kidney development, and cell cycle control.15,16 EYA proteins function as both tyrosine and threonine phosphatases and form bipartite transcriptional activators by interacting with a member of the sine oculis (Drosophila) family of proteins, termed SIX proteins in vertebrates. The SIX proteins provide the DNA binding domain to tether the EYA transcriptional activator to the promoter. EYA proteins activate genes essential for the development of multiple organs and the phosphatase function of EYA has been shown to be required for normal development. In breast cancer cells, EYA1 has been shown to induce transcription of Cyclin D1 dependent on its phosphatase function.17 EYA proteins are predominately expressed in progenitor cells where they regulate both cell proliferation and cell survival. Mice mutant for Eya1 demonstrate reduced proliferation and abnormal apoptosis of kidney, muscle, and ear, among other tissues. Recent data demonstrate that EYA1 is degraded during mitotic exit by the APC/cdh1 complex18 but the mechanism by which EYA proteins interact with the cell cycle to promote proliferation in normal cells is largely unknown (for review, see ref. 15).
The EYA proteins are overexpressed in several types of cancers including breast, lung, cervix, and kidney.15 EYA2 overexpression in breast cancer cells increased proliferation, transformation, and migration and correlated with poor prognosis/outcomes in ovarian and breast cancer.19,20 EYA2 expression has been shown to be upregulated in epithelial ovarian cancer, where it was associated with shortened overall survival.16 This study also revealed that EYA2 mRNA was highly expressed in ovarian, prostate, lung, and, to a lesser extent, breast and urinary tract cancers.16 Most recently, EYA overexpression has been observed in additional tumor types, including glioblastoma and other cancers of the brain,21 cervical cancer,21,22 colon and hematopoietic cancers,21,23 among others.21
To better understand CDK6-specific functions in differentiation and tumorigenesis, a yeast 2-hybrid screen was performed to identify targets that bound CDK6 but not its close homolog CDK4. This screen revealed an interaction between CDK6 and EYA2. Data presented here prove that this interaction occurs in cell lysates and identify a biologically important outcome of the interaction.
Results
CDK6 binds EYA2
To investigate non-redundant functions of the cell cycle kinases cdk4 and cdk6, a differential yeast 2-hybrid screen was performed to ascertain unique binding partners of CDK6 that were unable to bind to CDK4.24 Screens of both mouse and human fetal brain cDNA libraries (Gibco/Clontech) identified EYA2 as an interacting partner with CDK6 but not CDK4. It is important to note that this screen was performed with the human CDK6 cDNA that had been widely distributed in the field after being reported in a landmark paper.25 Later analysis of the human genome revealed that this circulated cDNA was not wild-type CDK6, but instead contained a point mutation at amino acid 224, changing D to Y. The aspartic acid residue is highly conserved at amino acid 224, being maintained in 8 of 8 species examined.26 The cDNA insert in the pCMVcdk6 plasmid had originated from a PCR amplification of conserved sequences in cdks. Sequences were amplified from HeLa cervical carcinoma cells and Nalm-6 pre-B leukemia cells.25,27 Thus, the interaction with EYA2 identified in our screen was isolated using CDK6D224Y. The CDK6 sequence has since been corrected to wild-type26 and is referred to throughout this work as CDK6WT. Previous work from several labs, unknowingly using the mutant CDK6, has demonstrated that the CDK6D224Y point mutation retains pRB kinase activity and does not affect the subcellular localization of the protein.5,26,28 However, neither the pRB kinase activity nor the ability to bind Cyclins, INK, and CIP/KIP family proteins have been rigorously compared between the wild-type and the D224Y mutant.
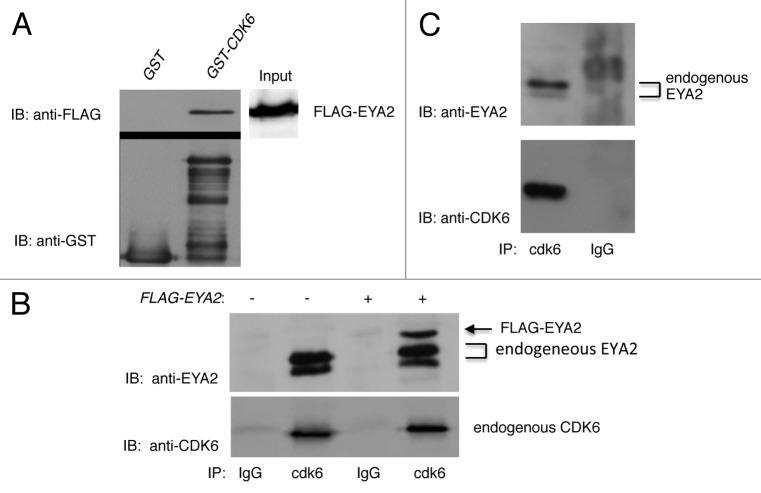
To determine if wild-type CDK6 bound to EYA2, GST-binding studies were performed. Figure 1A demonstrates that GST-CDK6WT was able to bind FLAG-EYA2 in lysates of transfected cells. Importantly, the same amount of input extract in the binding assay did not bind GST alone.
Figure 1. Cdk6 binds EYA2. (A) GST pull-down assay. Two hundred micrograms of FLAG-EYA2-expressing HEK293T cell extract was bound to GST or GST-CDK6. Top panels: Anti-FLAG immunoblot of binding assay. Bottom panel: anti-GST immunoblot of GST input extracts. Right panel: anti-FLAG immunoblot of 100µgs HEK extract expressing FLAG-EYA2. (B) Immunoprecipitation of untransfected (−) and FLAG-EYA2 transfected (+) HEK293T lysates with polyclonal CDK6 antibody precipitated EYA2 while immunoprecipitation with rabbit IgG control beads did not. Immunoprecipitations from transfected cells contained an additional slower migrating band correlating with the transfected FLAG-tagged protein. FLAG-EYA2 was detected using both anti-FLAG (not shown) and anti-EYA2 antibodies. (C) CDK6 immunoprecipitation (IP) of endogenous EYA2 from OVCAR3 cell lysates. IPs were immunoblotted with anti-EYA2 N-16 antibody (top panel) and anti-CDK6 polyclonal antibody (bottom panel).
In order to evaluate this interaction in cells, we transfected HEK293T cells with FLAG-EYA2-expressing plasmids. Human embryonic kidney cells were chosen as a relevant cell line, since EYA1 and EYA2 proteins are expressed in fetal (EYA1) and adult (EYA2) human kidneys,15 and because the EYA1 gene is responsible for Branchio-oto-renal syndrome (BOR), which causes malformations of the ears and kidney.15 Endogenous CDK6 is also expressed in HEK293T cells. Cells were lysed and immunoprecipitated with conjugated anti-CDK6 antibody or rabbit IgG as a control and immunoblotted with anti-EYA2 antibody. Results shown in Figure 1B demonstrate that CDK6 immunoprecipitated EYA2, while the control IgG did not. Importantly, CDK6 immunoprecipitated both endogenous and transfected EYA2, which were easily differentiated due to the FLAG-tag on transfected EYA2, visualized as a slower-migrating band. These results reveal CDK6 binding to both endogenous and ectopically expressed EYA2.
EYA2 has been shown to be overexpressed in epithelial ovarian cancers.16,20 Thus, the ovarian cancer cell line, OVCAR3 was chosen to further study the interaction of EYA2 and CDK6. Untransfected OVCAR3 cells were immunoprecipitated with polyclonal CDK6 antibody or IgG control and then immunoblotted with anti-EYA2 antibodies. Results in Figure 1C demonstrate that cdk6, but not an IgG control immunoprecipitation, co-precipitated endogenous EYA2. Together, these studies show that wild-type CDK6 binds both transfected and endogenous EYA2 in more than one cell type.
CDK6 mutants bind EYA2
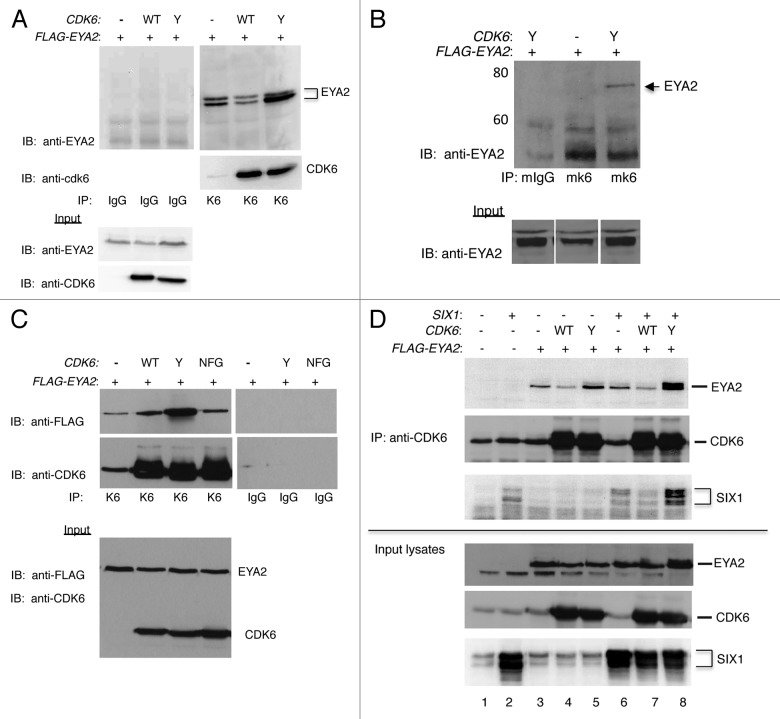
Binding studies shown in Figure 1 demonstrate that wild-type CDK6 binds EYA2. Because this interaction was first identified in yeast while inadvertently screening with a mutant version of CDK6, CDK6D224Y, we next compared binding of EYA2 to CDK6WT and CDK6D224Y. Figure 2A is a CDK6 immunoprecipitation of HEK cells transfected with FLAG-EYA2 and either no CDK6 (−), CDK6WT (WT) or CDK6D224Y (Y). EYA2 protein was precipitated from all 3 conditions, demonstrating that sufficient levels of endogenous CDK6 were available in these cells to bind and precipitate the transfected EYA2 protein. The use of multiple anti-EYA2 antibodies (noted in figure legends) ensured that the EYA band was specific for EYA2. Interestingly, we noted that overexpression of CDK6WT led to a decrease in the amount of EYA2 precipitated as compared with pull-downs of endogenous CDK6 or CDK6D224Y. This could be due to enhanced binding of CDK6D224Y or to an increase in the steady-state levels of EYA2 in the presence of CDK6D224Y, as suggested by the immunoblot of input lysates shown in the bottom panel of Figure 2A.
Figure 2. CDK6 MUTANTS bind EYA2. (A) Immunoprecipitation of HEK293T cells transfected with FLAG-EYA2 and wild-type CDK6 (WT), CDK6D224Y (Y), or empty vector (−). Lysates were immunoprecipitated with CDK6 antibody or rabbit IgG control and immunoblotted with anti-EYA2 (Abcam) and with anti-CDK6 (Pierce) antibody. Input lysates are shown in the bottom panels. (B) Monoclonal CDK6 Immunoprecipitation. Immunoblots of transfected HEK293T cells immunoprecipitated with monoclonal CDK6 antibody (Pierce) or monoclonal IgG control. Immunoblots of input lysates are shown in the bottom panel. Both panels probed with anti-EYA2 (Abcam) antibody. (C) HEK293T cells transfected with empty vector (−), CDK6WT (WT), CDK6D224Y (Y) or CDK6NFG (NFG) were immunoprecipitated with cdk6 antibody or rabbit IgG control, as noted. Immunoblots of IPs (top panels) and of input lysates (bottom panel) were probed with anti-FLAG and anti-cdk6 (Pierce) antibodies. (D) Complexes immunoprecipitated from HEK293T cells with or without the addition of vectors expressing FLAG-EYA2, SIX1, and CDK6 variants. Immunoprecipitations with polyclonal conjugated CDK6 antibody (Santa Cruz) in the top 3 panels. Input lysates shown in the bottom 3 panels. All blots probed with antibodies as indicated in the figure: EYA2 (Abcam), CDK6 (Novus), SIX1 (4–2, Ford).
To confirm and extend these binding studies, the interaction was next examined using an immunoprecipitation with monoclonal CDK6 antibody. In Figure 2B, the CDK6D224Y mutant was used to improve the EYA2 signal, since the monoclonal pull-down was found to be less effective than the polyclonal immunoprecipitation. Transfected cell lysates were precipitated using a monoclonal CDK6 antibody or mouse IgG control. Input lysates, shown in the bottom panel, reveal slightly more EYA2 protein in CDK6D224Y transfected cells than in cells not transfected with CDK6. Cells that had been transfected with CDK6D224Y and FLAG-EYA2 precipitated EYA2 that was not present in the IgG control precipitation.
To determine if a kinase-inactive CDK6 can bind EYA2, cells were transfected with the well-characterized, kinase-inactive CDK6 mutant, CDK6NFG.25 As originally cloned, the NFG mutant also contained the Y mutation at aa224, but the NFG mutant used here has been corrected to encode the wild-type D residue. Transfected cell lysates were immunoprecipitated with rabbit IgG or polyclonal CDK6 antibody and immunoblotted with anti-FLAG antibody, as indicated in Figure 2C. These experiments indicate that CDK6NFG binds FLAG-EYA2 only slightly less well than the CDK6WT protein and decidedly less well than does the CDK6224Y protein, suggesting that kinase activity is not required for EYA2 binding. In summary, CDK6 binds EYA2 when precipitated by polyclonal or monoclonal anti-CDK6 antibodies, and differences in the amount of EYA2 bound by CDK6 or CDK6 mutants in immunoprecipitations are observed regardless of the antibody used to detect EYA2 in the immunoblot.
The EYA2 protein forms a transcriptionally active complex with the SIX1 protein, which has been shown to localize EYA2 to the nucleus.29-31 Experiments were undertaken to investigate how the EYA2/CDK6 immune-complex is influenced by expression of SIX1. HEK293T cells were transfected with combinations of CDK6WT or CDK6D224Y, FLAG-EYA2, and SIX1 expressing plasmid, as noted. Immunoprecipitations were run on SDS–polyacrylamide gels and then immunoblotted with antibodies as indicated in Figure 2D. Input lysates of each protein are shown in the lower 3 panels, with CDK6-precipitated proteins shown in the upper 3 panels. These data indicate that co-expression of SIX1 increases the steady-state levels of EYA2 in the presence of overexpressed CDK6, especially when the mutant CDK6D224Y is co-expressed (input lysates, lane 8), supporting previous studies of Patrick et al. that suggested that SIX1 stabilized the EYA2 protein.31 The co-expression of SIX1 also resulted in more EYA2 being precipitated by CDK6D224Y (compare lane 5 and 8 of IP). Of note, lysates containing overexpressed wild-type CDK6 actually precipitate less EYA2 protein than those containing only endogenous CDK6. These results suggest that overexpressed CDK6 results in reduction of EYA2 being precipitated, regardless of the co-expression of SIX1 (lanes 3 vs. 4, lanes 6 vs. 7). Finally, either the expression of CDK6D224Y, or the presence of increased EYA2 results in a drastic increase in the levels of SIX1 being immunoprecipitated.
CDK6 promotes the degradation of the EYA2 protein
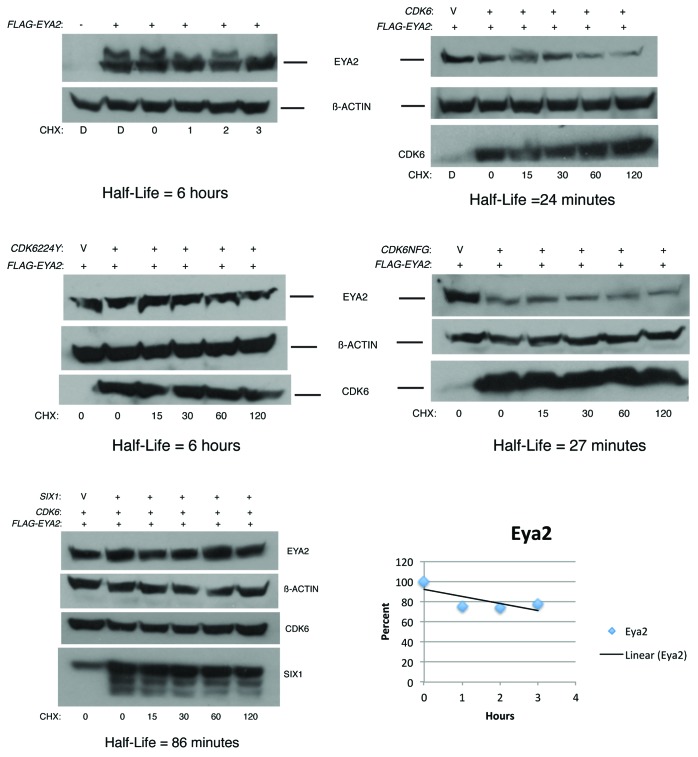
To determine the effect of CDK6 expression on the stability of the EYA2 protein, half-life experiments were performed using cycloheximide to block translation. HEK293T cells were transfected with CDK6 or mutant versions of CDK6, as indicated in Figure 3. After overnight transfection, cells were treated with cycloheximide, lysates collected at timepoints indicated, and EYA2 immunoblots performed.
Figure 3. CDK6 promotes degradation of EYA2. Immunoblots of HEK293T cells transfected with indicated plasmids and treated with cycloheximide (CHX) for indicated times in minutes, with the exception of the FLAG-EYA2 panel in which CHX treatment is noted in hours. D indicates DMSO (in which cycloheximide was dissolved) treatment for 2 hours. V in CDK6 lane indicates Vector, pCMV empty vector. Each transfection was equalized with empty vector to ensure that equal amounts of CMV promoter were included in each experiment. Loading controls and CDK6/SIX1 blots are also shown, as relevant, for each experiment. Pixels in the linear range were measured (Bio-Rad Chemi-Doc) for each band and normalized so that the number of pixels in the EYA2 alone condition was 100%. The percentage of pixels in all other EYA2 bands were plotted vs. time and the half-life calculated from a linear curve fit of the data points.
These experiments demonstrated that the half-life of EYA2 expressed in HEK293T cells was 6 hours. This is longer than that reported for stably expressed Flag-EYA1 in NIH3T3 cells, which has been reported to be 98 min.18 The addition of co-transfected CDK6WT reduced the EYA2 half-life 12-fold to 24 min. To determine the effect of SIX1 on the EYA2/CDK6 complex, half-life experiments were performed with lysates from cells transfected with all 3 plasmids. Interestingly, SIX1 expression partially stabilized the EYA2 protein against CDK6-mediated degradation. The calculated half-life of these experiments was 86 min, approximately 3 times longer than CDK6 alone, but well shy of the 6-h half-life of EYA2 without CDK6 expression. This finding is consistent with a previously published study that demonstrated that expression of the SIX1 protein stabilized the EYA2 protein.31
To understand some of the properties of CDK6 that facilitate EYA2 degradation, CDK6 mutants were examined in half-life experiments. Interestingly, expression of the Cdk6 mutant CDK6D224Y did not decrease the half-life of EYA2. In fact, experiments revealed that EYA2 half-life in the presence of CDK6D224Y was 6 hours, the same as EYA2 without CDK6 co-transfection. Importantly, an immunoblot for CDK6 demonstrated that the protein was indeed made and was relatively stable in these experiments. To determine if kinase activity of CDK6 was required for CDK6-mediated degradation of EYA2, the CDK6NFG mutant was tested. Transfection of CDK6NFG and EYA2 resulted in a half-life of 27 min, essentially indistinguishable from that of CDK6 wild-type.
Thus, CDK6 reduces EYA2 half-life significantly, and the SIX1 protein partially protects against this degradation. CDK6D224Y expression does not reduce the half-life of EYA2 despite a proven ability to bind the protein (Fig. 2). The kinase activity is not required for the degradation function of CDK6, since the EYA2 half-life was very similar when either CDK6 or CDK6NFG was co-expressed with EYA2.
Proteasome treatment stabilizes EYA2
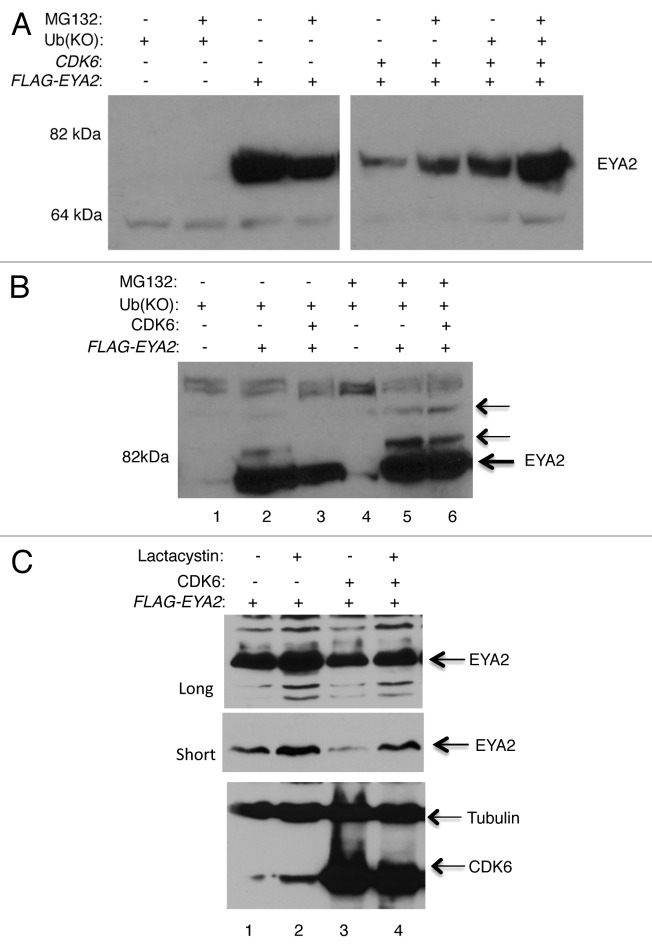
Next, experiments were undertaken to determine if CDK6-induced degradation of EYA2 involves the proteasome. HEK293T cells were transfected with FLAG-EYA2 with and without co-transfection of CDK6. Experiments were also co-transfected with a lysine-less ubiquitin construct. This ubiquitin mutant, UbKO, is conjugation-deficient due to mutation of all 7 lysines to arginines. Because UbKO does not contain a lysine as a point of attachment for additional ubiquitin molecules, it inhibits chain elongation and mono-ubiquitin intermediates are preferentially trapped. However, due to the large reservoir of endogenous ubiquitin, some longer Ub chains are formed in the presence of transfected UbKO.32 Figure 4A is an immunoblot that demonstrates that the addition of the proteasome blocker, MG132, stabilized the EYA2 protein even in the presence of CDK6 overexpression. This stabilization was further enhanced by the co-transfection of UbKO. Figure 4B provides additional evidence for proteasome-mediated degradation. In this figure, slower migrating bands that react with the EYA2 antibody can be visualized, suggesting ubiquitination of EYA2. Of note, in the presence of CDK6, the slower-migrating band is less prevalent, perhaps indicating that the mono-ubiquitinated form of EYA2 is reduced (compare lane 2 and 3, 5 and 6), as might be expected if CDK6 is causing the ubiquitinated forms to turnover. Figure 4C examines the effect of the proteasome inhibitor Lactacystin on EYA2 levels, also in HEK293T cells transfected with EYA2 or EYA2 and CDK6-expressing plasmids. Both short and long exposures of the immunoblot are shown for comparison, as are immunoblots of CDK6 and tubulin as a loading control. Together, these data indicate that both MG132 and Lactacystin stabilize the EYA2 protein against CDK6-mediated degradation, providing further support for a role for the proteasome in this process.
Figure 4. MG132 and lactacystin stabilize EYA2 against CDK6-mediated degradation. (A) MG132 stabilizes EYA2 protein even in the presence of CDK6. One hundred and fifty µg of transfected cell extract were loaded per lane and blotted with the Ford Lab19 anti-EYA2 antibody. Similar results were seen with the N-16 anti-EYA2 antibody. (B) Intentional overexposure of transfected cell extracts to allow detection of slower migrating species of EYA2. Each lane contains 75 µg of protein. (C) Lactacystin stabilizes EYA2 both without and with co-expression of CDK6. One hundred and fifty µg transfected cell extract per lane. Immunoblot was probed with the Ford lab anti-EYA2 antibody, CDK6 with C-21 antibody, and Tubulin with Sigma anti-Tubulin antibody.
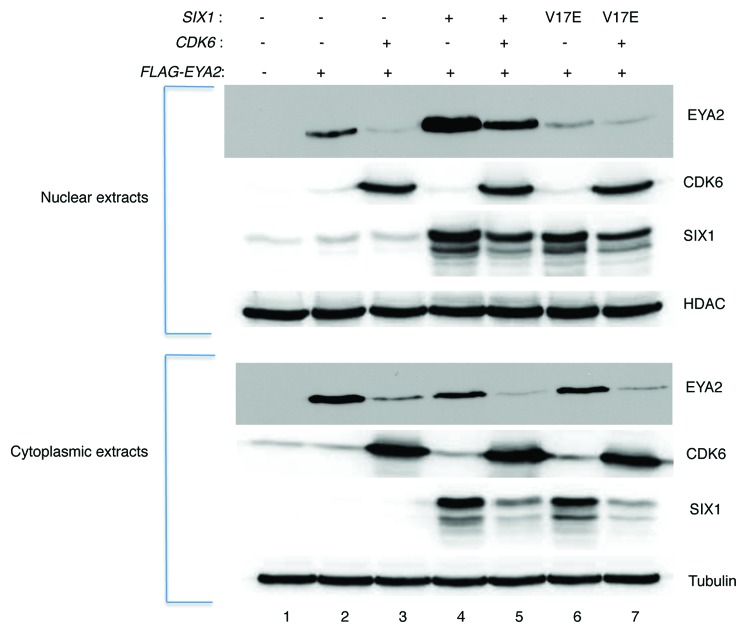
EYA2 degradation in nuclear and cytoplasmic compartments
Both nuclear and cytoplasmic expression of EYA2 are required for complete eye restoration in Drosophila,33 suggesting that EYA proteins function in both compartments. To investigate the influence of CDK6 on EYA2 localization in the cytoplasm and nucleus, fractionation experiments were performed. Previous work has demonstrated that SIX1 translocates EYA2 to the nucleus.29,30 Also, the SIX1V17E point mutant, which is unable to bind EYA2, abrogated nuclear translocation, and co-expression of the SIX1 protein resulted in stabilization of EYA2 protein.31
HEK293T cells were transfected with FLAG-EYA2, CDK6, and SIX1-expressing plasmids in combinations indicated in Figure 5. The addition of CDK6 to the cells resulted in a significant decrease in both the nuclear and cytoplasmic steady-state levels of EYA2 protein. The addition of SIX1 to the experiment resulted in a stabilization of EYA2, consistent with results in Figures 2D and 3. Data in Figure 5 suggest that expression of SIX1 may preferentially stabilize the nuclear fraction of EYA2 (compare lanes 2 and 4) and stabilize against cdk6-mediated degradation, especially in the nuclear fraction (lanes 3 vs. 5 nuclear and cytoplasmic fractions). Based on results with the V17E mutant of SIX1, stabilization appears to be dependent upon SIX1 binding or perhaps SIX1-directed nuclear localization of EYA2 (lanes 5 and 7).
Figure 5. Subcellular localization of EYA2. HEK293T cells transfected as indicated in the figure were lysed and fractionated. Eighty-five µg of cell extract was loaded in each lane of an SDS/PAGE gel. Resulting blots were then probed with EYA2 antibody (N-16), CDK6 antibody (Pierce), SIX1 antibody (4–2), or HDAC and tubulin antibodies as loading controls. Lane 1 is a vector-only control.
Upon addition of CDK6, the EYA2 protein is degraded, and decreased levels are seen in all lanes (lane 3, 5, 7). This CDK6-dependent loss of EYA2 protein is evident in both the nuclear and cytoplasmic fractions. Of interest, the SIX1 protein is also reduced in the presence of CDK6 expression (compare lanes 5 to 4, lane 7 to 6). This decrease is not simply due to the increase of CMV promoters transfected, because each transfection was equalized using the pCMV empty vector. Rather, the reduction in SIX1 protein in the presence of CDK6 may be secondary to the reduction in EYA2 protein levels or could reflect an independent effect of CDK6 on SIX1 steady-state levels.
Discussion
The EYA proteins are part of a conserved regulatory network that plays a key role in development of multiple organs, including the eye, muscle, ears, heart, lungs, pharyngeal pouches, craniofacial skeleton, and parathyroid. EYA proteins influence development and advance cancer progression through their function as pro-proliferative, anti-apoptotic factors (for review, see ref. 15). Despite a well-understood function of EYA in cell proliferation, its mechanism of action in the cell cycle is largely unknown. Of interest, the cell cycle regulators Cyclin D1, p27, and c-myc are target genes of the EYA transcriptional activators.
Here, we provide evidence of a novel interaction between EYA2 and the CDK6 cell cycle kinase. CDK6 was found to bind EYA2 in a yeast 2-hybrid screen, in a GST system, in transfected and untransfected HEK293T cells and in OVCAR3 cells. EYA2 binds 2 point mutants of CDK6, CDK6D224Y and CDK6NFG. We have here demonstrated a functional outcome of this interaction: CDK6 expression results in the degradation of the EYA2 protein. We have shown that the kinase activity of CDK6 is not required for this function, since the CDK6NFG mutant also leads to degradation of EYA2, but the CDK6D224Y mutant does not. Our data indicate that the SIX1 protein partially stabilized EYA2 against CDK6-mediated degradation. Data presented here are consistent with recent work by Sun et al., who demonstrated that EYA1 is degraded via ubiquitin-mediated proteolysis.18 Importantly, we find that both ectopic and endogenous CDK6 can bind ectopic and endogenous EYA2, supporting the physiological relevance of this interaction. Furthermore, we have shown that overexpressed CDK6 can degrade overexpressed EYA2 with functional specificity. However, it remains unkown if and when this degradation occurs physiologically; we hypothesize that this function of CDK6 is specific to cell cycle phase or differentiation status. Indeed, previous studies from our lab demonstrated that CDK6 is largely localized to the cytoplasm, suggesting that CDK6 could target EYA2 in the cytoplasm, limiting nuclear functions with the oncogenic SIX proteins. However, our studies demonstrated that EYA2 was localized to the nucleus in the presence of functional SIX1, regardless of CDK6 expression.
Regulated degradation of EYA2 may have relevance in cancer cells. EYA2 has been found to be overexpressed in epithelial ovarian cancers,16 cervical cancers22, and lung adenocarcinoma, and its overexpression correlated with poor prognosis.34 EYA2 is also overexpressed in breast cancers16,19,20 and hematopoietic cancers.21,23 In mammary epithelial cells, EYA2 promotes transformation, migration, and invasion, and EYA2 is required to mediate the prometastatic function of SIX1.19,20 Furthermore, the ability of SIX1 to mediate metastasis has been shown to be dependent on its interaction with EYA2.21
Given the well-established oncogenic roles of EYA proteins, it seems paradoxical that the CDK6 protein, which is itself oncogenic when activated by D Cyclins, promotes the degradation of the EYA2 protein. However, a model for EYA2 degradation that could rectify this seeming contradiction can be inferred from studies focused on the FoxM1 protein, which is synthesized and degraded in every cell division. Phosphorylation of FoxM1 by cell cycle kinases, including cdk6,12 activates its ability to promote cell cycle progression, and dephosphorylation of FoxM1 in late M phase leads to destruction by APC in the following G1 phase,35 thus allowing proper timing of FoxM1 function. In what may turn out to be a similar mechanism, EYA1 has been shown to be ubiquitinated by APC/cdh1 in mitotic exit/early G1 phase and degraded through the proteasome.18 Interestingly, SIX1 has also been shown to be degraded by APC in M phase.36,37 Our hypothesis is that EYA2 may be regulated in a manner similar to FoxM1, with the inactive pool of CDK6 functioning to constrain cell division by promoting degradation of the pro-mitotic EYA2 protein. In the experimental design employed here, the overexpression of CDK6 is expected to generate a large inactive pool, since the endogenous pool of activating Cyclin D molecules limits active complexes of CyclinD/CDK6. We demonstrate that inactive CDK6—either overexpressed CDK6WT or CDK6NFG—results in the degradation of EYA2. The model predicts that this inactive pool of Cdk6, mimicking the Cdk6 present in late M or early G1 phase, would constrain the cell cycle by promoting the degradation of the pro-mitotic EYA2 molecule.
Fortuitously, we have also found that the D224Y mutation of CDK6 prevents increased degradation of EYA2. Of interest, a 15 000 step combined Monte Carlo torsional variation and large-scale low mode method conformational search38-40 of the CDK6D224Y structure (pdb code = 1BLX) predicts a strong hydrogen bond between the phenolic proton on Tyr224 and the amide oxygen of Val190. As a consequence the Y224-I235 α helix is drawn closer to the E189-T198 loop. It remains to be determined how this structural change influences the ability of CDK6 to enhance the degradation of EYA2. This mutant provides an important tool with which we can now dissect CDK6 functions required to promote degradation of EYA2 and determine the involvement of Cyclin and SIX proteins in the complex.
Materials and Methods
Transfection of cells and derivation of plasmids
HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen/Gibco Life Sciences). pcDNA3FLAG-EYA2 constructs were a kind gift of Pascal Maire and Patrick Casey.41,42 In these vectors, the human EYA2 sequence used was identical to complete coding sequence (Accession Y10261). (Casey Lab, Duke). Human CDK6 and CDK6D224Y were expressed from the CMV promoter in the pCMV plasmid which has been previously described.26 The pCMVCDK6NFG was corrected using the Quick change site-directed mutagenesis kit (Agilent) to change the tyrosine at aa224 to aspartic acid in the CDK6NFG cDNA. The resulting cDNA was cloned back into the pCMV vector. The SIX1-expressing plasmid contains human SIX1 downstream of the CMV promoter in pCDNA3. The ubiquitin mutant UBKO was expressed from the CMV promoter in plasmid pGFP-UbKO.G76V, obtained from Addgene. This is a conjugation deficient mutant in which all 7 lysines are mutated to arginines. In experiments where EYA2 and CDK6 were co-transfected, pCMVNeoBam empty vector was used to equalize the CMV promoters in lanes lacking pCMVCDK6 transfection. HEK cells were carried in DMEM with 10% fetal bovine serum, 10% penicillin/streptomyocin with additional L-glutamine.
GST
HEK293T cells were transfected with 8 µg EYA2-expressing plasmid in a 60-mm dish. Cells were lysed in 200 µl ELB with 100 µl of 30% glycerol added to the lysate. GST and CDK6GST fusion proteins were extracted from the bacterial pellet in pull-down buffer (50 mM Tris 8.0, 100 mm NaCL, 1 mM EDTA, 1 mM DTT, with 1 mg/ml lysozyme, 0.3% sarkosyl, and 1.0% triton). 10 µl of glutathione 4B Sepharose (GE Healthcare) were added to approximately equivalent amounts, as indicated by immunoblot, of GST protein. 200 µg of HEK293T lysate expressing FLAG-EYA2 was bound with GST and CDK6-GST sepharose. 100 µg (50% of binding assay input) of extract was blotted in the input immunoblot. Antibodies: anti-GST, GE Healthcare; anti-FLAG, Pierce (DYKDDDDK epitope).
Immunoprecipitations and immunoblots
HEK293T cells were transfected with (+) or without (−) 3 µg FLAG-EYA2 plasmid, 3 µg CDK6, or CDK6 mutant plasmid, or 1 µg SIX1-expressing plasmids. Cells were lysed in E1A lysis Buffer (ELB) (250 mM NaCL, 50 mM HEPES, 5 mM EDTA, 0.1% NP40) with HALT protease inhibitors (Thermo Scientific). Five hundred to 800 µg of transfected cell extract were precipitated with agarose-conjugated anti-CDK6 antibody or control IgG pre-incubated with Protein A/G beads (Santa Cruz). Immunoprecipitations (IPs) were repeated 2 or 3 times and immunoblots of IPs were probed with several different antibodies to EYA2 to ensure specificity, as noted in figure legends. Anti-EYA2 antibodies used were as follows: N-16 anti-EYA2 antibody (Santa-Cruz Biotechnologies), RabMab anti-EYA2 (Abcam, rabbit monoclonal to a synthetic N-terminal peptide), or the Ford Lab anti-EYA2 (polyclonal, made to aa 17–3719). Antibodies to other proteins were C-21 anti-CDK6 antibody (Santa-Cruz), monoclonal CDK6 (Pierce Scientific), the 4–2 SIX1 antibody,37 anti-FLAG antibody (Pierce Scientific). OVCAR3 cells were lysed in ELB. The entire sample (13.4 mg) was pre-cleared with rabbit IgG and sepharose beads for 1 h at 4 °C. OVCAR3 immunoprecipitations and controls were performed using 3 mg of pre-cleared extract per reaction and precipitated with agarose beads in GST pull-down buffer for 3 h at 4 °C. Beads were washed 4 times in cold ELB.
Half-life experiments
HEK293T cells were transfected overnight with 1 µg pCDNA3FLAG-EYA2 and 3 µg pCMVCDK6, pCMVCDK6 mutants, or 3 µg pCMV empty vector. When used, 3 µg pCDNA3SIX1 was co-transfected in place of the pCMV vector. A total of 7 µg plasmid DNA were transfected onto 800 000 cells in a 6-well dish. After 20 h, media was removed and cells were fed fresh media. Media containing cycloheximide at 0.1 mg/ml was added to the monolayer, and cells were harvested at timepoints post-cycloheximide treatment, as indicated. Cells were lysed in ELB buffer, protein concentrations determined, and equal amounts of proteins loaded in each lane of an 8% SDS/PAGE gel. Proteins were transferred to PVDF membrane and probed with antibodies as indicated. Some blots were stripped with Thermo ReStore stripping reagent (Thermo/Fisher), as necessary for (size) overlapping signals. Blots were exposed to film and were also acquired on a Bio-Rad Chemi-Doc and analyzed with Quantity One 1-D analysis software (Bio-Rad), allowing quantitation of bands only in the linear range to allow accurate half-life calculations. Each experiment is representative of at least 3 independent repeats, and blots were probed with both N-16 and the Ford anti-EYA2 antibody, which has been shown to be specific for EYA2.19 Immunoblots of loading controls and co-expressed proteins are shown with each experiment.
Proteasome inhibitors
HEK293T cells were transfected as indicated in the figure using 1 µg of pCDNA3FLAG-EYA2 plasmid, 3 µg of pCMVCDK6 plasmid, and 4 µg of UbK0 plasmid, with the total of 8 µg being made up with pCMVneobam plasmid. Precipitates were left on for 20 h and MG132 was added to a concentration of 20 µM for 4 h prior to lysis. For lactacystin experiments, HEK293T cells were transfected as above and precipitates left on the cells for 18–20 h. Each transfected well was subsequently split equally into 2 wells, one of which was treated with 10 µM Lactacystin (Enzo) and one with DMSO control, both for 5 h. Cells were lysed in ELB with HALT protease inhibitors (Pierce) and 2 mM NEM (alfa Aesar). One hundred and fifty µg of extract was run on a 10% gel, transferred to nitrocellulose, and incubated with antibodies as noted. The short exposure was 1 min on film and the long exposure 12 min.
Fractionation
Cells were transfected, harvested 22 h later, lysed and fractionated according to the Nu-Per kit (Pierce), and 85 µg of protein were loaded in each lane. Fractionated extracts were loaded to 10% gels, transferred to PVDF membranes, and probed with the following antibodies: EYA2 with Abcam antibody, CDK6 with Pierce antibody, SIX1 with 4–2 antibody, polyclonal to HDAC (Pierce Scientific), and Tubulin (Sigma) antibody. Blots were stripped (Thermo ReStore) prior to exposure to SIX1 and tubulin.
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute(s) of the National Institutes of Health under award number R15CA125731-02A1. M Zimmer was supported under award number R15GM059108-04. HL Ford was supported by National Cancer Institute(s) award number R012RO1-CA095277.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26755
References
- 1.Hu MG, Deshpande A, Enos M, Mao D, Hinds EA, Hu GF, Chang R, Guo Z, Dose M, Mao C, et al. A requirement for Cyclin-dependent kinase 6 in thymocyte development and tumorigenesis. Cancer Res. 2009;69:810–8. doi: 10.1158/0008-5472.CAN-08-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malumbres M, Sotillo R, Santamaría D, Galán J, Cerezo A, Ortega S, Dubus P, Barbacid M. Mammalian cells cycle without the D-type Cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Matushansky I, Radparvar F, Skoultchi AI. Reprogramming leukemic cells to terminal differentiation by inhibiting specific Cyclin-dependent kinases in G1. Proc Natl Acad Sci U S A. 2000;97:14317–22. doi: 10.1073/pnas.250488697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matushansky I, Radparvar F, Skoultchi AI. Manipulating the onset of cell cycle withdrawal in differentiated erythroid cells with Cyclin-dependent kinases and inhibitors. Blood. 2000;96:2755–64. [PubMed] [Google Scholar]
- 5.Ericson KK, Krull D, Slomiany P, Grossel MJ. Expression of Cyclin-dependent kinase 6, but not Cyclin-dependent kinase 4, alters morphology of cultured mouse astrocytes. Mol Cancer Res. 2003;1:654–64. [PubMed] [Google Scholar]
- 6.Ogasawara T, Katagiri M, Yamamoto A, Hoshi K, Takato T, Nakamura K, Tanaka S, Okayama H, Kawaguchi H. Osteoclast differentiation by RANKL requires NF-kappaB-mediated downregulation of Cyclin-dependent kinase 6 (Cdk6) J Bone Miner Res. 2004;19:1128–36. doi: 10.1359/jbmr.2004.19.7.1128. [DOI] [PubMed] [Google Scholar]
- 7.Ogasawara T, Kawaguchi H, Jinno S, Hoshi K, Itaka K, Takato T, Nakamura K, Okayama H. Bone morphogenetic protein 2-induced osteoblast differentiation requires Smad-mediated down-regulation of Cdk6. Mol Cell Biol. 2004;24:6560–8. doi: 10.1128/MCB.24.15.6560-6568.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimoto T, Anderson K, Jacobsen SE, Nishikawa SI, Nerlov C. Cdk6 blocks myeloid differentiation by interfering with Runx1 DNA binding and Runx1-C/EBPalpha interaction. EMBO J. 2007;26:2361–70. doi: 10.1038/sj.emboj.7601675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu MG, Deshpande A, Schlichting N, Hinds EA, Mao C, Dose M, Hu GF, Van Etten RA, Gounari F, Hinds PW. CDK6 kinase activity is required for thymocyte development. Blood. 2011;117:6120–31. doi: 10.1182/blood-2010-08-300517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossel MJ, Hinds PW. From cell cycle to differentiation: an expanding role for cdk6. Cell Cycle. 2006;5:266–70. doi: 10.4161/cc.5.3.2385. [DOI] [PubMed] [Google Scholar]
- 11.Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RG, Schäfer M, Fajmann S, Schlederer M, Schiefer AI, et al. A Kinase-Independent Function of CDK6 Links the Cell Cycle to Tumor Angiogenesis. Cancer Cell. 2013;24:167–81. doi: 10.1016/j.ccr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, Zhai H, Vidal M, Gygi SP, Braun P, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell. 2011;20:620–34. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laman H, Funes JM, Ye H, Henderson S, Galinanes-Garcia L, Hara E, Knowles P, McDonald N, Boshoff C. Transforming activity of Fbxo7 is mediated specifically through regulation of Cyclin D/cdk6. EMBO J. 2005;24:3104–16. doi: 10.1038/sj.emboj.7600775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 15.Tadjuidje E, Hegde RS. The Eyes Absent proteins in development and disease. Cell Mol Life Sci. 2013;70:1897–913. doi: 10.1007/s00018-012-1144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Yang N, Huang J, Buckanovich RJ, Liang S, Barchetti A, Vezzani C, O’Brien-Jenkins A, Wang J, Ward MR, et al. Transcriptional coactivator Drosophila eyes absent homologue 2 is up-regulated in epithelial ovarian cancer and promotes tumor growth. Cancer Res. 2005;65:925–32. [PubMed] [Google Scholar]
- 17.Wu K, Li Z, Cai S, Tian L, Chen K, Wang J, Hu J, Sun Y, Li X, Ertel A, et al. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through Cyclin D1. Cancer Res. 2013;73:4488–99. doi: 10.1158/0008-5472.CAN-12-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Karoulia Z, Wong EY, Ahmed M, Itoh K, Xu PX. The phosphatase-transcription activator EYA1 is targeted by anaphase-promoting complex/Cdh1 for degradation at M-to-G1 transition. Mol Cell Biol. 2013;33:927–36. doi: 10.1128/MCB.01516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farabaugh SM, Micalizzi DS, Jedlicka P, Zhao R, Ford HL. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-β signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene. 2012;31:552–62. doi: 10.1038/onc.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey RN, Rani R, Yeo EJ, Spencer M, Hu S, Lang RA, Hegde RS. The Eyes Absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene. 2010;29:3715–22. doi: 10.1038/onc.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patrick AN, Cabrera JH, Smith AL, Chen XS, Ford HL, Zhao R. Structure-function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome. Nat Struct Mol Biol. 2013;20:447–53. doi: 10.1038/nsmb.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bierkens M, Krijgsman O, Wilting SM, Bosch L, Jaspers A, Meijer GA, Meijer CJ, Snijders PJ, Ylstra B, Steenbergen RD. Focal aberrations indicate EYA2 and hsa-miR-375 as oncogene and tumor suppressor in cervical carcinogenesis. Genes Chromosomes Cancer. 2013;52:56–68. doi: 10.1002/gcc.22006. [DOI] [PubMed] [Google Scholar]
- 23.Wang QF, Wu G, Mi S, He F, Wu J, Dong J, Luo RT, Mattison R, Kaberlein JJ, Prabhakar S, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossel MJ, Wang H, Gadea B, Yeung W, Hinds PW. A yeast 2-hybrid system for discerning differential interactions using multiple baits. Nat Biotechnol. 1999;17:1232–3. doi: 10.1038/70792. [DOI] [PubMed] [Google Scholar]
- 25.van den Heuvel S, Harlow E. Distinct roles for Cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–4. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 26.Kohrt DM, Crary JI, Gocheva V, Hinds PW, Grossel MJ. Distinct subcellular distribution of Cyclin dependent kinase 6. Cell Cycle. 2009;8:2837–43. doi: 10.4161/cc.8.17.9521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyerson M, Enders GH, Wu CL, Su LK, Gorka C, Nelson C, Harlow E, Tsai LH. A family of human cdc2-related protein kinases. EMBO J. 1992;11:2909–17. doi: 10.1002/j.1460-2075.1992.tb05360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossel MJ, Baker GL, Hinds PW. cdk6 can shorten G(1) phase dependent upon the N-terminal INK4 interaction domain. J Biol Chem. 1999;274:29960–7. doi: 10.1074/jbc.274.42.29960. [DOI] [PubMed] [Google Scholar]
- 29.Buller C, Xu X, Marquis V, Schwanke R, Xu PX. Molecular effects of Eya1 domain mutations causing organ defects in BOR syndrome. Hum Mol Genet. 2001;10:2775–81. doi: 10.1093/hmg/10.24.2775. [DOI] [PubMed] [Google Scholar]
- 30.Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19:6815–24. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patrick AN, Schiemann BJ, Yang K, Zhao R, Ford HL. Biochemical and functional characterization of six SIX1 Branchio-oto-renal syndrome mutations. J Biol Chem. 2009;284:20781–90. doi: 10.1074/jbc.M109.016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloom J, Pagano M. Experimental tests to definitively determine ubiquitylation of a substrate. Methods Enzymol. 2005;399:249–66. doi: 10.1016/S0076-6879(05)99017-4. [DOI] [PubMed] [Google Scholar]
- 33.Xiong W, Dabbouseh NM, Rebay I. Interactions with the Abelson tyrosine kinase reveal compartmentalization of eyes absent function between nucleus and cytoplasm. Dev Cell. 2009;16:271–9. doi: 10.1016/j.devcel.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo JT, Ding LH, Liang CY, Zhou NK, Ye QN. [Expression of EYA2 in non-small cell lang cancer] Zhonghua Zhong Liu Za Zhi. 2009;31:528–31. [PubMed] [Google Scholar]
- 35.Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–33. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen KL, Brennan JD, Aldridge CS, Ford HL. Cell cycle regulation of the human Six1 homeoprotein is mediated by APC(Cdh1) Oncogene. 2007;26:3406–14. doi: 10.1038/sj.onc.1210122. [DOI] [PubMed] [Google Scholar]
- 37.Ford HL, Landesman-Bollag E, Dacwag CS, Stukenberg PT, Pardee AB, Seldin DC. Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J Biol Chem. 2000;275:22245–54. doi: 10.1074/jbc.M002446200. [DOI] [PubMed] [Google Scholar]
- 38.Chang G, Guida C, Still WC. An internal-coordiante monte carlo method for searching conformational space. J Am Chem Soc. 1989;111:4379–86. doi: 10.1021/ja00194a035. [DOI] [Google Scholar]
- 39.Klossvary I, Keseru GM. Hessian-free low-mode conformational search for large-scale protein loop optimization: Application to c-jun N-terminal kinase JNK3. J Comput Chem. 2001;22:21–30. doi: 10.1002/1096-987X(20010115)22:1<21::AID-JCC3>3.0.CO;2-I. [DOI] [Google Scholar]
- 40.Saunders M, Houk K, Wu Y-D. Conformations of cycloheptadecane. A comparison of methods for confomational searching. J Am Chem Soc. 1990;112:1419. doi: 10.1021/ja00160a020. [DOI] [Google Scholar]
- 41.Embry AC, Glick JL, Linder ME, Casey PJ. Reciprocal signaling between the transcriptional co-factor Eya2 and specific members of the Galphai family. Mol Pharmacol. 2004;66:1325–31. doi: 10.1124/mol.104.004093. [DOI] [PubMed] [Google Scholar]
- 42.Fan X, Brass LF, Poncz M, Spitz F, Maire P, Manning DR. The alpha subunits of Gz and Gi interact with the eyes absent transcription cofactor Eya2, preventing its interaction with the six class of homeodomain-containing proteins. J Biol Chem. 2000;275:32129–34. doi: 10.1074/jbc.M004577200. [DOI] [PubMed] [Google Scholar]