Abstract
In our previous study, we have identified a PCBP2 siRNA that exhibits antifibrotic activity in rat hepatic stellate cells (HSCs) by inhibition of αCP2, a protein responsible for stabilization of the collagen α1 (I) mRNA in alcoholic liver fibrosis. This study aims to develop a streptavidin-based nanocomplex that can efficiently deliver the PCBP2 siRNA to HSCs. Biotin-siRNA and biotin-cholesterol were mixed with streptavidin to form the streptavidin-biotin complex, which was further condensed electrostatically with positively charged protamine to form the final multicomponent siRNA nanocomplex in the size range of 150-250 nm. The siRNA nanocomplex does not induce cytotoxicity in rat HSCs as compared to commercially available transfection agents. The cellular uptake efficiency of the siRNA nanocomplex is higher in rat HSCs than other cell lines, such as CaCO-2 and PC-3, indicating that receptor-mediated endocytosis mainly contributes to the cellular uptake of the siRNA nanocomplex. The siRNA nanocomplex exhibits more than 85% silencing effect on the PCBP2 mRNA in HSCs. Stability study indicates that the nanocomplex can efficiently protect siRNA from degradation in the serum. The streptavidin-based multicomponent siRNA nanocomplex provides a promising strategy to deliver the PCBP2 siRNA to HSCs. Moreover, the nanocomplex can be used as a platform for other diseases by changing the siRNA sequence and targeting ligand.
Keywords: siRNA delivery, cholesterol, nanocomplex, streptavidin, protamine, HSC-T6
Introduction
Alcohol abuse is a prominent cause of liver fibrosis in western developed countries, which accounts for more than 50% cirrhosis cases. The deposition of extracellular matrix (ECM) in hepatic stellate cells (HSCs) is a key characteristic of alcoholic liver fibrosis 1, 2. Excessive accumulation of type I collagen, the major component of the ECM, in fibrotic liver is attributed to the RNA binding protein αCP2, encoded by the poly (rC) binding protein 2 (PCBP2) gene, which binds to the 3′ end of the collagen α1(I) mRNA in activated HSCs. In our previous study, we have investigated one potent PCBP2 siRNA which can lead to destabilization of the collagen α1 (I) mRNA and eventually reduce the overexpressed type I collagen in HSCs 3. Specific knockdown of the target PCBP2 gene makes siRNA a considerable promise for therapeutic approach. However, numerous barriers, such as instability in the blood, off-target effect, and abundant negative charges significantly limit the clinical application of siRNA. Hence, there is an imperative need to develop a safe and efficient carrier that can address these barriers.
Among all the carriers that have been developed, complexes based on the streptavidin-biotin interaction have attracted a lot of attention because of safety and ease of construction. Moreover, the complex can be easily functionalized with various targeting ligands, making it a versatile platform for targeted delivery of therapeutic agents for a variety of life-threatening diseases 4. For example, Lee et al. demonstrated that a streptavidin-biotin complex decorated with recombinant human epidermal growth factor (EGF) selectively delivered DNA molecules to human epidermal carcinoma A431 cells overexpressing EGF receptor 5. In another study, an anti-prostate specific membrane antigen (anti-PSMA) aptamer was coupled to an siRNA via streptavidin, leading to specific uptake of the siRNA by PSMA-positive cells 6 .
Despite its promise, the streptavidin-biotin technology is not efficient alone to deliver negatively charged siRNAs due to the repulsion by negatively charged cell membrane. Therefore, cationic molecules such as polyethylene imine (PEI) have been utilized to condense anionic siRNAs 7. However, the severe cell toxicity associated with PEI limits its therapeutic applications. By contrast, protamine is an FDA approved cationic peptide that has been extensively investigated for DNA delivery. In recent years, several studies indicated similar capability of protamine to condense siRNA in combination with cationic liposomes 8. Taken together, a multicomponent siRNA nanocomplex made with streptavidin, biotin, and protamine could be a promising siRNA delivery system for the treatment of alcoholic liver fibrosis. In this study, siRNA nanocomplex targeting HSC-T6, a rat hepatic stellate cell line, was prepared by using cholesterol as the targeting ligand and the PCBP2 siRNA as the therapeutic agent. We demonstrated that the nanocomplex efficiently delivers the PCBP2 siRNA to the HSCs without inducing toxicity.
Materials and Methods
Protamine sulphate (salmon x grade) was purchased from Sigma-Aldrich (St. Louis, MO), Streptavidin (53 kDa), BCA protein assay kit and HABA assay kit were obtained from Pierce (Rockford, IL). Dulbecco’s phosphate buffered saline (DPBS), Dulbecco’s Modified Eagle’s Medium (DMEM), serum reduced OptiMem, penicillin-streptomycin, lipofectamine-2000, and goat anti-rabbit antibody were purchased from Invitrogen (Carlsbad, CA). GelRed™ was purchased from Biotium (Hayward, CA.). Anti- Low Density Lipoprotein Receptors (LDLR) antibody was obtained from Bio Vision Inc. (Milpitas, CA). Luciferase siRNA (sense sequence 5′-CUUACGCUGAGUACUUCGA-3′), the PCBP2 siRNA (sense sequence 5′-GUCAGUGUGGCUCUCUUAU-3′) and negative control siRNA were purchased from Invitrogen (Carlsbad, CA). Both FITC-siRNA and biotin-siRNA were also purchased from Invitrogen (Carlsbad, CA). FITC is conjugated to the 5′ end of the siRNA antisense strand, while biotin is conjugated to the 3′ end of the siRNA sense strand.
Synthesis of biotin cholesterol (NH2-Lys(Biotin)-Gly-Chol)
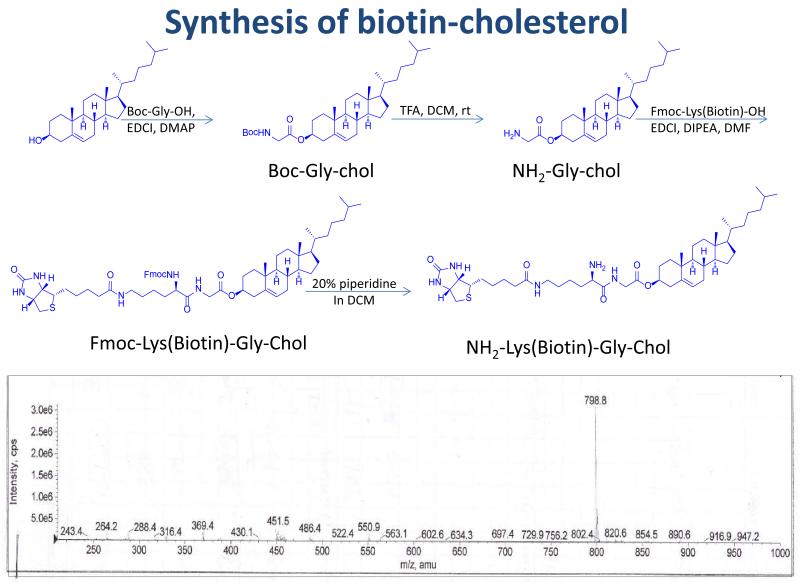
NH2-Gly-cholesterol was synthesized as reported 9 and then conjugated with biotin. Briefly, NH2-Gly-Chol (1mmol) and Fmoc-Lys(Biotin)-OH (1mmol) were dissolved in 2 mL of dry DMF, followed by adding EDCl (1.5 mmol) and DIPEA (1.5 mmol). The reaction mixture was stirred at room temperature overnight. After adding 5 ml of NH4Cl saturated solution to stop the reaction, the organic phase was separated, washed and concentrated under vacuum. The compound Fmoc-Lys(Biotin)-Gly-Chol was purified by silica gel chromatography. The purified compound (1 mmol) was then dissolved in 4 ml of DCM. One milliliter of piperidine was added into the reaction mixture to initialize deprotection of the Fmoc group. After 30 min, the organic solvent was evaporated under vacuum. The residue was washed with 20 ml of diethyl ether three times to yield pure NH2-Lys(Biotin)-Gly-Chol as white powder (overall yield ~ 50%). ESI-MS was calculated for C71H98N2O13 798.17; found 798.8 [M + H]+.
Fabrication of the streptavidin-based siRNA nanocomplex
Biotin-cholesterol was synthesized by addition of biotin to cholesterol via a GK (Glycine-Lysine) dipeptide linker. Biotin-siRNA containing a disulfide linker between the 3′ end of the sense strand and biotin was purchased from Invitrogen and used without any further purification. Disulfide linker was formed by the reaction of NHS esters in NHS-SS-biotin with the primary amine group at the 3′end of siRNA sense strand. The siRNA-Streptavidin-Cholesterol (SSC) complex was prepared by mixing biotin-siRNA, streptavidin and biotin-cholesterol in the 2:1:2 molar ratio. The complex was incubated at room temperature for 10 minutes and then condensed with varying amounts of protamine (N/P ratios of 1:1, 2:1, 3:1, 5:1, and 10:1). The final mixture was incubated at room temperature for 30 minutes to form the multicomponent siRNA-Streptavidin-Cholesterol-Protamine (SSCP) nanocomplex.
Characterization of the nanocomplex
Formation of the SSCrP nanocomplex was evaluated using the Gel-Red fluorescence quenching assay as described 10. Briefly, twenty microliters of the SSCP nanocomplexes containing 1 μM siRNA with varying N/P rations were added to black 96 well plates along with 80 μL Gel Red solution. The plates were incubated at room temperature in the dark for 5 min, and fluorescence was measured using a Beckman DTX 880 multimode detector (Beckman coulter, Inc., Brea, CA) at λexc 485 nm and λemi 625 nm.
Zeta potential and particle size of the nanocomplex were measured using a Malvern Zetasizer nano-ZS (Malvern Instruments, Westborough, MA).
Cell culture and siRNA transfection
HSC-T6 cells were grown in DMEM supplemented with 10% FBS, 100 units/mL penicillin-streptomycin at 37°C in a humidified atmosphere containing 5% CO2. The cells were seeded in 24-well plates at a density of 1×104 cells/well as described previously 3. The siRNA nanocomplex was prepared with N/P ratios of 5:1 and 10:1 as described above and added to each well along with 400 μL of Opti-Mem to produce a final concentration of 100 nM siRNA. The cells were then incubated for different time periods to evaluate cellular uptake or silencing activity. For the uptake experiment, FITC-labeled luciferase siRNA duplex was used. The PCBP2 siRNA was utilized in silencing experiment.
Lipofectamine-based transfection was carried out as we previously reported 3, 11. Briefly, the transfection mixture was prepared by mixing 2.5 μl of 20 μM biotin-siRNA and 2 μl Lipofectamine-2000 with two separate aliquots of 50 μl of serum free Opti-Mem and then combining the two mixtures at room temperature for 30 min to form a complex. Consequently, 100 μl of the mixture was added to each well along with 400 μl Opti-Mem to produce a final concentration of 100 nM of siRNA.
Real Time RT-PCR
Gene silencing effect was determined using real time RT-PCR as we described before 3. Briefly, total RNA was isolated using TRIzol and reverse transcribed into cDNA. Two hundred nanograms of the cDNA were amplified by real time PCR using Light Cycler 480 SYBR Green-1 Master mix on a Light Cycler 480 system (Roche, Indianapolis, IN). The primers used for the study were as follows: PCBP2: 5′-ACCAATAGCACAGCTGCCAGTAGA-3′ (forward primer) and 5′-AGTCTCCAACATGACCACGCAGAT-3′ (reverse primer); and 18s Ribosomal RNA (as internal control): 5′-GTCTGTGATGCCCTTAGATG-3′ (forward) and 5′-AGCTTATGACCCGCACTTAC -3′ (reverse).
Cytotoxicity assay
HSC-T6 cells were plated in 96-well plates at a density of 5×103 cells/well for 12 hours. An aliquot of 2 μl SSC complex or the SSCP nanocomplex with different N/P ratios was mixed with 23 μl Opti-Mem and added in each well along with 75 μl Opti-Mem medium to produce a final concentration of 100 nM siRNA. The cells were also transfected with the biotin-siRNA at a final concentration of 100 nM in the presence of Lipofectamine-2000. After 24-48 hours transfection, cell viability was measured using MTT assay.
Detection of LDLR by western blot
Western blot was conducted to analyze the LDLR expression in HSC-T6, PC-3, and CaCo-2 cells. Cell lysate was prepared as we reported previously 12, 13. The total protein content was measured using a BCA protein assay kit (Pierce, Rockford, IL). Equal amounts of protein samples (15 μg/well) were resolved on a 7% SDS PAGE gel. The separated proteins were electrophoretically transferred to a PVDF membrane, blocked with a 5% non-fat dry milk solution, and probed with anti-LDLR antibody. The protein was then visualized using horseradish peroxidase conjugated secondary antibody and a chemo-luminescence detection kit. The same membrane was re-probed with anti-β-actin antibody as an internal control.
Cellular Uptake Study
The FITC-labeled luciferase siRNA was used in cellular uptake study. HSC-T6, CaCO-2, and PC-3 cells were seeded in 96-well plates at a density of 5× 103 cells/well 24 h prior to transfection. The cells were transfected with the SSCP nanocomplex, SSC complex, protamine/siRNA complex, and naked siRNA as described above. After 6 h transfection, the cells were washed with DPBS and incubated with Opti-MEM containing 1 mg/ml heparin at 37°C for 30 min to remove nonspecific bound SSCP nanocomplex on the cell surface 14, 15. Following heparin treatment, the cells were washed with DPBS, and fluorescence images were obtained with a Leica DMI 3000B fluorescence microscope (Leica Microsystems Inc., Wetzlar, Germany).
To evaluate the role of cholesterol in the uptake of the SSCP nanocomplex, SSCP nanocomplex containing or not containing cholesterol were incubated with HSC-T6 cells at 37°C for 3, 4 and 6 h. The cells were then washed and imaged with a Leica DMI 3000B fluorescence microscope.
To further investigate the receptor-mediated uptake of the nanocomplex, HSC-T6 cells were incubated with 150 μg/ml puromycin, an effective LDLR inhibitor, for 1 h prior to transfection. The cells were washed three times with DPBS and then transfected with FITC-labeled siRNA alone or encapsulated in the SSCP nanocomplex. After transfection, the cells were lysed on ice for 1 h in the dark with RIPA buffer containing freshly added protease inhibitor cocktail (Roche, Indianapolis, IN). The relative fluorescence was measured using a Beckman plate reader (λexc 485 nm and λemi 535nm) and normalized to the total amount of protein for each sample.
Stability of the Nanocomplex
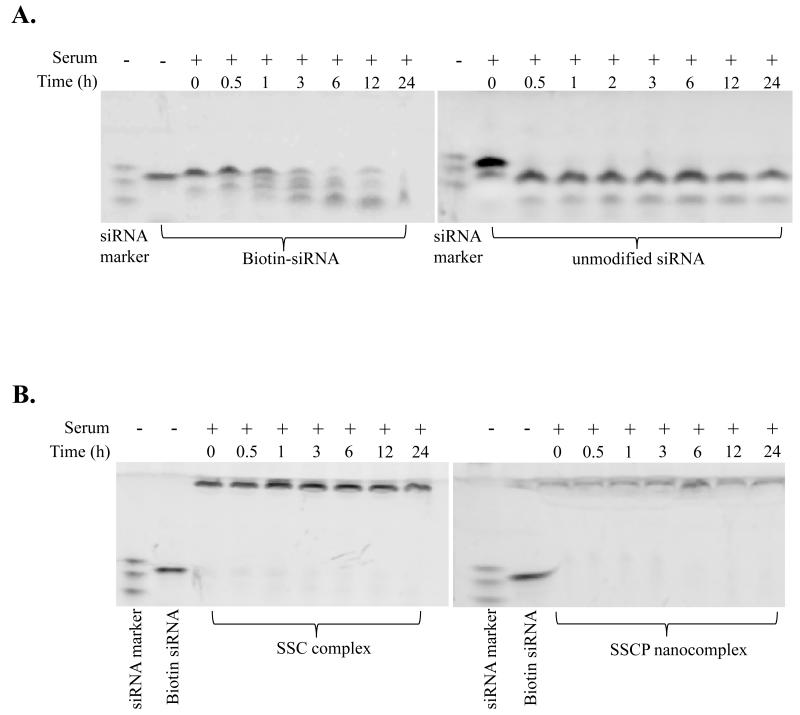
Biotin-siRNA, unmodified siRNA, SSC complex and SSCP nanocomplex were incubated in 50% rat serum at 37°C for different time intervals, followed by native polyacrylamide gel electrophoresis and staining with Gel Red™ to visualize the intact and degraded siRNA. Only free siRNA can enter the gel and be fully visualized by the dye. The siRNA encapsulated in the nanocomplex remains at the interface of the stacking and running gels and can only be partially stained.
To evaluate the effect of endogenous biotin in the serum on the stability of the SSC complex, a dissociation study was performed in the presence of biotin at the same concentration found in human blood. Briefly, 20 μl of the SSC complex containing 500 nM siRNA was incubated with biotin (1000 pM) for different time intervals, followed by native polyacrylamide gel electrophoresis and staining with Gel Red™.
Stability of the nanocomplex was further investigated using the heparin competition assay. The nanocomplexes were incubated with different concentrations of heparin (0, 2, 4, 8, 20, 40 and 80 μM) in PBS at 37°C for 15 min. Dissociated siRNAs were electrophoresed in 2% agarose gel and visualized using Gel Red™. Fluorescence quenching assay was also used to monitor the siRNA dissociation.
Reduction of disulfide bond
To confirm that cleavage of the disulfide bond can release siRNA, the SSCP nanocomplex was incubated with 100 mM DTT at 37°C for different time intervals. To further dissociate the released siRNA from protamine, the samples were also treated with heparin at 1 mM for 5 min, followed by native polyacrylamide gel electrophoresis and staining with Gel Red™. A similar cleavage study was conducted using glutathione, the most important cellular reducer, to mimic intracellular disulfide bond cleavage. SSC and SSCP nanocomplexes containing 0.5 μg siRNA were incubated with different concentrations of glutathione (10 and 50 mM) at 37°C for 2h as described 16.
Results
Fabrication of the streptavidin-based siRNA nanocomplex
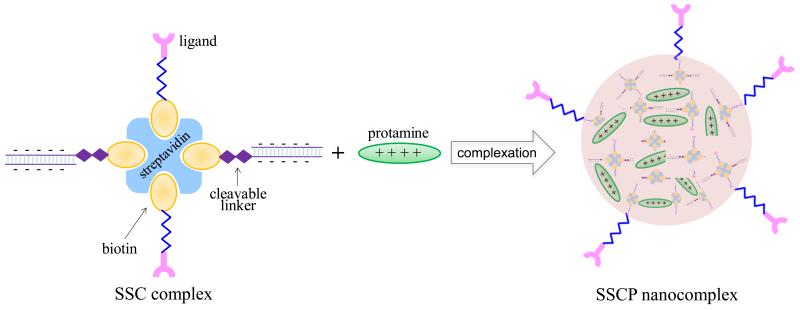
The streptavidin-based siRNA SSCP nanocomplex was fabricated as illustrated in Figure 1. The nanocomplex consists of four components: (i) streptavidin; (ii) biotin-cholesterol; (iii) biotin-siRNA; and (iv) protamine. The streptavidin, biotin-cholesterol, and biotin-siRNA can form the SSC complex via the streptavidin-biotin interaction, which is one of the strongest noncovalent interactions. Protamine was added to neutralize the negatively charged siRNA and compact the SSC complex to form the multicomponent SSCP nanocomplex.
Figure 1.
Schematic diagram of the SSCP nanocomplex using streptavidin-biotin technology. Biotin-siRNA and biotin-cholesterol were complexed with streptavidin to form the SSC complex, which is further condensed with protamine at different N/P ratios to form the final SSCP nanocomplex.
It has been shown by us and others that cholesterol can be employed as a liver-specific ligand for nucleic acids 17, 18, and the receptors that are responsible for the uptake are low density lipoprotein receptor (LDLR) and scavenger receptor class B1 (SR-B1) 19. Therefore, we use cholesterol as a liver-specific ligand to increase the cellular uptake of the SSCP nanocomplex in HSCs. As shown in Figure 2, biotin was conjugated to the phenolic hydroxyl group of cholesterol via the Gly-Lys dipeptide linker, which can increase the solubility of the biotinylated cholesterol. Biotin was also conjugated to the 3′ end of the sense strand of the PCBP2 siRNA via a disulfide bond, which can be cleaved by intracellular redox enzymes to release the siRNA inside cells 20.
Figure 2.
Synthetic scheme of biotin-cholesterol.
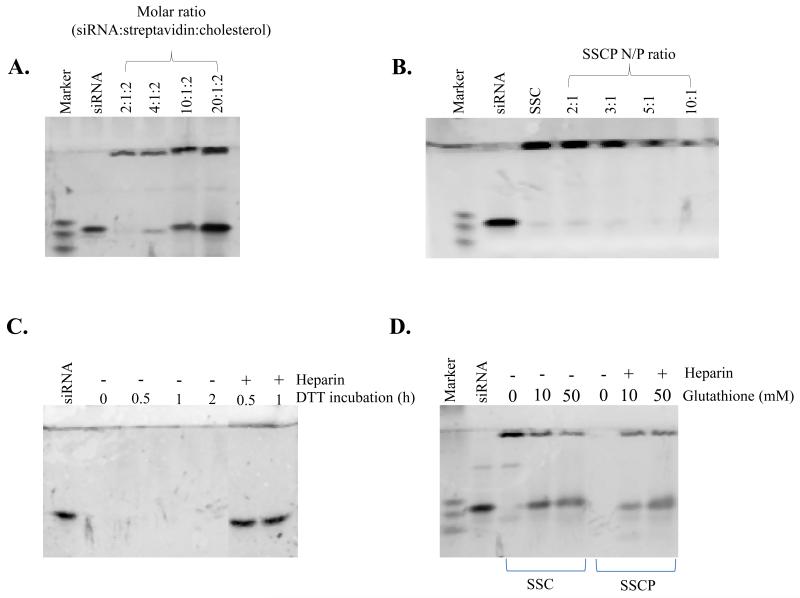
To investigate whether the biotin-siRNA and biotin-cholesterol have the same binding affinity for streptavidin, we prepared the SSC complex at different molar ratios of siRNA:streptavidin:cholesterol. As shown in Figure 3A, at the molar ratio 2:1:2, all biotin-siRNAs are complexed with streptavidin. When the molar ratio of biotin-siRNA was increased in the complexation, free biotin-siRNA was detected in the gel retardation assay, indicating that biotin-cholesterol has a comparable binding affinity as the biotin-siRNA. We chose 2:1:2 as the optimized ratio of the SSC complex for following studies so that each of the SSC complex contains equal number of siRNA and cholesterol. To further condense the negatively charged SSC complex, protamine was mixed with the SSC complex at different N/P ratios as illustrated in Figure 3B&C. Condensation of the SSC complex by protamine was monitored by gel retardation assay using Gel Red™ (Figure 3B). This study demonstrated that protamine can efficiently condense the SSC complex, and an increase of the N/P ratio increases the condensation.
Figure 3.
Optimization of the nanocomplex formulation. (A) Gel retardation assay of the SSC complexes at different molar ratios of siRNA:streptavidin:cholesterol. (B) Gel retardation assay of the SSCP nanocomplexes at different N/P ratios. The SSC complex with the molar ratio 2:1:2 (siRNA:streptavidin:cholesterol) was condensed with protamine at different N/P ratios and incubated for 30min. (C &D) Disulfide bond cleavage study. Heparin is added to dissociate cleaved siRNA from the nanocomplex prior to gel electrophoresis.
We next studied whether the cleavage of the disulfide bond can release siRNA from the SSCP nanocomplex. The SCCP nanocomplex was treated with DTT at 37°C for different time intervals, followed by native polyacrylamide gel electrophoresis and staining with Gel Red™. As Figure 3C revealed, DTT treatment alone cannot release siRNA from the SSCP complex due to the presence of protamine in the complex which binds to the cleaved siRNA. Further incubation of the DTT treated sample with heparin, a negatively charged molecule, can efficiently dissociate the cleaved siNRA from the SSCP nanocomplex.
We also studied the cleavage of the disulfide bond by glutathione, the most important cellular reducer. Two different concentrations of glutathione (10 and 50mM) were employed for this study, in which 10 mM resembles the cytosolic concentration of glutathione in cells 16, 21. As Figure 3D shows, glutathione cleaves disulfide bond and releases siRNA from the SSC complex and SSCP nanocomplex, indicating that siRNA can be cleaved from the SSCP nanocomplex inside cells.
Characterization of the SSCP nanocomplex
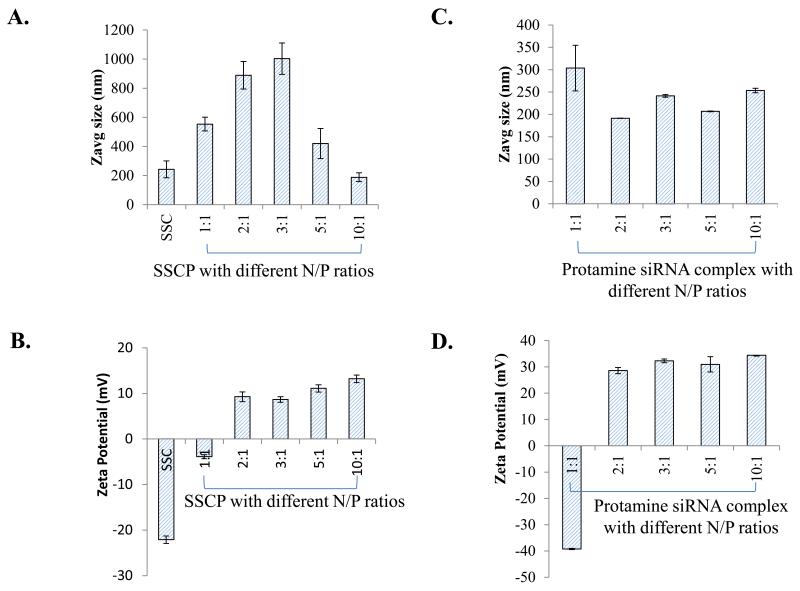
The particle size (Figure 4A) and zeta potential (Figure 4B) of the SSC complex and SSCP nanocomplexes with different N/P rations were evaluated by a light scattering particle sizer. The particle size of the SSC complex is around 230 nm, while the addition of protamine to the SSC complex increases the size at low N/P ratios but reduces the size to 170 nm at the N/P ratio 10:1. At low N/P ratios, the SSCP nanocomplex is not in a compact structure, leading to large particle size. By contrast, protamine could condense the SSC complex to a more compact structure at high N/P ratios. This result is in accordance with a recent report on siRNA peptide nanocomplexes which exhibit large particle size at low N/P ratios but small particle size at high N/P ratios 22. The zeta potential of the SSCP nanocomplexes at different N/P ratios were illustrated in Figure 4B. It is not surprising that the SSC complex exhibits a negative zeta potential due to the negatively charged siRNA in the complex. As the N/P ratio increases the zeta potential of the SSCP nanocomplex shifts from negative to positive owing to neutralization of the siRNA by protamine. Particle size and zeta potential of the protamine/siRNA complex were also examined (Figure 4 C&D). Unlike the SSCP nanocomplex, all the protamine/siRNA complexes at different N/P rations show similar particle size. Zeta potential of the protamine/siRNA complex shifts from negative to positive when the N/P ration is increased from 1:1 to 2:1 and slightly increases with N/P ratio afterward. This is consistent with findings in the SSCP nanocomplex (Fig. 4B).
Figure 4.
Characterization of the nanocomplex. Particle size (A) and zeta potential (B) of the SSCP nanocomplex at different N/P ratios. The SSC complex without protamine was used as the control. Similarly, particle size (C) and zeta potential (D) of the protamine/siRNA complex were evaluated. Data were shown as mean ± SD (n=3).
Cytotoxicity Assay
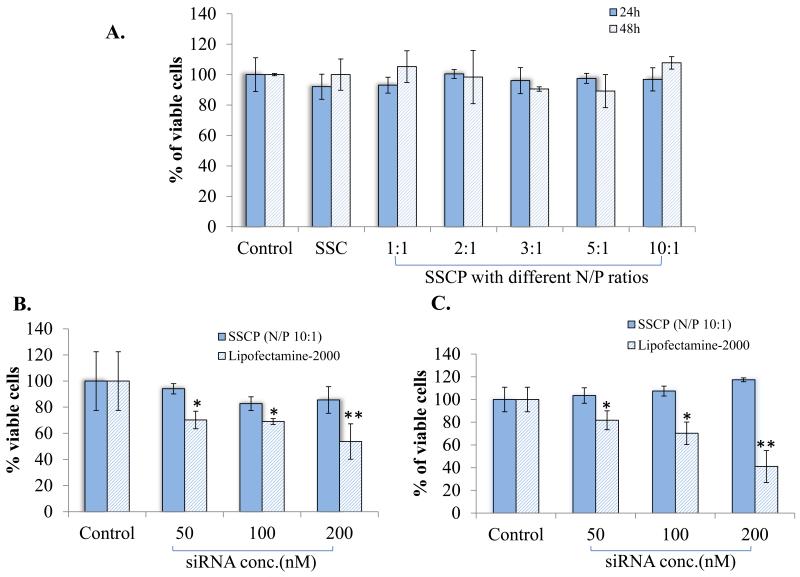
Cytotoxicity of the SSC complex and SSCP nanocomplexes with different N/P ratios were assessed in HSC-T6 cells after transfection for 24 and 48 hours. Figure 5A shows that both the SSC complex and SSCP nanocomplexes do not induce cytotoxicity in HSC-T6. Particularly, cationic protamine used in the SSCP nanocomplex does not affect cell viability even at the N/P ratio 10:1, indicating the nontoxic nature of all components used in the SSCP nanocomplex.
Figure 5.
Cytotoxicity of the nanocomplex. (A) HSC-T6 cells were incubated with the SSCP nanocomplexes at various N/P ratios for 24 and 48 h. Cytotoxicity of the SSCP nanocomplex with the N/P ratio 10:1 was compared with Lipofectamine-2000/siRNA complex at different siRNA concentrations 24 (B) and 48 (C) h post-transfection. Untreated cells were used as the control. Data were shown as mean ±SD (n=3). * p<0.05; ** p<0.01
Next, cytotoxicity of the SSCP nanocomplex with the N/P ratio 10:1 was compared with Lipofectamine-2000/siRNA complex 24 and 48 h post-transfection (Figure 5B & C). High siRNA concentration requires a large amount of biotin-cholesterol, streptavidin, and protamine to form the siRNA SSCP nanocomplex. This may lead to more significant effect on cell viability. However, we did not observe significant cytotoxicity of the SSCP nanocomplex even when the cells were transfected at a final siRNA concentration of 200 nM. By contrast, Lipofectamine-2000 induced significant cytotoxicity in the HSC-T6 cells. The cytotoxicity is siRNA concentration dependent, indicating that the cytotoxicity is associated with the amount of Lipofectamine-2000 used in the transfection.
Cellular Uptake Study
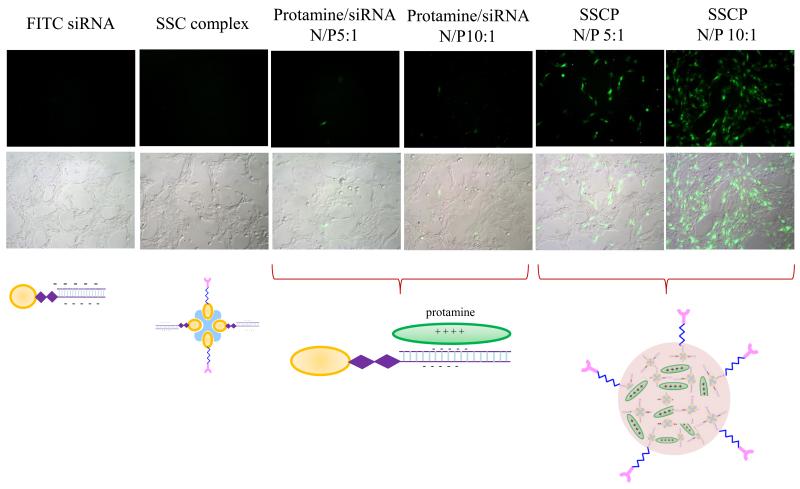
FITC-labeled siRNA was used in this study to evaluate cellular uptake of the naked siRNA, SSC complex, protamine/siRNA complex, and SSCP nanocomplex in HSC-T6 cells (Figure 6). Both the naked siRNA and SSC complex did not show any cellular uptake because they carry negative charge and are repelled away by negatively charged cell membrane. Moreover, protamine condensed siRNA complexes with two N/P ratios (5:1 and 10:1) only exhibited negligible cellular uptake, indicating that simple mixing siRNA with cationic material alone does not enhance its cellular uptake. This observation is consistent with our previous report, in which protamine alone cannot deliver siRNA into prostate cancer cells 23.
Figure 6.
Cellular uptake of FITC labeled siRNA in different formulations in HSC-T6 cells. HSC-T6 cells were incubated with the naked siRNA or its complexes for 6 h, followed by washing and imaging.
The SSCP nanocomplex showed significant cellular uptake, and the SSCP nanocomplex with the N/P ratio 10:1 is more efficient in delivering siRNA than the SSCP nanocomplex with the N/P ratio 5:1. As revealed above in Figure 4, high N/P ratios can form a more compact structure, which facilitates the cellular uptake of the SSCP nanocomlex.
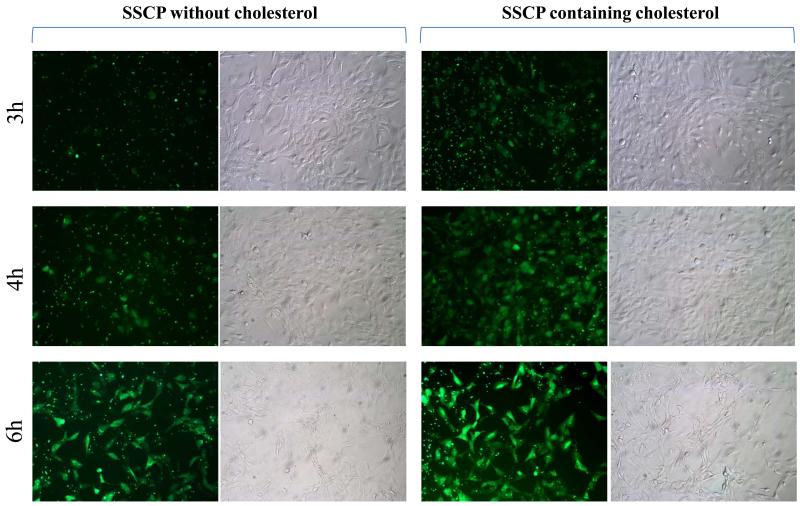
To study whether cholesterol mediates cellular uptake of the SSCP nanocomplex in HSC-T6 cells, cellular uptake of SSCP nanocomplexes containing or not containing cholesterol were evaluated at different incubation intervals. As Figure 7 shows, the SSCP nanocomplex containing cholesterol exhibits higher uptake than the SSCP nanocomplex that does not contain cholesterol. This is in accordance with our previous report in which cholesterol conjugated oligonucleotide shows higher cellular uptake than native oligonucleotide 17.
Figure 7.
Cellular uptake of SSCP nanocomplexes containing or not containing cholesterol in HSC-T6 cells.
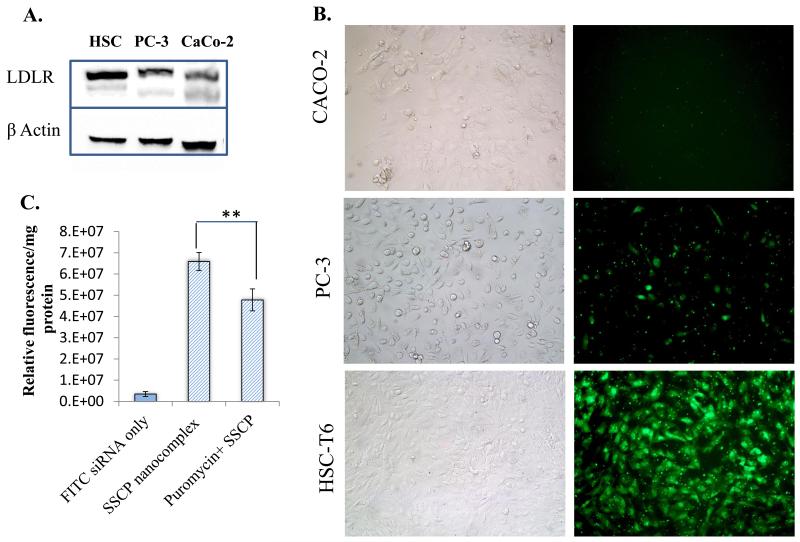
Our hypothesis is that the cellular uptake of the SSCP nanocomplex is mediated by cholesterol, which is a proved liver-specific ligand. Therefore, we compared the cellular uptake of the SSCP nanocomplex in three different cell lines, PC-3, CaCo-2, and HSC-T6. First, we performed western blot analysis of LDLR, which accounts for the cellular uptake of cholesterol. As illustrated in Figure 8A, HSC-T6 cells exhibit the highest LDLR expression among the three types of cells. This is in accordance with another study conducted by Srivastav et al., which shows higher expression of LDLR in HepG2 cells as compared to CaCO-2 cells 24. Accordingly, the cellular uptake of the SSCP nanocomplex in HSC-T6 cells is higher than that in PC-3 and CaCo-2 cells (Figure 8B). PC-3 cells show higher cellular uptake of the SSCP nanocomplex than CaCo-2 cells, which is consistent with the LDLR expression.
Figure 8.
Comparison of the uptake of the SSCP nanocomplex in different cell lines. (A) Western blot analysis of LDLR in HSC-T6, PC-3 and CaCO-2 cells. (B) Cellular uptake of the SSCP nanocomplex in HSC-T6, PC-3 and CaCO-2 cells. (C) Puromycin, an LDLR inhibitor, reduces the uptake of the SSCP nanocomplex in HSC-T6 cells. Data were shown as mean ±SD (n=3). * p<0.05; ** p<0.01
Furthermore, we treated HSC-T6 cells with puromycin, an LDLR inhibitor,25 to assess whether inhibition of LDLR decreases the cellular uptake of the SSCP nanocomplex. As shown in Figure 8C, puromycin treatment significantly inhibits the cellular uptake of the SSCP nanocomplex in HSC-T6 cells, indicating that LDLR is involved in the uptake of the SSCP nanocomplex.
Stability of the Nanocomplex
Poor stability is one of the major obstacles for therapeutic application of siRNA. Therefore, we evaluated the stability of naked siRNA, biotin-siRNA, SSC complex, and SSCP nanocomplex in rat serum. As illustrated in Figure 9A, naked siRNA without any modification is rapidly degraded within 0.5 h in rat serum. In contrast, biotin-siRNA is comparatively more stable in rat serum, which is attributed to the biotin modification at the 3′ end of the siRNA sense strand. siRNAs encapsulated in the SSC complex and SSCP nanocomplex are very stable and no degradation product was observed up to 24 h in rat serum (Figure 9B). All the siRNAs stay at the interface of the stacking and running gels. The siRNA in the SSCP nanocomplex is not efficiently stained by Gel Red due to the compact structure of the nanocomplex. The improved stability of siRNA in the SSC complex might be attributed to the complex three dimensional structure of streptavidin, which is not favorable for 3′ exonuclease degradation in the serum 26. The SSCP nanocomplex protects siRNAs from degradation because protamine condenses the siRNAs and shields them from nuclease degradation.
Figure 9.
Serum stability. (A) Serum stability of biotin-siRNA and unmodified siRNA. (B) Serum stability of the SSC complex and SSCP nanocomplex. The siRNA samples were incubated in 50% serum at 37°C for different time intervals, followed by native polyacrylamide gel electrophoresis and staining with Gel Red™ to visualize the intact and degraded siRNAs.
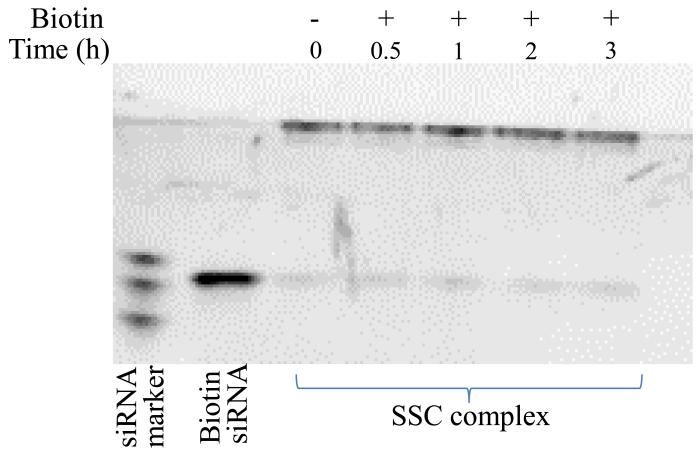
Both siRNA and cholesterol are anchored to streptavidin by the noncovalent streptavidin-biotin interaction. However, this interaction may be affected by the endogenous biotin present in human blood. Therefore, the stability of the SSC complex was evaluated in the presence of biotin at 1000 pM, which is similar as the endogenous biotin concentration (272-1050 pM) in human blood 27. As shown in Figure 10, no siRNA is dissociated in the presence of biotin up to 3 hours, indicating that the endogenous biotin in the blood will not disrupt the integrity of the SSC complex.
Figure 10.
Stability of the SSC complex in the presence of biotin at the same concentration found in human blood. The SSC complex was incubated with biotin (1000 pM) for different time intervals, followed by native polyacrylamide gel electrophoresis and staining with Gel Red™.
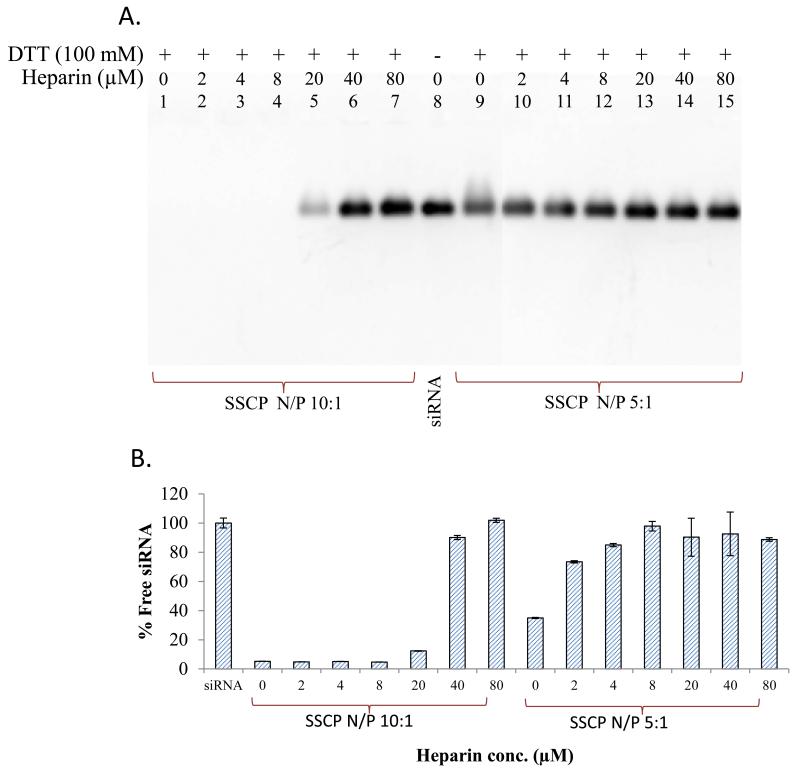
There are some logical arguments regarding the ability of cationic nanocarriers to reach the cytoplasm in the presence of polyanions, such as sulphated glycosaminoglycans in the exterior of cells 28. The anionic components present in the cell proximity (such as proteoglycans) 22 can compete with siRNA to bind with protamine (cationic component). Therefore, it is essential to evaluate whether these anions can disrupt the electrostatic condensation between siRNA and positively charged material used in the SSCP nanocomplex. The stability of the nanocomplex was assessed in the presence of heparin in this study. The SSCP nanocomplex was incubated with different concentrations of heparin and 100 mM dithiotheritol (DTT) at 37°C. DTT was used to cleave the disulfide bond so that siRNA condensed with protamine can be released from streptavidin. Results in figure 11A revealed better stability of the SSCP with the N/P ratio 10:1 as compared to the N/P ratio 5:1. The N/P ratio 10:1 is able to protect siRNA in the presence of heparin up to 5 μM. A similar result was observed in fluorescence quenching assay (Figure 11B). These observations indicate that the SSCP nanocomplex is stable in the presence of the anionic components present in the blood.
Figure 11.
Stability of the nanocomplex in the presence of polyanions. The SSCP nanocomplexes were incubated with various concentrations of heparin. (A) Dissociated siRNAs were electrophoresed in 2% agarose gel and visualized with Gel Red™. (B) Fluorescence quenching assay was also used to monitor the siRNA dissociation. Data were shown as mean ±SD (n=3).
Silencing Effect of the SSCP Nanocomplex
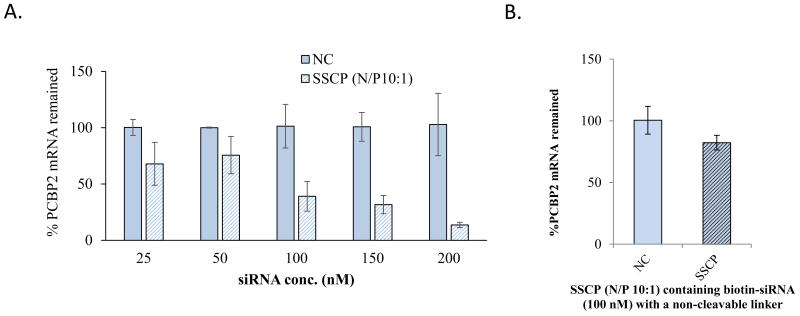
We finally evaluated the silencing activity of the SSCP nanocomplex with the N/P ratio 10:1 in HSC-T6 cells. HSC-T6 cells were incubated with SSCP nanocomplex containing various concentrations of PCBP2 siRNA or scrambled siRNA (Negative control, NC). As shown in Figure 12A, the SSCP nanocomplex showed a concentration-dependent silencing activity compared to the scrambled siRNA. This result indicates that the SSCP nanocomplex can efficiently deliver the siRNA into HSC-T6 cells and accordingly induce silencing activity.
Figure 12.
(A.) Silencing activity of the SSCP nanocomplex containing PCBP2 siRNA in HSC-T6 cells. (B.) Silencing activity of the SSCP nanocomplex containing non-cleavable biotin-siRNA. The results are expressed as the mean±SD (n=3).
Disulfide bond was introduced between siRNA and biotin to allow release of intact siRNA in the reductive environment of the cytoplasm. To prove this hypothesis, biotin-siRNA containing a non-cleavable linker was encapsulated in the SSCP nanocomplex and its silencing activity was evaluated in HSC-T6 cells. As Figure 12B shows, the biotin-siRNA containing a non-cleavable linker exhibits much lower silencing effect compared with the biotin-siRNA containing disulfide linker (Fig. 12A), indicating release of the intact siRNA from biotin is necessary to exert its silencing activity in cells. This finding is consistent with a previous report 29.
Discussions
Despite the great promise of siRNA in treating various diseases, lack of a safe and efficient delivery system is still the major hurdle for the successful clinical translation of siRNA. Although siRNA delivery systems using cationic lipids have been shown to have promise, these delivery systems are still facing several hurdles including cytotoxicity, stability and difficulty in large scale formulation 30-33. On the other hand, the streptavidin-biotin technology, characterized by tunable binding sites and a defined structure, presents a novel platform for siRNA delivery. Streptavidin has been found to be more effective in terms of its pharmacokinetic profile compared to its structurally resembled protein avidin. Moreover, low pI (~6) and lack of sugar molecules in streptavidin prevent its nonspecific interaction with leptin-like or negatively charged molecules present in the blood 34, 35.
Our current study demonstrates that the SSCP nanocomplex (Figure 1) made by the streptavidin-biotin technique can effectively deliver the PCPB2 siRNA to HSCs without causing toxicity. The siRNA is linked to biotin via a disulfide bond, which is expected to release free siRNA from the nanocomplex in the cytoplasm 36. This hypothesis was confirmed by siRNA cleavage study using DTT and glutahione. We demonstrated that cleavage of the disulfide bond allows release of siRNA from the nanocomplex (Figure 3C & 3D). In accordance with a previous study showing that conjugation at the 3′ or 5′ end of siRNA sense strand does not affect silencing activity, the biotin conjugation in our study was carried out at the 3′end of the siRNA sense strand 37.
One of the prime challenges of siRNA delivery is its poly-anionic nature, which impedes its reach to negatively charged cell vicinity and further retards its uptake. To overcome this limitation, several types of poly-cationic non-viral delivery systems have been developed 38-40. These systems utilize polycationic polymers/peptides that can form stable complex with siRNA through electrostatic interaction 36. In line with these findings, we utilize positively charged protamine in the nanocomplex preparation. Protamine is a cationic peptide (Mw~ 5 kDa) that has been widely used for gene delivery due to its inherent nature to condense nucleic acids present in sperm 41. The N/P ratio influences the final characteristics of the SSCP nanocomplex, such as particle size, zeta potential and stability, which determines the biological activity of the encapsulated siRNA. As a result, an optimum N/P ratio is essential to ensure the stability and activity of siRNA. It has to be noted that the particle size of the SSCP nanocomplex is not correlated inversely with the N/P ratio in our study (Figure 4). Instead, the particle size increases gradually, reaching up to a very large size when the N/P ratio is less than 5:1. This is due to the high tendency of the nanocomplex to aggregate at low N/P ratios 42. However, protamine can form a tight complexation with the SSC complex at high N/P ratios, leading to small particle size (Figure 4A) and better protection of the siRNA in the presence of serum (Figure 9) and polyanion (Figure 11).
We have previously shown that cholesterol conjugation at the 3′ end can significantly increase the liver uptake of a single-stranded oligonucleotide 17. Several other groups have also demonstrated enhanced liver uptake of cholesterol conjugated siRNA in vitro and in vivo 43-46. However, this direct conjugation strategy is relatively inefficient, requiring gram quantities of siRNA in regular infusions in humans to obtain measurable gene silencing effect 47. This inefficiency may be at least partly attributed to poor stability of the cholesterol conjugated siRNA in the blood because cholesterol conjugation alone cannot protect the siRNA from nuclease degradation. Therefore, we employed the streptavidin-biotin technology along with protamine complexation (Figure 1) to increase the stability of the PCBP2 siRNA.
The mechanism of hepatic uptake of cholesterol conjugated siRNA has been studied by Wolfrum et al. Cholesterol conjugated siRNA binds to LDL and HDL in the serum. The LDL-bound cholesterol-siRNAs are mainly taken up by LDLR, while HDL-bound cholesterol-siRNAs are taken up by SR-B1. Both LDLR and SR-B1 are highly expressed in the liver 19. In accordance with this finding, we observed that cellular uptake of the SSCP nanocomplex is correlated with the LDLR expression levels in HSC-T6, PC-3 and CaCO-2 cells (Figure 8), indicating the involvement of LDLR in the uptake of the nanocomplex. We also showed that the uptake of the nanocomplex in HSC-T6 cells is reduced by treating the cells with puromycin, which has been reported as an LDLR inhibitor in liver cells and can significantly inhibit LDLR expression 25. However, the uptake was only reduced by approximately 30% (Figure 8). This may be due to the fact that SR-B1 also plays an important role in the uptake of the nanocomplex. It is worthy to note that cholesterol is not an HSC-specific ligand and the SSCP nanocomplex can also be accumulated in other liver cells. As demonstrated in our previous study, cholesterol conjugation can enhance the cellular uptake of oligonucleotides in hepatocytes, kupffer cells and HSCs 17. In this study we used cholesterol as a model ligand to develop and validated the SSCP nanocomplex. In the future, we will replace cholesterol with other HSC-specific ligands, such as M6P and vitamin A, to specifically deliver the PCBP2 siRNA into HSCs.
The stability of siRNA in blood circulation is another major challenge for successful delivery of siRNA. Intravenously administered siRNAs are exposed to several blood components and subjected to nuclease degradation, resulting in poor stability and low cellular uptake in target tissues.48 Our result indicates that both the SSCP nanocomplex and SSC complex can efficiently protect siRNA from nuclease in the serum (Figure 9). It is not surprising that protamine in the SSCP nanocomplex can form electrostatic interaction with the siRNA and protect it from nucleases. The improved stability of the siRNA in the SSC complex might be attributed to the steric shielding of streptavidin. This finding is similar to a previous study in which an avidin-biotin system protects antisense oligonucleotides against serum nuclease 26.
Conclusion
In this study, we have developed a novel and safe SSCP nanocomplex made by streptavidin-biotin technology, which allows better control of the fabrication of the siRNA nanocomplex as well as easier attachment of targeting ligand without affecting siRNA activity. The results showed that the SSCP nanocomplex is not only stable in the serum, but also capable of entering cells and inducing silencing effect in HSC-T6 cells. These findings indicate that the SSCP nanocomplex can be used as a versatile platform for safe and effective delivery of siRNA to target cells.
References
- 1.Cheng K, Mahato RI. Gene modulation for treating liver fibrosis. Crit Rev Ther Drug Carrier Syst. 2007;24(2):93–146. doi: 10.1615/critrevtherdrugcarriersyst.v24.i2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegmund SV, Dooley S, Brenner DA. Molecular mechanisms of alcohol-induced hepatic fibrosis. Dig Dis. 2005;23(3-4):264–74. doi: 10.1159/000090174. [DOI] [PubMed] [Google Scholar]
- 3.Shukla RS, Qin B, Wan YJ, Cheng K. PCBP2 siRNA reverses the alcohol-induced pro-fibrogenic effects in hepatic stellate cells. Pharm Res. 2011;28(12):3058–68. doi: 10.1007/s11095-011-0475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lesch HP, Kaikkonen MU, Pikkarainen JT, Yla-Herttuala S. Avidin-biotin technology in targeted therapy. Expert Opin Drug Deliv. 2010;7(5):551–64. doi: 10.1517/17425241003677749. [DOI] [PubMed] [Google Scholar]
- 5.Lee H, Kim TH, Park TG. A receptor-mediated gene delivery system using streptavidin and biotin-derivatized, pegylated epidermal growth factor. Journal of controlled release: official journal of the Controlled Release Society. 2002;83(1):109–19. doi: 10.1016/s0168-3659(02)00166-9. [DOI] [PubMed] [Google Scholar]
- 6.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic acids research. 2006;34(10):e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng X, Sun YX, Zhang XZ, Zhuo RX. Biotinylated disulfide containing PEI/avidin bioconjugate shows specific enhanced transfection efficiency in HepG2 cells. Organic & biomolecular chemistry. 2009;7(20):4201–10. doi: 10.1039/b910831a. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Rizzo MA, Bhattacharya S, Huang L. Characterization of cationic lipid-protamine-DNA (LPD) complexes for intravenous gene delivery. Gene Ther. 1998;5(7):930–7. doi: 10.1038/sj.gt.3300683. [DOI] [PubMed] [Google Scholar]
- 9.Ha W, Wu H, Wang X-LW, Peng S-L, Ding L-S, Zhang S, Li B-J. Self-aggregates of cholesterol-modified carboxymethyl konjac glucomannan conjugate: Preparation, characterization, and preliminary assessment as a carrier of etoposide. Carbohydrate Polymers. 2011;86(2):513–519. [Google Scholar]
- 10.Haberland A, Zaitsev S, Waldofner N, Erdmann B, Bottger M, Henke W. Structural appearance of linker histone H1/siRNA complexes. Mol Biol Rep. 2009;36(5):1083–93. doi: 10.1007/s11033-008-9282-8. [DOI] [PubMed] [Google Scholar]
- 11.Mahato R, Qin B, Cheng K. Blocking IKKalpha expression inhibits prostate cancer invasiveness. Pharm Res. 2011;28(6):1357–69. doi: 10.1007/s11095-010-0351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tai W, Chen Z, Cheng K. Expression profile and functional activity of peptide transporters in prostate cancer cells. Mol Pharm. 2013;10(2):477–87. doi: 10.1021/mp300364k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin B, Cheng K. Silencing of the IKKepsilon gene by siRNA inhibits invasiveness and growth of breast cancer cells. Breast cancer research: BCR. 2010;12(5):R74. doi: 10.1186/bcr2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNaughton BR, Cronican JJ, Thompson DB, Liu DR. Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. Proc Natl Acad Sci U S A. 2009;106(15):6111–6. doi: 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren Y, Hauert S, Lo JH, Bhatia SN. Identification and characterization of receptor-specific peptides for siRNA delivery. ACS Nano. 2012;6(10):8620–31. doi: 10.1021/nn301975s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. PEG conjugated VEGF siRNA for anti-angiogenic gene therapy. J Control Release. 2006;116(2):123–9. doi: 10.1016/j.jconrel.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Cheng K, Ye Z, Guntaka RV, Mahato RI. Enhanced hepatic uptake and bioactivity of type alpha1(I) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. J Pharmacol Exp Ther. 2006;317(2):797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- 18.Lorenz C, Hadwiger P, John M, Vornlocher HP, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett. 2004;14(19):4975–7. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 19.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25(10):1149–57. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Kristensen J, Ruffner DE. Delivery of antisense oligonucleotides using HPMA polymer: synthesis of A thiol polymer and its conjugation to water-soluble molecules. Bioconjugate chemistry. 1998;9(6):749–57. doi: 10.1021/bc980034k. [DOI] [PubMed] [Google Scholar]
- 21.Singh N, Agrawal A, Leung AK, Sharp PA, Bhatia SN. Effect of nanoparticle conjugation on gene silencing by RNA interference. J Am Chem Soc. 2010;132(24):8241–3. doi: 10.1021/ja102132e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Asbeck AH, Beyerle A, McNeill H, Bovee-Geurts PH, Lindberg S, Verdurmen WP, Hallbrink M, Langel U, Heidenreich O, Brock R. Molecular Parameters of siRNA-Cell Penetrating Peptide Nanocomplexes for Efficient Cellular Delivery. ACS Nano. 2013 doi: 10.1021/nn305754c. [DOI] [PubMed] [Google Scholar]
- 23.Qin B, Tai W, Shukla RS, Cheng K. Identification of a LNCaP-specific binding peptide using phage display. Pharmaceutical research. 2011;28(10):2422–34. doi: 10.1007/s11095-011-0469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava RA, Ito H, Hess M, Srivastava N, Schonfeld G. Regulation of low density lipoprotein receptor gene expression in HepG2 and Caco2 cells by palmitate, oleate, and 25-hydroxycholesterol. J Lipid Res. 1995;36(7):1434–46. [PubMed] [Google Scholar]
- 25.Moon J, Lee SM, Do HJ, Cho Y, Chung JH, Shin MJ. Quercetin Up-regulates LDL Receptor Expression in HepG2 Cells. Phytotherapy research: PTR. 2012 doi: 10.1002/ptr.4646. [DOI] [PubMed] [Google Scholar]
- 26.Boado RJ, Pardridge WM. Complete protection of antisense oligonucleotides against serum nuclease degradation by an avidin-biotin system. Bioconjug Chem. 1992;3(6):519–23. doi: 10.1021/bc00018a010. [DOI] [PubMed] [Google Scholar]
- 27.Bogusiewicz A, Mock NI, Mock DM. A biotin-protein bond with stability in plasma. Anal Biochem. 2005;337(1):98–102. doi: 10.1016/j.ab.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 28.Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP. Sticky overhangs enhance siRNA-mediated gene silencing. Proc Natl Acad Sci U S A. 2007;104(41):16050–5. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Targeted quantum dot conjugates for siRNA delivery. Bioconjug Chem. 2007;18(5):1391–6. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- 30.Leng Q, Woodle MC, Lu PY, Mixson AJ. Advances in Systemic siRNA Delivery. Drugs of the future. 2009;34(9):721. doi: 10.1358/dof.2009.034.09.1413267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Lu Z, Wientjes MG, Au JL. Delivery of siRNA therapeutics: barriers and carriers. The AAPS journal. 2010;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehead KA, Dahlman JE, Langer RS, Anderson DG. Silencing or stimulation? siRNA delivery and the immune system. Annual review of chemical and biomolecular engineering. 2011;2:77–96. doi: 10.1146/annurev-chembioeng-061010-114133. [DOI] [PubMed] [Google Scholar]
- 33.Bouxsein NF, McAllister CS, Ewert KK, Samuel CE, Safinya CR. Structure and gene silencing activities of monovalent and pentavalent cationic lipid vectors complexed with siRNA. Biochemistry. 2007;46(16):4785–92. doi: 10.1021/bi062138l. [DOI] [PubMed] [Google Scholar]
- 34.Lesch HP, Kaikkonen MU, Pikkarainen JT, Yla-Herttuala S. Avidin-biotin technology in targeted therapy. Expert Opin Drug Deliv. 2010;7(5):551–64. doi: 10.1517/17425241003677749. [DOI] [PubMed] [Google Scholar]
- 35.Rosebrough SF. Pharmacokinetics and biodistribution of radiolabeled avidin, streptavidin and biotin. Nucl Med Biol. 1993;20(5):663–8. doi: 10.1016/0969-8051(93)90037-u. [DOI] [PubMed] [Google Scholar]
- 36.Kim SH, Jeong JH, Kim TI, Kim SW, Bull DA. VEGF siRNA delivery system using arginine-grafted bioreducible poly(disulfide amine) Mol Pharm. 2009;6(3):718–26. doi: 10.1021/mp800161e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia CF, Zhang Y, Zhang Y, Boado RJ, Pardridge WM. Intravenous siRNA of brain cancer with receptor targeting and avidin-biotin technology. Pharmaceutical research. 2007;24(12):2309–16. doi: 10.1007/s11095-007-9460-8. [DOI] [PubMed] [Google Scholar]
- 38.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharm. 2006;3(5):579–88. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 39.Zhao ZX, Gao SY, Wang JC, Chen CJ, Zhao EY, Hou WJ, Feng Q, Gao LY, Liu XY, Zhang LR, Zhang Q. Self-assembly nanomicelles based on cationic mPEG-PLA-b-Polyarginine(R(15)) triblock copolymer for siRNA delivery. Biomaterials. 2012 doi: 10.1016/j.biomaterials.2012.05.067. [DOI] [PubMed] [Google Scholar]
- 40.Yang XZ, Dou S, Wang YC, Long HY, Xiong MH, Mao CQ, Yao YD, Wang J. Single-Step Assembly of Cationic Lipid-Polymer Hybrid Nanoparticles for Systemic Delivery of siRNA. ACS Nano. 2012;6(6):4955–65. doi: 10.1021/nn300500u. [DOI] [PubMed] [Google Scholar]
- 41.Sorgi FL, Bhattacharya S, Huang L. Protamine sulfate enhances lipid-mediated gene transfer. Gene Ther. 1997;4(9):961–8. doi: 10.1038/sj.gt.3300484. [DOI] [PubMed] [Google Scholar]
- 42.Gao J, Liu W, Xia Y, Li W, Sun J, Chen H, Li B, Zhang D, Qian W, Meng Y, Deng L, Wang H, Chen J, Guo Y. The promotion of siRNA delivery to breast cancer overexpressing epidermal growth factor receptor through anti-EGFR antibody conjugation by immunoliposomes. Biomaterials. 2011;32(13):3459–70. doi: 10.1016/j.biomaterials.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 43.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nature biotechnology. 2007;25(10):1149–57. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama T, Butler JS, Sehgal A, Severgnini M, Racie T, Sharman J, Ding F, Morskaya SS, Brodsky J, Tchangov L, Kosovrasti V, Meys M, Nechev L, Wang G, Peng CG, Fang Y, Maier M, Rajeev KG, Li R, Hettinger J, Barros S, Clausen V, Zhang X, Wang Q, Hutabarat R, Dokholyan NV, Wolfrum C, Manoharan M, Kotelianski V, Stoffel M, Sah DW. Harnessing a physiologic mechanism for siRNA delivery with mimetic lipoprotein particles. Molecular therapy: the journal of the American Society of Gene Therapy. 2012;20(8):1582–9. doi: 10.1038/mt.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong SC, Klein JJ, Hamilton HL, Chu Q, Frey CL, Trubetskoy VS, Hegge J, Wakefield D, Rozema DB, Lewis DL. Co-injection of a targeted, reversibly masked endosomolytic polymer dramatically improves the efficacy of cholesterol-conjugated small interfering RNAs in vivo. Nucleic acid therapeutics. 2012;22(6):380–90. doi: 10.1089/nat.2012.0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–8. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 47.Rossi JJ. Medicine: a cholesterol connection in RNAi. Nature. 2004;432(7014):155–6. doi: 10.1038/432155a. [DOI] [PubMed] [Google Scholar]
- 48.Takakura Y, Nishikawa M, Yamashita F, Hashida M. Influence of physicochemical properties on pharmacokinetics of non-viral vectors for gene delivery. Journal of drug targeting. 2002;10(2):99–104. doi: 10.1080/10611860290016694. [DOI] [PubMed] [Google Scholar]