Abstract
It has recently been established that exosomes can mediate intercellular cross-talk under normal and pathological conditions through the transfer of specific miRNAs. As muscle cells secrete exosomes, we addressed the question of whether skeletal muscle (SkM) exosomes contained specific miRNAs, and whether they could act as “endocrine signals” during myogenesis. We compared the miRNA repertoires found in exosomes released from C2C12 myoblasts and myotubes and found that 171 and 182 miRNAs were exported into exosomes from myoblasts and myotubes, respectively. Interestingly, some miRNAs were expressed at higher levels in exosomes than in their donor cells and vice versa, indicating a selectivity in the incorporation of miRNAs into exosomes. Moreover miRNAs from C2C12 exosomes were regulated during myogenesis. The predicted target genes of regulated exosomal miRNAs are mainly involved in the control of important signaling pathways for muscle cell differentiation (e.g., Wnt signaling pathway). We demonstrated that exosomes from myotubes can transfer small RNAs (C. elegans miRNAs and siRNA) into myoblasts. Moreover, we present evidence that exosome miRNAs secreted by myotubes are functionally able to silence Sirt1 in myoblasts. As Sirt1 regulates muscle gene expression and differentiation, our results show that myotube–exosome miRNAs could contribute to the commitment of myoblasts in the process of differentiation. Until now, myokines in muscle cell secretome provided a conceptual basis for communication between muscles. Here, we show that miRNA exosomal transfer would be a powerful means by which gene expression is orchestrated to regulate SkM metabolic homeostasis.
Keywords: exosomes, skeletal muscle, differentiation, microRNAs, Sirtuin 1
Introduction
microRNAs (miRNAs) are a class of evolutionally conserved non-coding RNAs of 19–22 nucleotides that function as negative regulators of gene expression. Originally discovered in C. elegans, these small RNAs regulate fundamental cellular processes in diverse organisms.1,2 miRNAs are encoded within the genome and are initially transcribed as primary transcripts (pri-miR) that can be several kilobases in length. Primary transcripts are successively cleaved by 2 RNase III enzymes, Drosha in the nucleus and Dicer in the cytoplasm, to produce 70 nucleotides long precursor miRNAs (pre-miR) and then mature miRNAs, respectively. Mature miRNAs regulate gene expression post-transcriptionally by binding to target mRNAs in association with the multiprotein RNA induced silencing complex (RISC).3 Three mechanisms have been described for gene regulation via miRNA (1) translation repression, (2) direct mRNA degradation, and (3) miRNA-mediated mRNA decay. Recent data have suggested that the mechanism of repression is predominantly via decrease in mRNA target stability.4 miRNA activity and abundance is also regulated at various levels, ranging from transcription and processing to target site binding and miRNA stability.5 Bioinformatic analyses indicate that miRNAs can regulate multiple target mRNAs, and individual mRNA can be targeted by several miRNAs, providing enormous complexity.6
Induction of the loss of all miRNAs in mice by depleting the enzyme Dicer causes arrested development during gastrulation before the body plans are fully configured.7 In addition, the levels of individual miRNAs are dramatically altered in different cell types and different developmental stages, confirming that miRNAs play a major role in cell growth, differentiation, and programmed cell death. As a consequence of this widespread influence, it is not surprising that miRNA deregulation is a hallmark of several pathological conditions including, cancer,8 inflammation,9 neurological disorders,10 and metabolic disorders.11
All these data largely assume that miRNAs reside and elicit their regulatory actions within their cell of origin. However, studies during the past 5 y have demonstrated that miRNAs are found not only intracellularly, but are also detectable outside of cells, including in various bodily fluids (e.g., serum, plasma, saliva, urine, and milk).12 This phenomenon raises questions about the biological function of such extracellular miRNAs. They are enclosed in small membranous vesicles (e.g., in exosomes, shedding vesicles and apoptotic bodies) or packaged with RNA-binding proteins (e.g., high-density lipoprotein, Argonaute 2, and nucleophosmin 1).13-16 In the latter case, extracellular miRNAs may represent byproducts of dead/dying cells that persist due to their stability within the protein complex. The extent to which free miRNAs act in a paracrine manner remains to be investigated. For miRNAs exported in exosomes, compelling evidence supports their role in a broad range of physiological and pathological processes.
Exosomes are 50–150-nm diameter membranous vesicles derived from the late endosomal system that are released from cells both constitutively and upon induction, under normal and pathological conditions.17 The endosomal system controls the uptake and processing of various types of macromolecules from the extracellular milieu and the plasma membrane into the cell. It consists of interconnected vesicular organelles, i.e., the primary endocytic vesicles, the early endosomes, the recycling endosomes, the late endosomes, and the lysosomes.18 Inward budding of endosomal membranes results in the accumulation of intraluminal vesicles. These late endosomes are called multivesicular bodies (MVBs). MVBs can either fuse with lysosomes to degrade their intraluminal cargo or fuse with the plasma membrane to release their intraluminal vesicles, as exosomes, into the extracellular milieu. This process of directed transport relies on several components of the endocytic machinery, such as Rab GTPases (Rab11, Rab27a and b), cytoskeleton regulatory proteins, molecular motors such as myosin, and SNAREs.19 Exosomes bear surface receptors/ligands of the original cells and have the potential to selectively interact with specific target cells.17,20,21
In addition to numerous lipids and proteins, exosomes contain mRNAs and miRNAs.22-24 Previous studies have demonstrated that exosomes can horizontally transfer mRNAs to other cells, which can then be translated into functional proteins in the new location.22,23,25 Similarly, miRNAs can be transferred by an exosomal route and further exert gene silencing in the recipient cells.22 These findings shed new light on the physiological relevance of secretory genomic information by exosomes, and indicate a role of exosomes as new mediators of intercellular cell signaling between neighboring cells and between distant tissues, which could act independently, but synergistically, with soluble growth factors and hormones.
Skeletal muscle (SkM), the largest organ in the human body, is responsible for whole-body metabolism, energy homeostasis and locomotion, and serves as a body protein pool. It is a highly adaptable tissue responding to numerous environmental and physiological challenges by changing its phenotypic profile in terms of size as well as composition. During the last decade, SkM-secreted proteins have been identified with important roles in intercellular communications.26-28 Among them, a large number of soluble peptide hormones and cytokines, called myokines, are capable of triggering homeostasis adaptations in other peripheral organs (e.g., pancreas, adipose tissue),26 or are involved in the process of myogenesis (e.g., IL-4, IL-7, and IL-13).27,28 Recently, it has been demonstrated that muscle cells secrete nanoparticles with exosomal properties.29,30 Because exosomes released from donor cells can be taken up by recipient cells, where an array of biological processes, including cell proliferation and differentiation, can be affected,31,32 we examined whether skeletal muscle exosomes contained specific miRNAs, and whether these miRNAs could act as endocrine signals during myogenesis. Using qRT-PCR, we analyzed and compared the miRNA repertoires within exosomes from C2C12 myoblasts and myotubes. We then determined whether exosomes could transfer miRNAs from myotubes to myoblasts and thus regulate gene expression in the recipient cells. By using Sirtuin 1 (silent mating type information regulation 2 homolog) (Sirt1), the reporter gene, we present evidence that exosome miRNAs secreted by myotubes are able to silence Sirt1 in myoblasts. As Sirt1 regulates muscle gene expression and differentiation,33 our results show that myotube-exosome miRNAs could contribute to the commitment of myoblasts in the process of differentiation.
Results
In this study we purified exosomes from conditioned media of C2C12 myoblasts and myotubes by differential centrifugations coupled with membrane filtration (0.2 μm) to eliminate large contaminating extracellular vesicles. We found that myoblasts and myotubes secreted, respectively, 0.37 + 0.15 and 0.41 + 0.23 μg /ml/24h exosomes in exosome-free medium (results from 6 independent extractions). Electron microscopic analysis showed that the resulting pellet contained a homogenous population of nanovesicles at the expected size of 50–150 nm, which expressed the exosomal marker CD63, CD81, and Alix (Fig. 1).34 We purified total RNA from 3 independent preparations of exosomes, and analyzed their miRNA populations by real-time quantitative PCR.
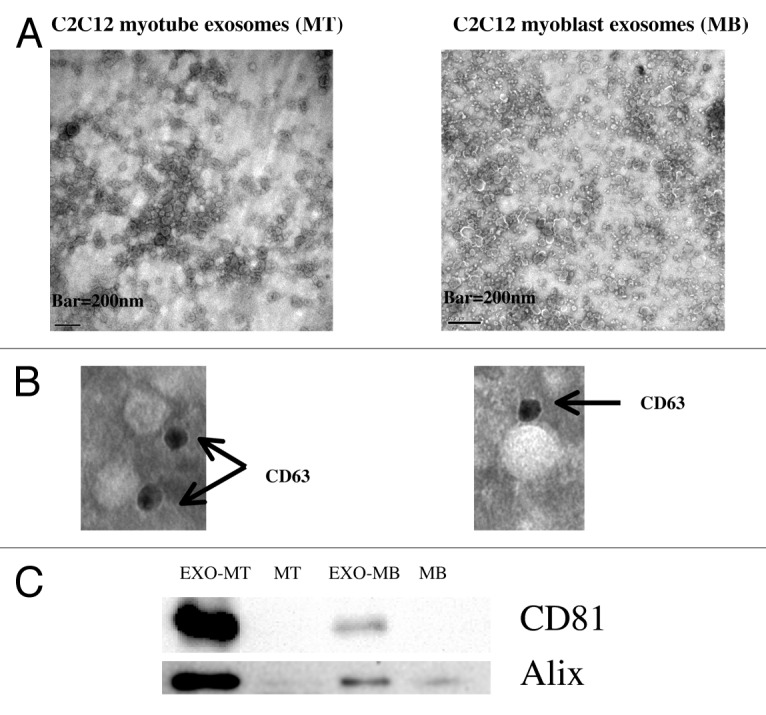
Figure 1. Characterization of nanovesicles isolated from C2C12 myoblast- and myotube-conditioned medium. (A) Transmisson electron microscopic images of purified secreted nanovesicles from myoblasts or myotubes. Bar = 200 nm. (B) These vesicles were labeled with anti-CD63 gold particles to confirm that they expressed this exosomal protein at their membranes. (C) Equal protein amounts of extracts prepared from cells or exosomes were subjected to western blot analysis. The multivesicular body marker Alix (ALG2-interacting protein 1) and the tetraspanin CD81 were strongly enriched in exosome preparations compared with cell lysates.
miRNA profiling in C2C12 cells and in released exosomes
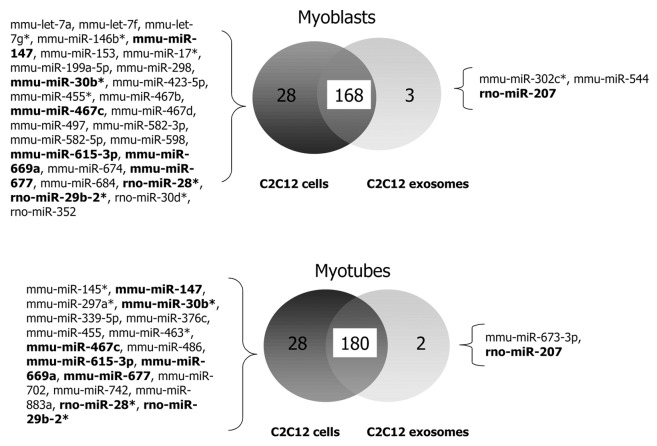
For each miRNA, the threshold cycle (Ct) was calculated by the ABI 7900 Sequence Detection System software. Raw Ct values considered “undetermined” by the software or at a level ≥40 cycles were excluded from analysis. For each TaqMan Low-Density Array, quality controls were performed on the raw data by checking internal controls and using box plot diagrams. Since the currently used normalization factor mammU6 plotted in each card was not stably expressed in our different samples, we used the mean expression level of all fully observed (fully detected) miRNAs for normalization.35,36 Comparisons between groups were made by using the Student t myotubes, or cells vs. exosomes). Using these criteria, 171 and 182 miRNAs were detected in myoblasts and myotubes, respectively (Fig. 2). The majority of the miRNAs expressed in C2C12 cells were sorted into exosomes (Fig. 2). In addition, exosomes from C2C12 cells also contained snoRNA135 and snoRNA202, small RNA molecules that primarily guide chemical modifications of other RNAs (e.g., rRNAs and tRNAs), and scRNA Y1, a small cytoplasmic RNA and component of the Rho ribonucleoprotein complex.
Figure 2. Venn diagrams showing miRNA profile overlaps between C2C12 myoblasts and myotubes and exosomes. miRNAs commonly detected in C2C12 myoblasts and myotubes are indicated in bold.
Comparisons of the subsets of miRNAs exported in exosomes with the total population of miRNA expressed in C2C12 cells enabled identification of 2 groups of 28 miRNAs which were not detected in exosomes secreted from myoblasts and myotubes (Fig. 2). Among them, miR-147, miR-30b*, miR-467c, miR-615–3p, miR-669a, miR-677, miR-28*, and miR-29–2* were never found in C2C12-secreted exosomes. Conversely, 3 and 2 miRNAs, respectively, were not detected in C2C12 muscle cells by qRT-PCR but were found in myoblast- and/or myotube-secreted exosomes. In addition, 23 and 20 miRNAs, respectively, displayed higher expression in myoblast- and myotube-secreted exosomes compared with C2C12 cells, estimated by the mean of Ct values after data normalization (Table S1).
Together, these results indicate that C2C12 muscle cells exported only a sub-population of miRNAs into exosomes. In addition, some miRNAs could not be identified by qRT-PCR in C2C12 cells even using a pre-amplification step before the PCR reaction. To determine whether these results were specific to the C2C12 murine muscle cells, we compared this population of miRNAs to the population of miRNAs identified previously in exosomes from other murine cell lines (i.e., immature and mature bone marrow [BM] cells, and mast cells [MC9]).22,37 We identified a sub-group of 21 miRNAs (let-7b, let-7c, let-7d, let-7i, miR-106b, miR-130b, miR-146b, miR-150, miR-15b, miR-16, miR-191, miR-20a, miR-21, miR-222, miR-23b, miR-24, miR-26a, miR-27b, miR-29a, miR-30c, miR-320) that were always exported, regardless cell type and state of differentiation.22,37 On the other hand, miR-147, miR-467c, and miR-669a were never exported in murine exosomes.
miRNAs in C2C12 exosomes are regulated during myogenesis
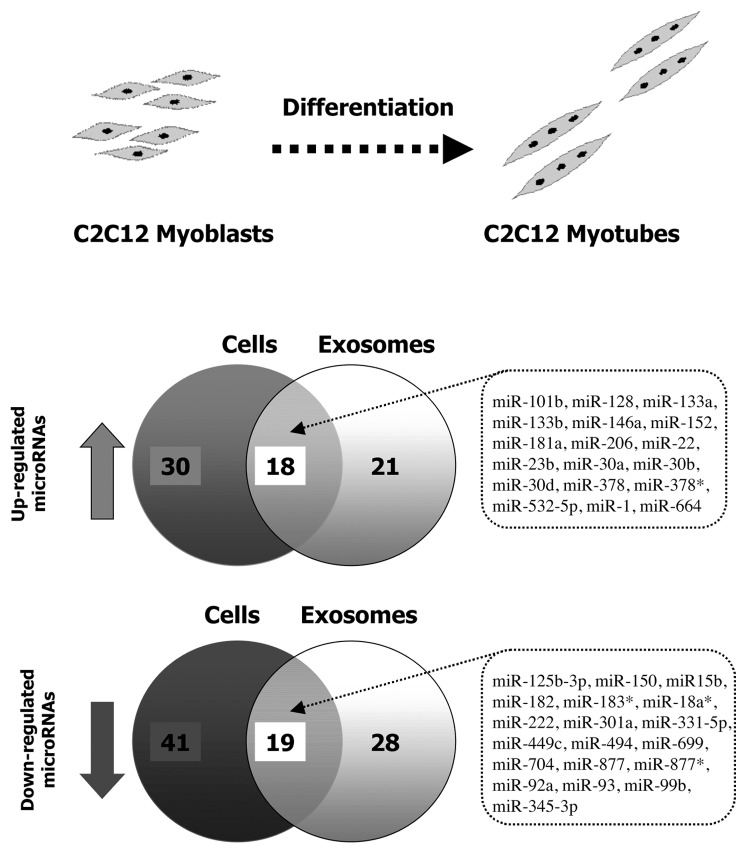
miRNAs are regulated during muscle cell myogenesis.38 We predicted that miRNAs exported into exosomes might be similarly regulated. As shown in Figure 3, 108 miRNAs were differentially expressed between myoblasts and myotubes, (i.e., 48 were up- and 60 were downregulated during the process of C2C12 differentiation). Among these were miR-1, miR-133a, miR-133b, miR-206, miR-222, miR-223, and miR-126, all previously identified for their important roles during muscle cell differentiation.39-41 We found that 86 miRNAs expressed in exosomes were also differentially expressed between exosomes from C2C12 myoblasts and exosomes from C2C12 myotubes (i.e., 39 were up- and 47 were down regulated) (Fig. 3). Among them, 18 were up- and 19 were downregulated both in C2C12 cells and exosomes during differentiation (Fig. 3). One miRNA, let-7d, was inversely regulated in exosomes compared with cells during differentiation (down vs. up).
Figure 3. miRNAs regulations in C2C12 cells and in secreted exosomes during differentiation.
Functional analysis of target genes of exported miRNAs
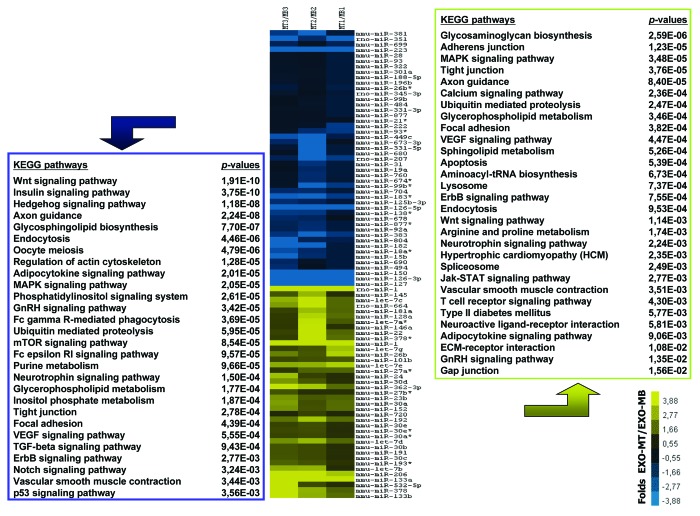
Target genes of the 86 exosome-secreted miRNAs regulated during myoblast differentiation were predicted by bioinformatics using TargetScan 6.1 (http://www.targetscan.org/). We focused this study on miRNA binding sites conserved among species in order to reduce the number of false positive target genes. Among the 47 downregulated miRNAs 31 had predicted target genes (n = 4641 genes). Among the 39 upregulated miRNAs 31 also had predicted target genes (n = 4232 genes). Genes that were targeted by both up- and downregulated exosome-miRNAs were removed to functional analysis. Functional analysis of exosome-secreted miRNA target genes were thus determined on 2021 genes for downregulated miRNAs and 1612 genes for the upregulated miRNAs, by using Babelomics 4.3 (http://babelomics.bioinfo.cipf.es). For the functional analysis of the predicted target genes, we analyzed all miRNA target genes collectively in order to have a general picture of their cellular functions. Figure 4 shows KEGG pathways that are most significant in terms of containing more genes than expected (P < 0.05). The majority of the significant categories were involved in signaling pathways.
Figure 4. KEGG pathways significantly enriched in target genes for upregulated miRNAs (blue) or downregulated miRNAs (yellow) in exosomes from C2C12 muscle cells during differentiation. Only pathways containing at least 10 genes are considered. Hierarchical clustering groups the miRNAs according to their fold changes during myogenesis (expression in myotubes vs. expression in myoblasts: MT/MB). Three independent preparations of exosomes were analyzed.
Myotube exosomes can transfer small RNA in myoblasts
As exosome miRNA profiles from myoblasts and myotubes were different, we postulated that these secreted miRNAs would have specific roles during the process of myogenesis, and that transfer of specific miRNAs could occur between myotubes and myoblasts. Because exosomes can promote the transfer of exogenous siRNAs,42 which are similar in length to endogenous miRNAs, we decided to use C2C12 EXO-MT as a vehicle for siRNAs. We transfected differentiated myotubes with siRNA against Itga7, a muscle-expressed integrin.43 We verified that Itga7 expression was reduced in C2C12 myotubes at the protein and mRNA level (Fig. S1) and purified the exosomes released. C2C12 myoblasts were incubated in the presence of 2 μg EXO-MT from siRNA Itga7-treated cells or with control exosomes, in one ml of culture medium. As shown in Figure 5A, the mRNA level of Itga7 in myoblasts was significantly reduced in the presence of EXO-MT from siRNA Itga7-treated cells, indicating that these exosomes could transfer siRNA Itga7 from myotubes to myoblasts and, as a consequence, could decrease the expression of Itga7 in myoblasts. We performed a similar experiment but with a synthetic C. elegans miRNA, cel-miR-238, which shows no sequence homology to any known human miRNAs and has no binding sites on human mRNAs. C2C12 myoblasts were incubated either with EXO-MT from cel-miR-238-transfected myotubes or with control myotube-exosomes. As shown in Figure 5B, cel-miR-238 was detected by qRT-PCR in exosomes released by cel-miR-238-transfected myotubes and in myoblasts incubated with these exosomes.
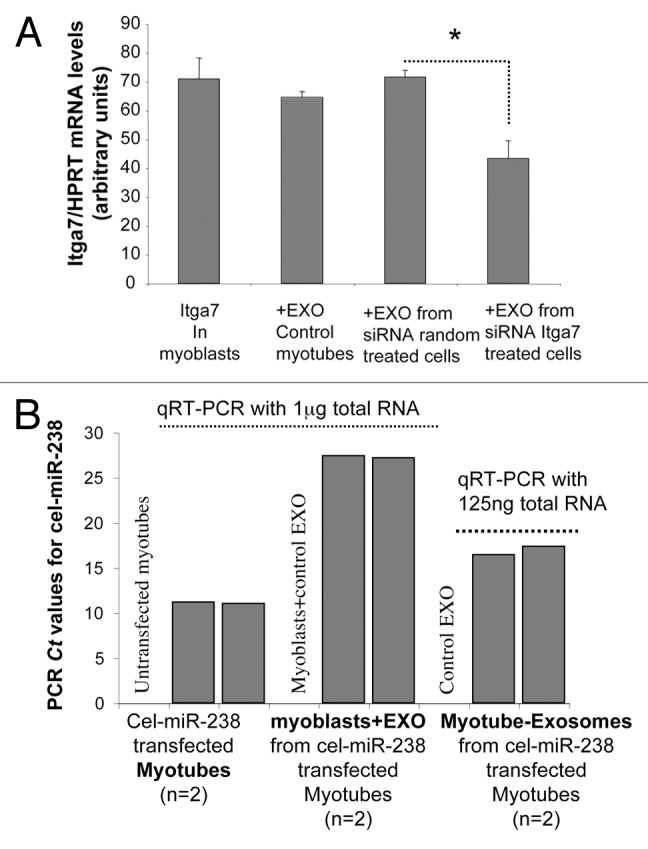
Figure 5. Exosomes from myotubes can transfer small RNAs in proliferating myoblasts. (A) Levels of Itga7 mRNA in C2C12 myoblasts, treated for 48 h with exosomes (2 μg/ml of culture medium) released either from myotubes transfected with siRNA Itga7 or with random siRNAs. Exosomes from untransfected myotubes were also used as control. Data are expressed as mean ± SEM. Results were normalized with the gene encoding HPRT used as the reference. Comparisons were analyzed using Student t test. Significance was defined as P value of ≤0.05. (B) Ct (cycle threshold) level of synthetic cel-miR-238 in transfected myotubes or in their released exosomes, and in myoblasts incubated with myotube-exosomes from cel-miR-238-transfected myotubes (n = 2 independent biological replicates). The Ct is defined as the number of cycles required for the fluorescent signal to cross the threshold (i.e., exceeds background level). Ct levels are inversely proportional to the amount of target nucleic acid in the sample (i.e., the lower the Ct level the greater the amount of target nucleic acid in the sample).
These data indicate that exosomes can horizontally transfer siRNAs and miRNAs from myotubes to myoblasts. This data supports a recent published paper (Le Bihan et al. 2012),30 which showed the incorporation of fluorescent myotube-exosomes into myoblasts, and further demonstrated that exosome small RNAs are not degraded in the recipient cells as they are detectable by qRT-PCR and are functional.
Myotube miRNAs exported in exosomes can regulate Sirt1 in myoblasts
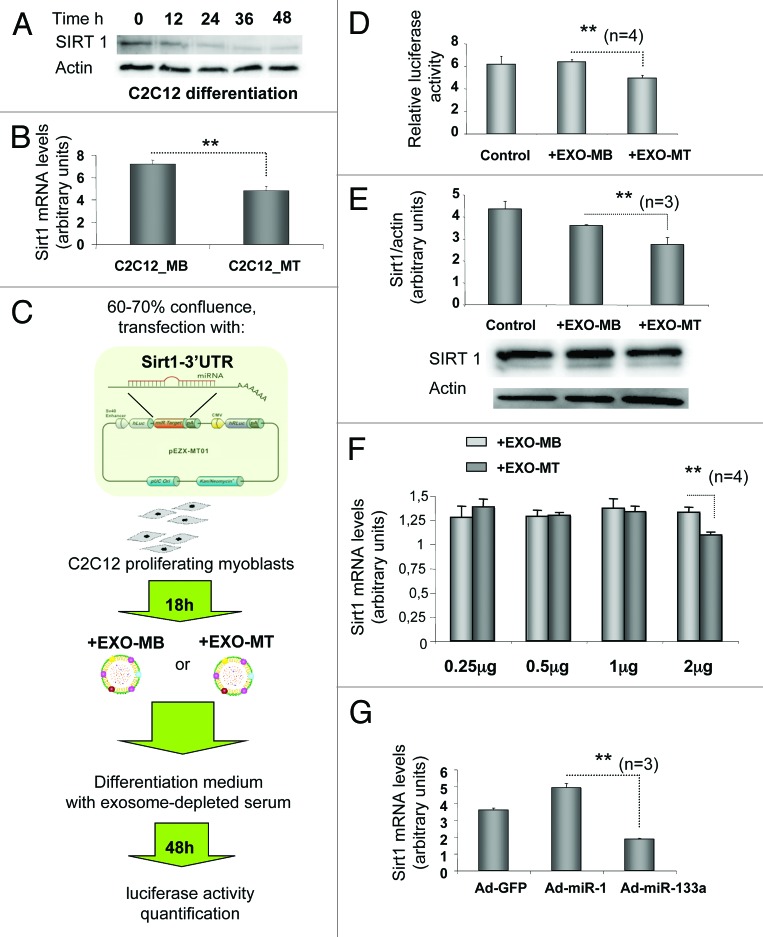
We then determined whether EXO-MT miRNAs could regulate the expression of genes in myoblast cells. We previously found that combinations of different miRNA binding sites in 3′-UTRs have synergistic effects on gene expression;44 thus, we focused this study on the genes targeted by multiple exosome miRNAs. The list of the top 20 genes putatively targeted by different miRNAs, which are regulated during myogenesis (Fig. 3), is given in Table 1. Sirt1, a NAD+-dependent histone deacetylase enzyme which is an important protein for muscle precursor cell proliferation,45 myoblast survival,46 and muscle differentiation33 was included in this list. We found that Sirt1 is decreased during C2C12 differentiation at both protein and mRNA levels (Fig. 6A and B), confirming previously published results in C2C12s.33 Numerous miRNAs can regulate Sirt1 and, among them, miR-22 and miR-181a were expressed in EXO-MT (Fig. 4).47 To demonstrate that EXO-MT miRNAs could collectively regulate gene expression in myoblasts, we performed luciferase reporter assays with full-length 3′-UTR constructs of Sirt1 (Fig. 6C). As shown in Figure 6D luciferase activity was significantly reduced when myoblasts were incubated with EXO-MT (2 μg/ml of culture medium). This result correlated with a reduced SIRT1 expression at both protein and mRNA levels in myoblasts (Fig. 6E and F). By contrast, addition of EXO-MB at the same concentration had no effect on Sirt1 3′-UTR luciferase activity. Since miR-133a is one of the most upregulated miRNA in exosomes during myoblast differentiation (Fig. 4) and has a conserved predicted binding site in Sirt1 3′UTR-region (www.targetscan.org), we determined whether this miRNA could participate in the down-reduction of SIRT1. Overexpression of miR-133a in C2C12 cells led to the downregulation of Sirt1 mRNA (Fig. 6F). miR-1, which is also highly expressed in EXO-MT, but with no predicted binding site for SIRT1, did not induce this downregulation. These results indicate that miR-133a, contained in EXO-MT, may participate in the downregulation of Sirt1 in myoblasts.
Table 1. List of the 20 genes targeted by at least 6 exosome microRNAs, up- or downregulated during C2C12 myoblast differentiation.
| Target genes of downregulated microRNAs | Number of microRNA binding sites in 3′-UTR | Name |
|---|---|---|
| MYT1L | 8 | myelin transcription factor 1-like protein |
| FAT3 | 8 | FAT tumor suppressor homolog 3 |
| SPRY3 | 8 | sprouty homolog 3 |
| CACNB1 | 7 | calcium channel, voltage-dependent, β 1 subunit |
| FAM116A | 7 | family with sequence similarity 116, member A |
| KIF1B | 7 | kinesin family member 1B |
| TACC1 | 7 | transforming, acidic coiled-coil containing protein 1 |
| MYO1C | 6 | myosin IC |
| MTMR3 | 6 | myotubularin related protein 3 |
| RAPGEF5 | 6 | Rap guanine nucleotide exchange factor (GEF) 5 |
| PTPRE | 6 | protein tyrosine phosphatase, receptor type, E |
| C9orf69 | 6 | chromosome 9 open reading frame 69 |
| GTDC1 | 6 | glycosyltransferase-like domain containing 1 |
| FBXO10 | 6 | F-box protein 10 |
| FGF7 | 6 | fibroblast growth factor 7 |
| MAP2K1 | 6 | mitogen-activated protein kinase kinase 1 |
| SNTB2 | 6 | syntrophin, β 2 |
| SMURF1 | 6 | SMAD specific E3 ubiquitin protein ligase 1 |
| WAC | 6 | WW domain containing adaptor with coiled-coil |
| WDR32 | 6 | DDB1 and CUL4 associated factor 10 |
| Target genes of upregulated microRNAs | Number of microRNA binding sites in 3′-UTR | Name |
|---|---|---|
| RAB15 | 11 | Ras-related protein Rab-15 |
| GATM | 11 | glycine amidinotransferase |
| P4HA2 | 10 | prolyl 4-hydroxylase, α polypeptide II |
| MTX3 | 10 | metaxin 3 |
| PDP2 | 10 | pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| PICALM | 10 | phosphatidylinositol binding clathrin assembly protein |
| KREMEN1 | 10 | kringle containing transmembrane protein 1 |
| SURF4 | 10 | surfeit 4 |
| UBXD8 | 10 | Fas associated factor family member 2 |
| SCYL3 | 10 | SCY1-like 3 (S. cerevisiae) |
| RFXDC1 | 10 | regulatory factor X, 6 |
| SLC30A4 | 10 | solute carrier family 30 (zinc transporter), member 4 |
| COL9A3 | 10 | collagen, type IX, α 3 |
| B3GAT1 | 10 | β-1,3-glucuronyltransferase 1 |
| CASP3 | 10 | caspase 3, apoptosis-related cysteine peptidase |
| MXD1 | 9 | MAX dimerization protein 1 |
| PDCD10 | 9 | programmed cell death 10 |
| HOXB4 | 9 | homeobox B4 |
| MAB21L1 | 9 | mab-21-like 1 (C. elegans) |
| SIRT1 | 9 | sirtuin 1 |
Target genes were determined by using the software TargetScan 6.0.
Figure 6. Myotube-exosome miRNAs can silence Sirtuin1 in proliferating myoblasts. (A) SIRT1 and Actin protein detection during C2C12 differentiation (from 0 to 48 h post-induction). (B) mRNA level of Sirt1 during C2C12 differentiation (from 0 to 48 h post-induction). Data are expressed as mean ± SEM. Results were normalized with the gene encoding HPRT used as the reference (2 μg of total RNA was used for qRT-PCR analysis). (C) Summary of the protocol used to transfect proliferating C2C12 myoblasts with the plasmid containing the Sirt1 3′-UTR region. (D) Sirt1 relative luciferase activity 48 h after induction of differentiation in C2C12 myoblasts pre-treated with EXO-MB or EXO-MT (2 μg/ml of medium). (E) Western blot analysis of SIRT1 and Actin protein expressions in C2C12 myoblasts pre-treated with EXO-MB or EXO-MT (2 μg/ml of medium), 48 h after induction of differentiation. Quantification was realized with Image Lab Software 3.0 (Bio-Rad Laboratories). (F) mRNA level of Sirt1 48 h after induction of differentiation. Cells had been pre-treated with different concentrations of exosomes (μg/ml of medium) from either myoblasts or myotubes, at the begining of proliferation (0.7 μg of total RNA was used for qRT-PCR analysis). (G) mRNA level of Sirt1 transfected with adenovirus expressing mature miR-133a or miR-1, or GFP as control (2 μg of total RNA was used for qRT-PCR analysis). EXO-MB, myoblast-secreted exosomes collected during proliferation; EXO-MT, exosomes from 1-wk-differentiated myotubes. Comparisons were analyzed using Student t test. Significance was defined as P value of ≤0.05 (**).
Discussion
Almost all aspects of skeletal muscle (SkM) development and homeostasis are regulated by miRNAs either directly or indirectly.38 Several muscle-specific miRNAs have been found to be important for normal myoblast differentiation, proliferation, and muscle remodeling in response to stress and hormones.44,48 In addition to these intracellular regulations, the data presented in this study establish for the first time that miRNAs from muscle cells can also be sorted into exosomes and are likely to be involved in muscle cell differentiation.
Among the exported miRNAs, 37 were regulated in the same way as in the C2C12 cells during myoblast differentiation (18 up- and 19 downregulated). Exosome-upregulated miRNAs included miR-181a, 146a, miR-145, miR-1, miR-24, miR-206, miR-133a, miR-133b, and miR-378, which are well known for their intracellular actions in control of muscle cell differentiation,39,49-54 along with let-7d and let-7e, which are implicated in cell growth arrest.55,56 The population of exosome-downregulated miRNAs included miR-381 and miR-93, involved in cell proliferation.57,58 As exosome miRNA profiles changed during muscle cell differentiation, we postulated that these exported miRNAs would have a role in the process of myogenesis. It has been previously found that exosomes can transfer miRNAs between different cell types and may silence gene expression in the recipient cells.22,37,59,60 Our results showed that myotube exosomes can transfer small RNAs into myoblasts, strongly suggesting that during myogenesis, myotubes and myoblasts could transfer regulatory genetic information.
Importantly, our data also demonstrated that not all miRNAs can be incorporated into exosomes. Surprisingly, some miRNAs were inversely regulated in exosomes compared with their cytoplasmic regulation during myogenesis. Similar observations have been made by Montecalvo and coworkers,37 who demonstrated that exosomes isolated from murine dendritic cell (DC) culture media enclosed >200 miRNAs, with 5 uniquely detected in exosomes from immature DCs and 58 exclusively present in exosomes from mature DCs. Among them, we identified 21 miRNAs that were also exported in exosomes from C2C12 muscle cells. In addition, we found 3 miRNAs that were never exported in exosomes from C2C12 and dendritic cells (i.e., miR-147, miR-467c, and miR-669a) indicating that a common mechanism for selective miRNA export may exist. Recent data confirmed that selective miRNA export exists also for human cell lines, and thus it is evolutionarily conserved between distant cell types.59,61,62 Presently, it is not known how this export is regulated.
The earliest role proposed for exosomes was to shed unwanted proteins from cells undergoing terminal differentiation, in order to reduce their intracellular concentrations. This could also be the case for miRNAs, as we observed that miR-133a, miR-133b, miR-1, and miR-206 have the highest increase of expression concomitantly in cytoplasm and in exosomes during C2C12 differentiation (data not shown). It could also be possible that cells export unwanted miRNAs. For example, the highly expressed liver-specific miR-122 is weakly detected in human renal HEK293T cells, but is highly enriched in their released exosomes.63 Other studies support the concept that association with proteins of the RNA-induced silencing complex (RISC) controls the packaging of miRNAs in exosomes,64 because components of the RISC complex have been detected in exosomes isolated from monocytes64 and reticulocytes65 (e.g., AGO2). In addition, association of miRNAs with AGO2 complexes plays a critical role in stabilizing miRNAs in cell-released exosomes.63 However, of the 146 studies reported in the Exosome database (http://www.exocarta.org/), only one reported the identification of AGO2 using a proteomic approach.65 Moreover, we recently performed a comparative proteomic analysis of myoblast and myotube exosomes and did not detect any protein related to the RISC complex in exosomes from C2C12 muscle cells (data not shown). A recent study demonstrated that ceramide, whose biosynthesis is tightly controlled by neutral sphingomyelinase 2 (nSMase2), regulated the secretion of exosomal miRNAs. It also provided evidence that the endosomal sorting complex required for the transport system (ESCRT) was unnecessary for the release of miRNAs.59,66
Bioinformatic analysis revealed that predicted target genes of the miRNAs regulated in exosomes during differentiation were mainly involved in the control of signaling pathways. The Wnt signaling pathway was predicted as the most significant targeted pathway by those exosome-miRNAs that were downregulated during myogenesis. It is known that several Wnt signaling components are upregulated during the transition from cell proliferation to myogenic differentiation,67 and that some miRNAs are implicated.68 Based on our data, we suggest that part of this upregulation would come from the concomitant decreased expression of both intracellular and exosome repressor miRNAs. To corroborate this hypothesis, we demonstrated that exported miRNAs from muscle cells were collectively functional. Using an “in silico” approach, we determined the target genes of the myotube-secreted miRNAs and focused on those with putative multiple binding sites in their 3′-UTR regions. Sirt1, a gene involved in muscle cell proliferation33 and predicted to be targeted by 6 different miRNAs present in myotube exosomes (2 of them being experimentally validated47), was used as reporter gene. Our data showed that miRNAs from myotube exosomes negatively regulated the expression of Sirt1 in myoblasts. As reduction of the level of endogenous Sirt1 augments muscle gene expression and causes increased cell differentiation,33 our result supported the concept that the exosomal transfer of silencing RNAs may potentially be a new powerful means of orchestrating gene expression during myogenesis. Besides its important role in myogenesis,33 Sirt1 is also involved in mitochondrial biogenesis and fatty acid oxidation and provides a link between energy homeostasis and SkM growth and development.69
Finally, we have found that exosomes from myotubes are able to transfer other small RNAs, like siRNA or synthetic miRNAs, in myoblasts, raising very exciting possibilities for therapeutic uses. Current techniques for small RNA transfer use viruses or synthetic compounds as delivery vehicles. The use of exosomes to deliver siRNA and miRNA would potentially be better tolerated by the immune system and would also be useful to target a specific tissue. Recently, Alvarez-Erviti and coworkers (2011) were able to target exosome transfer of siRNAs from dendritic cells to the brain, by expressing specific neuron-targeting proteins at the dendritic-secreted exosome surface.42 In our study, we observed that the muscle-specific integrin ITGA7 was exported in exosomes from myotubes (Fig. S1). This integrin has never been described in exosomes from other tissues or other cell types (see Exocarta, http://www.exocarta.org/). Recently it has been found that the exosomal tetraspanin web contributes to target cell selection.21,70 It is thus tempting to speculate that expressing specific muscle integrin on the surface of exosomes might permit specific targeting of the skeletal muscle for delivering either miRNAs or siRNAs.
Conclusions
It is well known that SkM is an endocrine organ, which, through secretion of hormone-like factors, may influence metabolism in tissues and organs.26-28 Until now, myokines from the muscle cell secretome provided a conceptual basis to explain how muscles communicate with other organs.71 In this study, we demonstrated for the first time that SkM secreted exosomes, which contain specific and transferable miRNAs, can act as “endocrine-like” signals. Previous studies on cancer cells have shown that exosomal miRNA expression profiles have signatures related to tumor classification, diagnosis, and disease progression.72 It is thus tempting to speculate that during insulin-resistance, or other metabolic disorders affecting muscle physiology, SkM would release a new class of exosomes containing specific populations of miRNAs different to those in normal muscle cells, which could modify the inter-cellular message between SkM and other tissues, or could locally amplify the disease. Additional studies are now required to demonstrate the role of exosome miRNAs in muscular pathologies.
Material and Methods
C2C12 culture conditions
C2C12 mouse myoblasts were maintained in DMEM (4.5 g/l glucose) supplemented with 10% heat-inactivated FBS, 1000 UI/ml penicillin, 1000 UI/ml streptomycin, and 2 mM L-glutamine at 37 °C in humidified air containing 5% CO2. Differentiation was induced with 2% horse serum (HS). FBS and HS were depleted of their exosomes by centrifugation at 110 000 g overnight at 4 °C. The supernatant was passed through a 0.22-μm filter and diluted with sterile DMEM. The exosome-depleted serum was used to grow the cells when conditioned media were collected for exosome purification.
Isolation of myoblasts and myotubes exosomes
Exosomes were purified from 200 ml of the C2C12 myoblast or myotube conditioned media. Briefly, cell debris and organelles were eliminated at 2000 g for 20 min and at 10 000 g for 30 min. The resulting supernatant was filtered through a 0.22-μm filter. Exosomes were pelleted by ultracentrifugation at 100 000 g for 70 min +4 °C (Beckman-Coulter, Optima™ L-80-XP ultracentrifuge, type 50–2Ti rotor). The nanovesicle pellet was washed with 25 ml of cold PBS in order to minimize sticking and trapping of non-exosome materials and resuspended in 50 μl PBS. Exosomal protein content was quantified using the Bradford protein assay, and in this study, exosome concentrations are expressed as μg of protein-containing exosomes.
Transmission electron microscopy
Exosomes in PBS were adsorbed on 200 mesh nickel grids coated with formar-C. Immunogold labeling was performed by flotation of grids on drops of reactive media. Non-specific sites were coated with 1% BSA in 50 mM Tris–HCL, pH 7.4 for 10 min at RT. Antibody incubation was performed for 4 h at 4 °C in a wet chamber with mouse monoclonal antibody raised against CD63 (sc-15363) antibodies (Santa Cruz Biotechnology) (dilution 1/50) in 1% BSA, 50 mM Tris-HCL, pH 7.4. Grids were successively washed once in 50 mM Tris–HCL, pH 7.4, and pH 8.2 at RT. They were then preincubated with 1% BSA in 50 mM Tris–HCL, pH 8.2 for 10 min at RT and labeled with a goat anti mouse IgG gold-conjugated 10 nm, (Tebu bio) diluted 1/80 in 1% BSA-, 50 mM Tris-HCL, pH 8.2 in a wet chamber for 45 min. Grids were successively washed once in 50 mM Tris–HCL, pH 8.2 then pH 7.4, and in filtrated distilled water at RT. Grids with suspensions were colored with 2% phosphotunstic acid for 2 min and examined using a JEM Jeol 1400 transmission electron microscope equipped with a Orius 600 camera.
Western blotting
Proteins from exosomes or from cellular lysates were migrated on SDS-PAGE gels (30 μg). Following electrophoresis, proteins were transferred onto nitrocellulose PVDF membranes blocked at room temperature with 4% BSA in Tris-buffered saline/0.3% Tween20 and incubated overnight at 4 °C, with gentle shaking with anti-CD81 (sc-166028) or anti-Alix (sc-49268) antibodies from Santa Cruz Biotechnology. The signal was detected by using a horseradish peroxidase-conjugated secondary antibody and revealed with the enhanced chemiluminescence system (Pierce).
Total RNA extraction from C2C12 cells and exosomes
Total RNA was extracted from either C2C12 myoblasts and myotubes or exosomes, in triplicate, by using TriPure Isolation Reagent (Roche Applied Science). RNA was quantified with a NanoDrop spectrophotometer (Labtech).
qRT-PCR
Real-time RT-PCR was performed using ABsolute QPCR SYBR Green ROX Mix (Abgene, Courtaboeuf) with a Rotor-Gene 6000 system (Corbett Life Science). ITGA7 primers were purchased from Qiagen (Quantitect Primer Assay, QT10003135), and SIRT1 primers were S-GATAAGACGT CATCTTCAGA G and AS-TGAGAAAATG CTGGCCTAAT A. Results were normalized with the gene encoding HPRT (Hypoxanthine phosphoribosyltransferase 1) (S-AGTTGAGAGA TCATCTCCA and AS-TTGCTGACCT GCTGGATTAC). Data are expressed as mean ± SEM. Comparisons were analyzed using Student t test. Significance was defined as P ≤ 0.05. Cel-miR-238 was quantified by using TaqMan® miRNA Assays (LifeTechnologies).
Quantification of mature miRNAs
Expression of mature miRNAs was measured in triplicate by using the TaqMan® Low-Density Arrays V2 with Applied Biosystems 7900HT Fast Real-time PCR system. Briefly, 100 ng of total RNA were used for each multiplex reverse transcription (RT). Each RT reaction was diluted 62.5-fold and mixed with 50 μl TaqMan Universal PCR Master mix (2X). The 100 μl were loaded into the corresponding fill port. Individual singleplex PCR reactions were performed in 384-well reaction plates with Applied Biosystems 7900HT Fast Real-Time PCR system. The level of miRNA expression was measured using Ct (threshold cycle) determined by RQ Manager. Included on each array were 3 TaqMan miRNA endogenous controls, and 1 Taqman miRNA assay not related to rodent.
Endogenous silencing of Itga7
Endogenous silencing of Itga7 in C2C12 was performed using siRNA (#SI02733290, Qiagen) to transfect differentiated myotubes for 36 h. In parallel, transfections were realized with negative control siRNAs that had no homology to any known mammalian gene (AllStars Negative Controls # 1027280, Qiagen) to pinpoint the effects of Itga7 knockdown. After 48 h transfection, decreased Itga7 expression in C2C12 cells was validated at the mRNA level by qRT-PCR (primers QT00136990 from QIAGEN) and at the protein level by western blot ([ITA7 {sc-50431} and TSG101 {sc-6037} are from Santa Cruz Biotechnology]) (Fig. S1).
To extract siRNA, Itga7-containing exosomes from Itga7 knockdown C2C12 cells, conditioned medium was collected 48 h post-transfection, and released exosomes were extracted as described above. Then proliferating C2C12 cells were incubated for 48 h with 2 μg per ml of culture medium of either siRNA Itga7-containing exosomes or control exosomes (from non-transfected cells or from cells transfected with negative control siRNAs). Then Itga7 expression in C2C12 recipient cells was quantified by qRT-PCR.
Transfection assay
C2C12 myoblasts were plated on 6-well plates. At 60–70% confluence myoblasts were transfected with 2 μg of pEZX-MT01 vector containing the full-length 3′-UTR segment of Sirt1 (Sirtuin [silent mating type information regulation 2 homolog] 1) (GeneCopoeia, LabOmics, Belgium, clone MmiT032125-MT01). After 18 h, transfected myoblasts were incubated in 1 ml of exosome-depleted DMEM, containing 2 μg of either EXO-MB or EXO-MT. At confluence, transfected C2C12 myoblasts were incubated in 1 ml of exosome-depleted differentiation medium, for 48 h. Then, activities of firefly (Photinus pyralis) and Renilla (Renilla reniformis) luciferases were measured sequentially from each cell lysate by using the Dual-Luciferase® Reporter Assay System (Promega, #E1910). Firefly luciferase activities were normalized with Renilla luciferase activities to minimize experimental variabilities caused by differences in cell viability or transfection efficiency.
Because we have found that C2C12 cells released around 0.4 μg exosomes/ml per 24 h in DMEM exosome-free medium (see “Results” section), it was thus necessary to use higher quantities of exosomes in order to detect the biological effect of myotube-released exosomes on myoblasts. We have decided to use 2 μg exosomes per ml of medium, which is less than the previous studies demonstrating the biological effect of exosomes (from 10–1000 μg /ml in22,73 and less than the concentration of exosomes detected in plasma74.
Overexpression of miRNAs in C2C12
C2C12 cells were plated on 6-well plates and infected for 48 h with 108 particules/ml of recombinant adenoviruses expressing either GFP (control), pre-miR-133a-1, or pre-miR-1-1.
To collect myotube-exosomes containing cel-miR-238 (C. elegans RNA synthesized by Qiagen), differentiated C2C12 cells were transfected with cel-miR-238 (120 νg/ml) for 36 h. Then the medium was removed, and an exosome-depleted medium was added for 48 h, and exosomes were extracted from this medium.
Supplementary Material
Acknowledgments
This work was supported by grants from Fondation pour la Recherche Médicale (FRM), Association Française de recherche sur les Myopathies (AFM), Association Française de Diabétologie (SDF), and INRA-specific grant (ANSSD).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
AF is PhD student who participated in the design of the study and to the manuscript. SR conceived the study, and coordinated and helped to draft the manuscript. AJ, SP, and VE are technicians who performed western blot analysis, TLDA and microRNA amplifications and in vitro studies. EE performed Transmission Electron Microscopy (TEM). HV and KC helped to draft the manuscript and gave constructive comments. EL participated in luciferase assay analysis. All authors read and approved the final manuscript.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/26808
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26808
References
- 1.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. microRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and function. Thromb Haemost. 2012;107:605–10. doi: 10.1160/TH11-12-0836. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 8.Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel JP. microRNAs: molecular features and role in cancer. Front Biosci (Landmark Ed) 2012;17:2508–40. doi: 10.2741/4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 10.Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012;11:189–200. doi: 10.1016/S1474-4422(11)70286-1. [DOI] [PubMed] [Google Scholar]
- 11.Lorenzen J, Kumarswamy R, Dangwal S, Thum T. microRNAs in diabetes and diabetes-associated complications. RNA Biol. 2012;9:820–7. doi: 10.4161/rna.20162. [DOI] [PubMed] [Google Scholar]
- 12.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Köppel T, Jahantigh MN, Lutgens E, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 16.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 18.Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463:464–73. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- 19.Hurley JH, Odorizzi G. Get on the exosome bus with ALIX. Nat Cell Biol. 2012;14:654–5. doi: 10.1038/ncb2530. [DOI] [PubMed] [Google Scholar]
- 20.Rana S, Yue S, Stadel D, Zöller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol. 2012;44:1574–84. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 21.Rana S, Zöller M. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem Soc Trans. 2011;39:559–62. doi: 10.1042/BST0390559. [DOI] [PubMed] [Google Scholar]
- 22.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 23.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr., Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aliotta JM, Pereira M, Johnson KW, de Paz N, Dooner MS, Puente N, Ayala C, Brilliant K, Berz D, Lee D, et al. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp Hematol. 2010;38:233–45. doi: 10.1016/j.exphem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aliotta JM, Pereira M, Johnson KW, de Paz N, Dooner MS, Puente N, Ayala C, Brilliant K, Berz D, Lee D, et al. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp Hematol. 2010;38:233–45. doi: 10.1016/j.exphem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouzakri K, Plomgaard P, Berney T, Donath MY, Pedersen BK, Halban PA. Bimodal effect on pancreatic β-cells of secretory products from normal or insulin-resistant human skeletal muscle. Diabetes. 2011;60:1111–21. doi: 10.2337/db10-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–94. doi: 10.1016/S0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- 28.Haugen F, Norheim F, Lian H, Wensaas AJ, Dueland S, Berg O, Funderud A, Skålhegg BS, Raastad T, Drevon CA. IL-7 is expressed and secreted by human skeletal muscle cells. Am J Physiol Cell Physiol. 2010;298:C807–16. doi: 10.1152/ajpcell.00094.2009. [DOI] [PubMed] [Google Scholar]
- 29.Guescini M, Guidolin D, Vallorani L, Casadei L, Gioacchini AM, Tibollo P, Battistelli M, Falcieri E, Battistin L, Agnati LF, et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp Cell Res. 2010;316:1977–84. doi: 10.1016/j.yexcr.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Le Bihan MC, Bigot A, Jensen SS, Dennis JL, Rogowska-Wrzesinska A, Lainé J, Gache V, Furling D, Jensen ON, Voit T, et al. In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. J Proteomics. 2012;77:344–56. doi: 10.1016/j.jprot.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Abusamra AJ, Zhong Z, Zheng X, Li M, Ichim TE, Chin JL, Min WP. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol Dis. 2005;35:169–73. doi: 10.1016/j.bcmd.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Yu S, Liu C, Su K, Wang J, Liu Y, Zhang L, Li C, Cong Y, Kimberly R, Grizzle WE, et al. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178:6867–75. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- 33.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/S1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 34.Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–18. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 35.Deo A, Carlsson J, Lindlöf A. How to choose a normalization strategy for miRNA quantitative real-time (qPCR) arrays. J Bioinform Comput Biol. 2011;9:795–812. doi: 10.1142/S0219720011005793. [DOI] [PubMed] [Google Scholar]
- 36.Ng YH, Rome S, Jalabert A, Forterre A, Singh H, Hincks CL, Salamonsen LA. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS One. 2013;8:e58502. doi: 10.1371/journal.pone.0058502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–66. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge Y, Chen J. microRNAs in skeletal myogenesis. Cell Cycle. 2011;10:441–8. doi: 10.4161/cc.10.3.14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townley-Tilson WH, Callis TE, Wang D. microRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42:1252–5. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardinali B, Castellani L, Fasanaro P, Basso A, Alemà S, Martelli F, Falcone G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4:e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamrick MW, Herberg S, Arounleut P, He HZ, Shiver A, Qi RQ, Zhou L, Isales CM, Mi QS. The adipokine leptin increases skeletal muscle mass and significantly alters skeletal muscle miRNA expression profile in aged mice. Biochem Biophys Res Commun. 2010;400:379–83. doi: 10.1016/j.bbrc.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Burkin DJ, Kaufman SJ. Increasing alpha 7 beta 1-integrin promotes muscle cell proliferation, adhesion, and resistance to apoptosis without changing gene expression. Am J Physiol Cell Physiol. 2008;294:C627–40. doi: 10.1152/ajpcell.00329.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Granjon A, Gustin MP, Rieusset J, Lefai E, Meugnier E, Güller I, Cerutti C, Paultre C, Disse E, Rabasa-Lhoret R, et al. The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes. 2009;58:2555–64. doi: 10.2337/db09-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rathbone CR, Booth FW, Lees SJ. Sirt1 increases skeletal muscle precursor cell proliferation. Eur J Cell Biol. 2009;88:35–44. doi: 10.1016/j.ejcb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saini A, Al-Shanti N, Sharples AP, Stewart CE. Sirtuin 1 regulates skeletal myoblast survival and enhances differentiation in the presence of resveratrol. Exp Physiol. 2012;97:400–18. doi: 10.1113/expphysiol.2011.061028. [DOI] [PubMed] [Google Scholar]
- 47.Yamakuchi M. microRNA Regulation of SIRT1. Front Physiol. 2012;3:68. doi: 10.3389/fphys.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104:17016–21. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol. 2011;11:34. doi: 10.1186/1471-213X-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. microRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–26. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. microRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem. 2011;286:19431–8. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–84. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 53.Panguluri SK, Bhatnagar S, Kumar A, McCarthy JJ, Srivastava AK, Cooper NG, Lundy RF, Kumar A. Genomic profiling of messenger RNAs and microRNAs reveals potential mechanisms of TWEAK-induced skeletal muscle wasting in mice. PLoS One. 2010;5:e8760. doi: 10.1371/journal.pone.0008760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G, Wang J, Sun Y, Zhang P, Fan M, et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36:2690–9. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu ML, Wang JF, Wang GK, You XH, Zhao XX, Jing Q, Qin YW. Vascular smooth muscle cell proliferation is influenced by let-7d microRNA and its interaction with KRAS. Circ J. 2011;75:703–9. doi: 10.1253/circj.CJ-10-0393. [DOI] [PubMed] [Google Scholar]
- 56.Drummond MJ, McCarthy JJ, Sinha M, Spratt HM, Volpi E, Esser KA, Rasmussen BB. Aging and microRNA expression in human skeletal muscle: a microarray and bioinformatics analysis. Physiol Genomics. 2011;43:595–603. doi: 10.1152/physiolgenomics.00148.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang H, Liu X, Wang Z, She X, Zeng X, Deng M, Liao Q, Guo X, Wang R, Li X, et al. Interaction of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma growth. Brain Res. 2011;1390:21–32. doi: 10.1016/j.brainres.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 58.Xu D, He XX, Chang Y, Sun SZ, Xu CR, Lin JS. Downregulation of miR-93 expression reduces cell proliferation and clonogenicity of HepG2 cells. Hepatogastroenterology. 2012;59:2367–73. doi: 10.5754/hge12458. [DOI] [PubMed] [Google Scholar]
- 59.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MA, Bernad A, Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katakowski M, Buller B, Wang X, Rogers T, Chopp M. Functional microRNA is transferred between glioma cells. Cancer Res. 2010;70:8259–63. doi: 10.1158/0008-5472.CAN-10-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li L, Zhu D, Huang L, Zhang J, Bian Z, Chen X, Liu Y, Zhang CY, Zen K. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One. 2012;7:e46957. doi: 10.1371/journal.pone.0046957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–9. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 65.Carayon K, Chaoui K, Ronzier E, Lazar I, Bertrand-Michel J, Roques V, Balor S, Terce F, Lopez A, Salomé L, et al. Proteolipidic composition of exosomes changes during reticulocyte maturation. J Biol Chem. 2011;286:34426–39. doi: 10.1074/jbc.M111.257444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka S, Terada K, Nohno T. Canonical Wnt signaling is involved in switching from cell proliferation to myogenic differentiation of mouse myoblast cells. J Mol Signal. 2011;6:12. doi: 10.1186/1750-2187-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anton R, Chatterjee SS, Simundza J, Cowin P, Dasgupta R. A systematic screen for micro-RNAs regulating the canonical Wnt pathway. PLoS One. 2011;6:e26257. doi: 10.1371/journal.pone.0026257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ryall JG. The role of sirtuins in the regulation of metabolic homeostasis in skeletal muscle. Curr Opin Clin Nutr Metab Care. 2012;15:561–6. doi: 10.1097/MCO.0b013e3283590914. [DOI] [PubMed] [Google Scholar]
- 70.Rana S, Yue S, Stadel D, Zöller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol. 2012;44:1574–84. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 71.Ge X, McFarlane C, Vajjala A, Lokireddy S, Ng ZH, Tan CK, Tan NS, Wahli W, Sharma M, Kambadur R. Smad3 signaling is required for satellite cell function and myogenic differentiation of myoblasts. Cell Res. 2011;21:1591–604. doi: 10.1038/cr.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taylor DD, Gercel-Taylor C. microRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 73.Deng ZB, Poliakov A, Hardy RW, Clements R, Liu C, Liu Y, Wang J, Xiang X, Zhang S, Zhuang X, et al. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes. 2009;58:2498–505. doi: 10.2337/db09-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Azevedo LC, Janiszewski M, Pontieri V, Pedro MdeA, Bassi E, Tucci PJ, Laurindo FR. Platelet-derived exosomes from septic shock patients induce myocardial dysfunction. Crit Care. 2007;11:R120. doi: 10.1186/cc6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.