Abstract
Objective
The goal of the present study was to identify differences in gene expression between SAT, VAT, and EAT depots in Class III severely obese individuals.
Design
Human subcutaneous (SAT) and visceral (VAT) adipose tissues exhibit differential gene expression profiles. There is little information, however, about the other proximal white adipose tissue, epigastric (EAT) in terms of its function and contribution to metabolism.
Subjects and Methods
Using RNA from adipose biospecimens obtained from Class III severely obese patients undergoing open Roux-en-Y gastric bypass surgery, we compared gene expression profiles between SAT, VAT, and EAT, using microarrays validated by real time quantitative PCR.
Results
The three depots were found to share 1,907 genes. VAT had the greatest number of genes [66] expressed exclusively in this depot, followed by SAT [23], and then EAT [14]. Moreover, VAT shared more genes with EAT [65] than with SAT [38]. Further analyses using ratios of SAT/EAT, VAT/EAT, and SAT/VAT, identified specific as well as overlapping networks and pathways of genes representing dermatological diseases, inflammation, cell cycle and growth, cancer, and development. Targeted analysis of genes playing a role in adipose tissue development and function, revealed that Peroxisome proliferator-activated receptor Gamma Coactivator 1-alpha (PGC1-α) that regulates the precursor of the hormone Irisin (FNCD5), were abundantly expressed in all three fat depots, along with fibroblast growth factors (FGF) FGF1, FGF7, and FGF10, whereas, FGF19 and FGF21 were undetectable.
Conclusions
These data indicate that EAT has more in common with VAT suggesting similar metabolic potential. The human epigastric adipose depot could play a significant functional role in metabolic diseases and should be further investigated.
Keywords: epigastric, visceral, subcutaneous, adipose, FGF19, microarrays
Introduction
Human adipose is a diversified tissue-organ with different depots secreting a variety of hormones.1 Subcutaneous (SAT) and visceral (VAT) adipose tissues are two major types of adipose in adult humans, whereas, a small amount of brown adipose plays a role in temperature control in newborns.2 Differences in gene expression between pre-adipocytes and adipocytes3 and between subcutaneous and visceral human adipose tissue have been reported previously.4, 5 VAT is associated with insulin resistance, diabetes, hypertension, atherosclerosis, and hepatic steatosis, while, SAT responds better to insulin and secretes more adiponectin and less inflammatory cytokines.6 These differences may be expected given that subcutaneous and visceral adipocytes have arisen from different progenitor cells that exhibit different gene expression patterns.6,7
Epigastric adipose tissue (EAT), on the other hand, is not well-studied in human physiology. Only a few studies have determined functional differences between SAT, VAT, and EAT adipose. EAT is considered to be more closely related to abdominal SAT than visceral adipose, but there is fat cell size variation between genders.8 Epigastric fat cell size is closely related to depots from femoral and gluteal regions, while, SAT adipose is more sensitive than epigastric adipose to hormonal and nutritional factors.9 Furthermore, there is significant correlation between hyperinsulinemia and lipolytic activity of EAT in obese patients, independently from fat distribution, while in visceral adipose such a relationship was found only in patients with predominant visceral adiposity.10 In general, however, there is little information regarding the functional importance of EAT and its relationship with other depots in the region of the waist that is closely associated with metabolic diseases.11-14 Previous studies using microarrays have used expression profiling in SAT to predict weight loss in non-responders consuming low fat diet.15 In addition, gene expression profiling using VAT revealed an upregulation of mitogen-activated protein kinases in obese compared to lean individuals.16
Here, we tested the hypothesis that SAT, VAT, and EAT share gene expression profiles, networks, and pathways, but are also characterized by unique genetic signatures. We used RNA prepared from adipose biospecimens obtained from Class III obese (i.e., body mass index - BMI > 40 kg/m2) patients undergoing open Roux-en-Y gastric bypass (RYGB) surgery. Global gene expression profiling was performed using the Affymetrix GeneChip Exon 1.0ST Arrays followed by validation of gene expression using the sensitive real time quantitative PCR methods. In addition we determined the expression profile of a select number of fibroblast growth factors, FNDC5 (Fibronectin type III domain containing 5) which encodes the newly discovered hormone Irisin17, and the Peroxisome proliferator-activated receptor Gamma Coactivator 1-alpha (PGC1-α) which regulates FNDC5.18
Materials and Methods
Study-subject characteristics
Three types of white adipose tissue [subcutaneous (SAT), visceral (VAT), epigastric (EAT)] were obtained from patients with extreme (or Class III) obesity (mean BMI: 56.8 kg/m2) undergoing open Roux-en-Y gastric bypass (RYGB) at Geisinger Medical Center, as we have previously described.19-21 SAT was obtained approximately 2 cm deep to the abdominal skin. VAT was obtained from the omental apron several centimeters from the transverse colon. EAT was obtained from partial excision of the gastric fat pad at the angle of His. All adipose tissue biopsies were obtained after completion of the bypass procedure and right before the closing phase.
Two different groups of patients with available electronic health records were used in these experiments: (a) a group of six patients whose adipose was used for the microarray experiments, and (b) a confirmation group of 10 patients that were used for validation of genes from the microarray by real time qPCR, which included five randomly selected patients out of the six whose RNA was used for the microarray component, and five new RYGB patients with similar characteristics (Table 1). These studies were approved by the Geisinger Medical Center Institutional Review Board (IRB) for research. All participants provided informed written consent.
Table 1. Characteristics of patients used for the microarray and real-time qPCR experiments.
The numbers represent the sample size (N), mean Age (years old), ethnic background as Black to White ratio (B/W), male to female ratio (M/F), and mean body mass index (BMI: kg/m2) for each group of patients. The group of the 10 patients used for the qPCR confirmation experiments was comprised of five randomly selected patients that were used in the microarrays and five new patients that were entirely different.
| N | Age | B/W | M/F | BMI | |
|---|---|---|---|---|---|
| Adipose (microarrays) |
6 | 56 | 1/5 | 2/4 | 56.8 |
| Adipose (qPCR confirmation) |
10 | 51 | 0/10 | 3/7 | 50.8 |
RNA preparation
Human SAT, VAT, and EAT biopsies were collected from six patients undergoing open gastric bypass surgery. Samples were immediately placed in RNAlater and stored at −80 °C until analysis. RNA from biopsy tissue was prepared using the RNeasy Lipid Tissue Mini Kit (Qiagen, Valencia, CA). RNA quantitation was performed using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE).
Microarrays
The Human GeneChip Exon 1.0ST Array from Affymetrix (Santa Clara, CA) was used to generate RNA profiles. RNA samples from two individuals, from the same adipose type, were pooled for analysis on a single Genechip, allowing for three chips for each type of adipose. Combining RNA from two individuals per chip served to reduce inter-individual variation on gene expression. A total of nine chips were used (three for SAT, three for VAT, and three for EAT). RNA was labeled and hybridized in a GeneChip Hybridization Oven 640 according to the manufacturer’s protocol (Affymetrix, Santa Clara, CA). The hybridized arrays were washed, and stained using the Affymetrix GeneChip Fluids Station 480. Each array was scanned using the Affymetrix GeneChip Scanner 3000 7G and the Affymetrix GeneChip Operating Software (GCOS) was used to compute the expression levels from the probe level data. Quality control analysis was completed with RMA Sketch (Affymetrix).
Microarray data analysis
Primary analysis of microarrays was completed with Spotfire Decision Site version 9.1.2 using Decision site for functional genomics (TIBCO, Palo Alto, CA). A fluorescence signal level of “50” or below was considered to be the lowest threshold to determine presence or absence of expression. This level of “50” was confirmed by qPCR (discussed below). Ratios of SAT/EAT, VAT/EAT, and SAT/VAT were used to compare fold-changes among pairs of adipose types. ANOVA was used to determine significance of differences between ratios, at P-value < 0.05. A secondary analysis was completed with IPA (Ingenuity Systems, Inc. Redwood City, CA). The fold changes were converted to logbase 2 and uploaded to IPA. A core analysis was completed and the software was used to model and analyze the data.
All microarray expression data were analyzed under MIAME guidelines. Data were submitted and accepted to Gene Expression Omnibus (GEO) with accession number “GSE42715” and available with the following WWW link:http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ltijjyoesmswqtc&acc=GSE42715
Real Time qPCR
Microarray data were confirmed using the real time quantitative polymerase chain reaction (qPCR) method. Human adipose samples from SAT, VAT, and EAT were obtained from an additional five patients undergoing open gastric bypass surgery. Total RNA was prepared as described above. RNA was reverse transcribed using a high capacity cDNA reverse transcription with RNase inhibitor kit (Applied Biosystems-Life Technologies, Carlsbad, CA). Quantitative PCR was performed in duplicate on the ABI 7500 Fast Plate (Applied Biosystems). Pre-designed assays were obtained from Applied Biosystems for the following genes: AOX1 (Hs00154079_m1), AQP9 (Hs01035888_m1), AREG (Hs00950669_m1), CFB (Hs00156060_m1), CFI (Hs00989715_m1), DLR1 (Hs01552593_m1), EGFL6 (Hs01556006_m1), FGF1 (Hs00265254_m1), FGF7 (Hs00940253_m1), FGF9 (Hs00181829_m1), FGF10 (Hs00610298_m1), FGF19 (Hs00192780_m1), FGF21 (Hs00173927_m1), Fibronectin type III domain containing 5 - FNDC5 (Hs00401006_m1), HPR (Hs00750565_s1), IL6 (Hs00985639_m1), IL8 (Hs00174103_m1), ITLN1 (Hs00914745_m1), LDLR (Hs00181192_m1), PGC1-alpha (Hs01016719_m1), PTX3 (Hs00173615_m1), SELE (Hs00950401_m1), SERPINB (Hs01010736_m1), SYT4 (Hs01086433_m1), and TCF21 (Hs00162646_m1). GAPDH (Hs02758991_m1) was used as the calibrator gene. Analysis was conducted by subtracting the Ct value of GAPDH from the Ct value of the gene of interest. This delta value was adjusted by subtracting from 40, as previously described shown 22,23. The threshold of gene detection using this equation [40-Δ(Ct-Ct of GAPDH)] was set at “30” which corresponds to, approximately, a Ct value for any given gene, at ~35.
Tissue specific and co-expressed genes
Only genes that had already met the criterion of P<0.05 in the initial microarray data ANOVA analysis were considered. As described above, genes whose fluorescence on the microarrays was less than 50 arbitrary units were considered not expressed. Genes with fluorescence greater than 50 arbitrary units were considered to be expressed and were analyzed for expression in the three adipose depots.
Statistical analyses
Comparisons of fold changes after Spotfire analysis were performed by ANOVA with the significance set at P-value < 0.05. Correlation analysis between the microarray and qPCR data was performed using the non-parametric Spearman’s rho analysis. Pearson correlation coefficients were used to assess associations between qPCR data with metabolic variables. Analysis of qPCR data was performed by ANOVA followed by pairwise comparisons by using the least square differences test. Two-Way ANOVA analysis was used to determine if gender modified the association between fat type and qPCR data. JMP Pro 10 and SAS version 9.3 (SAS Institute, Cary, NC) were used for statistical analysis. P-values < 0.05 were considered significant.
Results
Gene expression profiling in SAT, VAT, and EAT
Total RNA was prepared from three types of adipose tissues [subcutaneous (SAT), visceral (VAT), epigastric (EAT)] from six patients with extreme obesity undergoing open RYGB surgery (Table 1). Gene expression profiling was performed using the Human GeneChip Exon 1.0ST Arrays from Affymetrix. Of the 22,011 transcripts represented on the Exon 1.0ST Array, a total of 2,354 transcripts with designated gene identities met the significance level of P<0.05, by ANOVA and were used for all the analyses described here (Figure 1A).
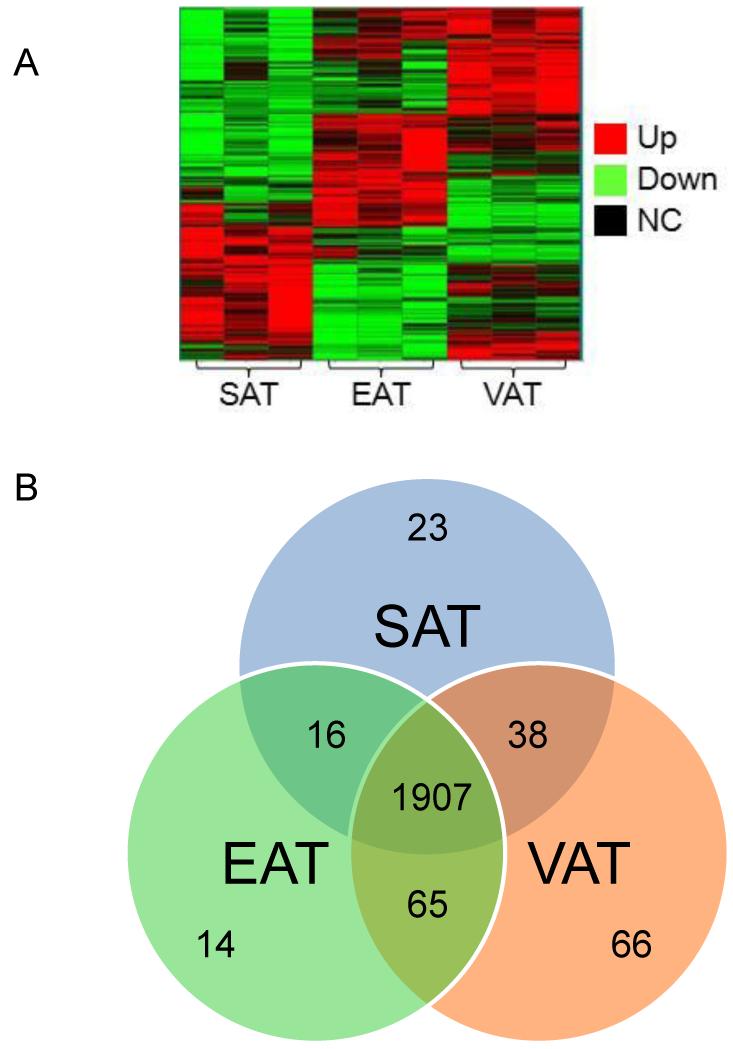
Figure 1. Unique and co-expressed genes in SAT, VAT, and EAT.
(A) Hierarchical cluster analysis of microarray data for genes at P<0.05 by ANOVA (NC: no change). (B) Using as a cut-off for gene expression in the microarrays the value of “50” (corresponding to a Ct-value of “35” by qPCR), the number of unique and co-expressed genes were determined. Each depot is represented by a circle in the Venn diagram. The numbers in the various regions of the three overlapping circles indicate the unique and co-expressed genes among pairs of depots and among all three depots. 1,907 genes were co-expressed in all three depots. EAT had the lowest number (14) of unique genes (i.e., genes expressed only in this depot), while it shared 65 genes with VAT and 16 genes with SAT. SAT and VAT shared 38 genes, while each depot was represented by 23 and 66 unique genes, respectively.
Unique and co-expressed genes
Almost 90% (1,907/2129) of the genes were co-expressed in all three adipose depots (Supplemental Information, Table S1). Hierarchical cluster analysis of these genes revealed distinct patterns of gene expression in the three fat depots (Figure 1A). The three pools of two patients were grouped by fat depot, with SAT having the most differences in gene expression relative to VAT and EAT. VAT had the highest number of unique genes (66), followed by SAT (24) and EAT (14) (Figure 1B, Table 2). VAT had the highest number of co-expressed genes with EAT (65) and then with SAT (38). SAT and EAT shared the expression of 16 genes that were not expressed in VAT (Figure 1B, Table 3).
Table 2. Unique genes for SAT, VAT, EAT, and combinations of depots.
Primary analysis of microarrays from SAT, VAT, and EAT depots was performed with the Spotfire Decision Site version 9.1.2 software. Using as minimum value in the microarrays the value of “50” (corresponding to a Ct-value of “35” by qPCR), a total of 103 genes were found to be expressed (23 in SAT, 66 in VAT, and 14 in EAT).
| SAT (N = 23) |
VAT (N = 66) |
EAT (N = 14) |
|---|---|---|
|
C13orf29; CXCL3; DMRT3;
HOXA9; HOXC9; IL1RL2; KRTAP12-3; LCE3C; MIR17HG; MLF1; MYEOV; NRCAM; NRIP3; ODZ1; PNLDC1; PROKR2; QPCT; RTP3; SERPINA5; SIM1; SLC26A8; SSTR5; STMN2 |
ADAMTS3; ANKS1B; BCHE;
BCO2; BNC1; C21orf62; C6orf203; C7orf46; CACNB2; CDH3; CENPJ; CMYA5; COL8A1; CPA4; CXADR; DLG2; DLG2; DPYS; DSC3; DYNC2LI1; ELMOD2; ENOX1; ENPP5; EXTL2; FAM150B; FLRT3; FRAS1; GPRASP2; HHIP; HOMER1; HSD17B6; IL20; IQCA1; ISL1; ITGB8; ITLN2; KCNT2; KCTD1; KIAA1009; KLHDC10; KLHL4; LARP1B; LCA5; LPAR4; LRP2; LXN; MORN4; MYO5B; NEK11; NME7; PKHD1L1; PNMA2; PRELID2; RHPN2; SERTAD4; SGOL2; SLC28A3; SUCNR1; SULT1C4; THUMPD2; TMEM136; TRPM3; UPK1B; VSNL1; WDR35; ZNF99 |
BTN2A3; C3orf35; FLJ11710;
FLJ13197; KCNA10; LOC100130950; LOC100131492; MYCN; PPP1R14C; TMEM81; ZKSCAN4; ZNF23; ZNF235; ZNF441 |
Table 3. Co-expressed genes for SAT, VAT, EAT, and combinations of depots.
Primary analysis of microarrays from SAT, VAT, and EAT depots was performed with the Spotfire Decision Site version 9.1.2 software. Using as minimum value in the microarrays the value of “50” (corresponding to a Ct-value of “35” by qPCR), 2,026 genes were found to be co-expressed (16 in SAT and EAT, 65 in VAT and EAT, 38 in SAT and VAT, and 1,907 in all three depots).
| SAT & EAT (N = 16) |
VAT & EAT (N = 65) |
SAT & VAT (N = 38) |
|---|---|---|
|
C6orf126; CEBPE;
COMMD9; EMX2; FMR1NB; IGFL4; KCNG1; LCN10; NBPF4; NEU4; OSTalpha; RWDD1; TBX5; TMIGD2; TSPAN11; WFDC13 |
ANKRA2; ANKRD26;
ARMCX2; ARMCX4; ARMCX5; BTBD8; C5; C8orf31; CACNA2D3; CARNS1; CASP6; CCDC111; CGNL1; CHODL; CHRM3; EDA; FAM63B; FGF9; GIN1; GPR64; GRIN1; IL18; ITLN1; LAYN; LOC100132356; LRFN5; MREG; MRPL48; MSH2; MTERFD2; NEK1; PDE1C; PM20D1; PON3; PTN; RARB; RCHY1; REEP1; RG9MTD3; RMND1; RPL17; SASS6; STXBP4; TBX19; THNSL1; TSGA10; TTC5; VPS54; WDSUB1; WT1-AS; ZFP30; ZMYM1; ZNF197; ZNF248; ZNF253; ZNF347; ZNF37BP; ZNF397; ZNF44; ZNF483; ZNF487P; ZNF558; ZNF568; ZNF610; ZNF616 |
ACPP; ARHGEF26; BMP3;
C3orf52; C6orf97; CASS4; CLEC12A; CLEC4E; CREM; CXCL2; DKK1; EIF4E; FPR2; HAL; HDC; HP; IL1RL1; LILRA2; LOC100131826; LRRN1; MT1JP; NLRC4; NLRP12; OR13J1; OR52N1; RGS4; SELE; SEMA3D; SLC22A4; SLCO4C1; SPTY2D1; TAF1A; TREM1; TRIM29; TRPM6; VAT1L; VNN3; VSTM1 |
|
SAT & EAT & VAT (N = 1907) (Complete list of gene provided in Supplementary Information, Table S1) | ||
Relative gene expression levels were analyzed using gene expression ratios (SAT/EAT, VAT/EAT, SAT/VAT). Values of the ratios were logarithmically (base of 2) converted and analyzed using IPA Ingenuity software. Table 4 provides a synopsis of the genes displaying upregulation or downregulation with respect to the depot in the denominator. Selectin (SELE) displayed the highest levels of expression in SAT relative to EAT (30.6-fold, before log2 conversion) and the third highest levels of expression relative to VAT adipose (3.6-fold before log2 conversion). Intelectin 1 (ITLN1), also known as Omentin 1, had the highest levels of expression in VAT adipose relative to EAT (30.7-fold, before log2 conversion) as well as relative to SAT adipose (44.4-fold, before log2 conversion) (Table 4).
Table 4. Fold-difference of gene expression for pairs of adipose depots.
Secondary gene expression analysis was performed with IPA (Ingenuity Systems) software, after logarithmic (log2) conversion of the data. Relative gene expression for each of the indicated pairswas ranked by the fold-increase or decrease. The “+” sign represents higher expression with reference to the tissue in the denominator, and the “−” sign represents lower expression.
| SAT/EAT | VAT/EAT | SAT/VAT |
|---|---|---|
| Fold increase | ||
|
SELE (+4.935); IL6 (+4.734); AREG/ARGEB (+4.502); TREM1 (+4.159); TNFAIP6 (+3.586); EGFL6 (+3.573); AQP9 (+3.443); S100A12 (+3.356); IL8 (+3.304); MMP19 (+3.282) |
ITLN1 (+4.944); CLDN1 (+4.544); IL6 (+4.266); BTG3 (+4.100); FLRT3 (+3.845); AREG/AREGB (+3.715); TREM1 (+3.550); DSC3 (+3.481); BCHE (+3.415); UPK1B (+3.350) |
EGFL6 (+2.863); NRCAM (+2.081); SELE (+1.857); DMRT3 (+1.717); CLIC6 (+1.637); HMOX1 (+1.591); TNFAIP6 (+1.581); PNP (+1.552); TNC (+1.520); STMN2 (+1.478); |
| Fold decrease | ||
|
SPTBN1 (−1.388); MEIS1 (− 1.407); ZNF33B (−1.418); PTGDS (−1.435); PCSK6 (− 1.478); PLAT (−1.483); WT1- AS (−1.589); MYOC (−1.763); C7 (−2.278); GREM1 (−2.595) |
HIST1HA4 (−0.640); EMX2 (− 0.652); MYOC (−0.897); CHS1/CSH2 (−1.504) |
BNC1 (−3.106); WT1-AS (− 3.267); UPK1B (−3.323); DSC3 (−3.437); GREM1 (− 3.577) BCHE (−3.697); FLRT3 (−3.952); CLDN1 (− 4.922); BTG3 (−5.014); ITLN1 (−5.473) |
Network and Pathway analysis
Second level analysis was performed using the IPA Ingenuity Systems application to identify the networks and pathways represented by genes that were differentially expressed among the three adipose depots (Table 5, and Supplemental Information Table S2). Analysis using genes differentially expressed in SAT relative to EAT identified networks that are characteristic of endocrine system development, connective tissue and inflammatory disease, skeletal muscle development, endocrine and gastrointestinal disorders, and cell morphology-embryonic development. The analysis using genes differentially expressed in VAT relative to EAT identified networks of genes reflecting cardiovascular system development, cancer-reproductive disease, embryonic development, tumor morphology and cell cycle, and cancer-connective tissue disorders. The analysis using genes differentially expressed in SAT relative to VAT identified networks of genes that play a role in connective tissue and dermatological disorders, cancer/skeletal muscle disorders, cellular-embryonic development, DNA replication-repair, and cell morphology/embryonic development.
Table 5. Top associated Networks of function and top canonical Pathways for the indicated ratios (pairings) of SAT, VAT, and EAT.
Networks and pathways for each ratio of adipose depots were configured by IPA (Ingenuity Systems) software analysis, using the logarithmically (to the base of 2) converted microarray data. The networks for each pairing are ranked from top to bottom, as determined by the IPA analysis. Complete list of genes is provided in the Supplemental Information, Tables S2 & S3. (RA: rheumatoid arthritis; HMGB1: high mobility group box 1; TREM1: Triggering Receptor Expressed on Myeloid cells 1; HER-2: epidermal growth factor receptor 2).
| SAT/EAT | VAT/EAT | SAT/VAT |
|---|---|---|
| TOP NETWORKS | ||
| Endocrine System Development | Cardiovascular System Development |
Connective Tissue Disorders, Dermatological Disease |
| Connective Tissue Disorders, Inflammatory Disease |
Cancer, Reproductive System Disease |
Cancer, Skeletal and Muscular Disorders |
| Skeletal and Muscular System Development |
Embryonic Development) | Cellular Development, Embryonic Development |
| Endocrine, and Gastrointestinal Disease |
Tumor Morphology, Cell Cycle | DNA Replication, Repair |
| Cell Morphology, Embryonic Development |
Cancer, Connective Tissue Disorders |
Cell Morphology, Cellular Development |
| TOP PATHWAYS | ||
| IL-10 Signaling | Macrophages, Fibroblasts in RA | Glioma Invasiveness Signaling |
| IL-8 Signaling | IL-10 Signaling | Macrophages, Fibroblasts in RA |
| IL-6 Signaling | IL-6 Signaling | Osteoblasts, Osteoclasts in RA |
| Macrophages, Fibroblasts in RA | IL-7A Signaling in Fibroblasts | Pancreatic Adenocarcinoma Signaling |
| HMGB1 Signaling | TREM1 Signaling | HER-2 Signaling in Breast Cancer |
IPA analysis also identified top canonical pathways that were particularly conserved among the ratios of depots (Table 5). Pathways represented by genes differentially expressed in SAT and VAT relative to EAT included IL-10, IL-6, IL8, Macrophage and fibroblasts in rheumatoid arthritis, Osteoblasts and osteoclasts in rheumatoid arthritis, and IL-7A pathways, which collectively reflect the presence of inflammation and immune response to inflammation (Table 5). SAT relative to VAT identified different pathways including glioma invasiveness signaling, osteoblasts-osteoclasts in rheumatoid arthritis, pancreatic adrenocarcinoma, and HER-2 signaling in breast cancer (Table 5 and Supplemental Information Table S3).
Validation of microarray gene expression by qPCR
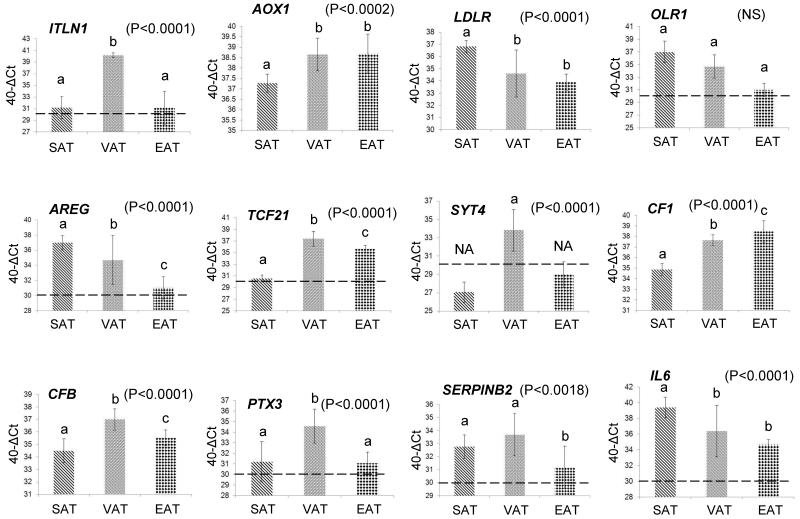
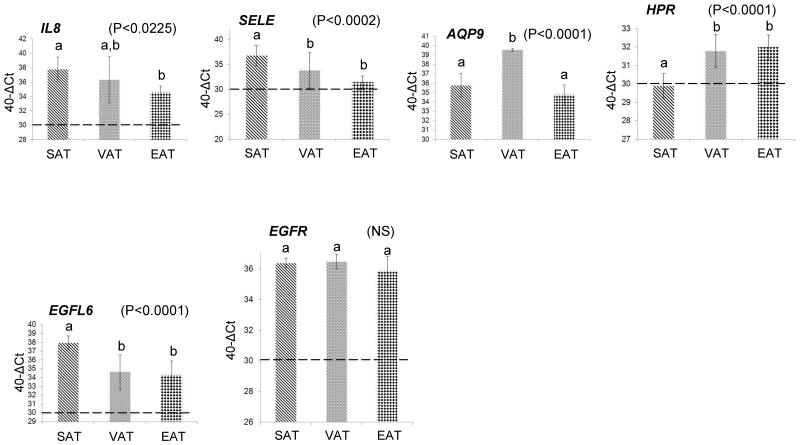
In order to quantify more precisely the differential expression found by microarray analysis, 18 genes were selected for validation by quantitative qPCR. The criteria for gene selection were based on the degree of differential expression of each gene and its known functional roles in metabolic diseases. RNA from fat biopsies obtained from an independent group of five patients undergoing RYGB surgery was used in the qPCR experiments, along with the RNA from five randomly selected patients out of the six patients that had been used for the microarray analysis (Table 1). The qPCR data were also stratified according to gender which did not reveal significant gender-specific differences. Most of the genes showed similar patterns of expression compared to the microarrays (GEO accession number: GSE42715). For example, ITLN1 displayed the highest expression in visceral adipose (Figure 2), which was identical to the pattern of expression identified by the microarrays. Similarly, AOX1, LDLR, OLR1, AREG, TCF21, SYT4, CF1, CFB, PTX3, SERPINB2, IL6, IL8, SELE, AQP9, HRP, EGFL6, EGFR (Figure 2) displayed almost identical patterns of expression in all three adipose depots as determined by the microarrays (Figures 2 & 3, Table S4). Correlation analysis between the microarray and qPCR data from all patients identified significant correlations in all depots, as follows: [SAT r-value: 0.86 (P-value < 0.0001); VAT r-value: 0.62 (P < 0.0008); EAT r-value: 0.74 (P < 0.0001)]. Further correlation analysis using the qPCR data only from the patients that had been used in the microarrays, provided comparable results: [SAT r-value: 0.85 (P-value < 0.0001); VAT r-value: 0.57 (P < 0.002); EAT r-value: 0.75 (P < 0.0001)].
Figure 2. Confirmation of microarray gene expression levels using quantitative real time qPCR.
Genes displaying the highest fold differences (Table 2) were selected for confirmation by qPCR using RNA from SAT, VAT, and EAT isolated from 10 patients undergoing open Roux-en-Y gastric bypass. The following genes were tested: ITLN1, AOX1, LDLR, OLR1, AREG, TCF21, SYT4, CF1, CFB, PTX3, SERPINB2, IL6, IL8, SELE, AQP9, HRP, EGFL6, EGFR. The Y-axis values represent the difference (Δ) of the Ct value of GAPDH from the Ct value of the gene of interest which was further adjusted by subtracting from 40. The horizontal dotted line on each figure represents the threshold of gene detection using the above equation [40-Δ(Ct-GAPDH Ct)], set at “30” which corresponds to a gene Ct value of, approximately, 35. Data are presented as mean ± SD. The different low-case letters above the columns indicate pairs that were statistically different from each other. The same letter indicates no statistical significance between adipose depots in terms of gene expression. Significance was set at P < 0.05 and determined by ANOVA and post-hoc analysis by the least square difference test (NS: Not significant, NA: Not applicable – below detection level).
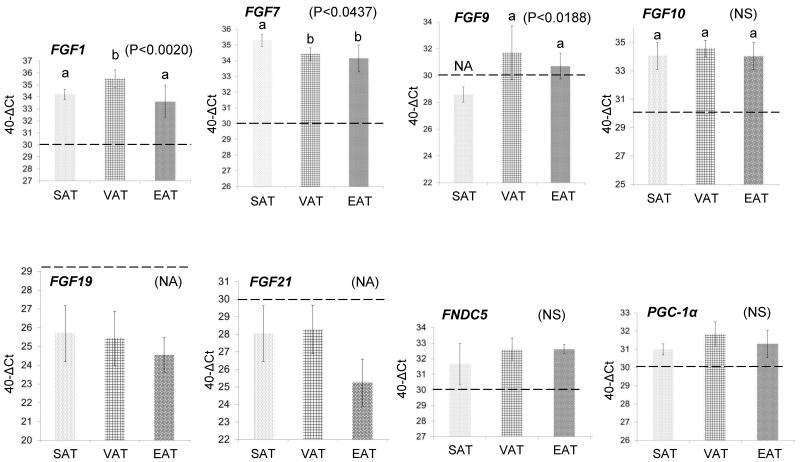
Figure 3. Confirmation of microarray gene expression levels of a select number of genes of interest, using quantitative real time qPCR.
The microarray expression levels of six fibroblast growth factors (FGF1, FGF7, FGF9, FGF10, FGF19, FGF21), the precursor gene of Irisin (Fibronectin type III domain containing 5 - FNDC5), and the transcription factor Peroxisome proliferator-activated receptor Gamma Coactivator 1-alpha (PGC1-α) were confirmed by qPCR, using SAT, VAT, and EAT isolated from 10 patients undergoing open Roux-en-Y gastric bypass. The Y-axis values represent the difference (Δ) of the Ct value of GAPDH from the Ct value of the gene of interest which was further adjusted by subtracting from 40. The horizontal dotted line on each figure represents the threshold of gene detection using the above equation [40-Δ(Ct-GAPDH Ct)], set at “30” which corresponds to a gene Ct value of, approximately, 35. Data are presented as mean ± SD. The different low-case letters above the columns indicate pairs that were statistically different from each other. The same letter indicates no statistical significance between adipose depots in terms of gene expression. Significance was set at P < 0.05 and determined by ANOVA and post-hoc analysis by the least square difference test (NS: Not significant, NA: Not applicable - below detection level).
Other genes with functional significance in human adipose
We were particularly interested in adipose-expressed genes that play a role in adipocyte development as well as the pathophysiology of obesity and diabetes. We examined the expression levels of fibroblast growth factors (FGF1, FGF7, FGF9, FGF10, FGF19, FGF21), the precursor of Irisin (Fibronectin type III domain containing 5 - FNDC5), and the transcription factor Peroxisome proliferator-activated receptor Gamma Coactivator 1-alpha (PGC1-α). The qPCR experiments again confirmed the microarray data in terms of magnitude and pattern of expression. For example, the microarrays showed expression values that were at background levels for FGF19 and FGF21 (<50) which was confirmed by qPCR which showed that these two genes were undetectable (Figure 3). FGF1, FGF7, FGF10, FNCD5, PGC-1α, on the other hand, showed robust expression in all three depots (Figure 3), while, FGF9 was marginally expressed.
Correlations of gene expression with clinical variables
Correlation analyses were performed using the qPCR data of the genes in Figures 2 and 3, in all 10 patients, with the following metabolic phenotypes: fasting glucose levels, HDL, LDL, total cholesterol, triglycerides, systolic blood pressure, diastolic blood pressure, and body mass index using Pearson correlation coefficients. We found strong correlations (correlation co-efficient > 0.70 or < −0.70) between some of these clinical features with gene expression in VAT or EAT (but not in SAT), as follows: SERPINB2 in VAT vs systolic BP: r = −0.814; LDLR in EAT vs diastolic BP: r = −0.801; OLR1 in VAT vs systolic BP: r = −0.729; LDLR in VAT vs diastolic BP: r = −0.717; EGFL6 in VAT vs BMI: r = 0.714; AQP9 in EAT vs glucose: r = 0.704; IL6 in VAT vs systolic BP: r = 0.700. None of these correlations were significant following Bonferroni corrections for multiple testing.
Discussion
Adipose tissue is anatomically, functionally, and metabolically heterogeneous and is thought to contribute to the pathophysiology of a variety of medically important conditions, particularly those related to obesity. Here, we conducted gene expression profiling using RNA obtained from three types of human white adipose depots: subcutaneous, visceral, and epigastric. Differences between abdominal SAT and VAT adipose24, and between abdominal SAT and gluteal adipose25 were recently reported in humans, though not in the severely obese or Class III obese patients.
Validation of the microarray data in each depot was performed by qPCR using 26 genes which showed significantly high correlations between the two methodologies, confirmed by using either the qPCR data from all patients or just the patients that were used for the microarrays. We found that the majority of genes were expressed in all three depots. Only 66 genes were exclusively expressed in VAT, 23 in SAT, and 14 in EAT. These depot-specific genes were predominantly not known to have functional roles (e.g., as leptin and adiponectin do) in metabolic diseases but had documented roles in other diseases. For example, IL1RL2 expressed exclusively in SAT has been shown to play a role in skin inflammation26, ADAMTS3 expressed only in VAT plays a role in myocardial infarction, and KCNA10 expressed in EAT has been associated with cardiomyocyte activation27. The fact that VAT had the highest number of genes expressed exclusive in this depot is indicative of more specialized lineage and/or specific functional properties by this depot. VAT shared with EAT more than twice as many genes than it shared with SAT. This suggests that VAT and EAT may have more in common in terms of lineage and/or functional properties than SAT does with EAT. We should emphasize that only genes displaying significantly differential expression among the three depots and, in addition, could potentially be validated by qPCR were used, leaving out ~90% of the total number of genes available on the microarrays. Using less stringent inclusion criteria, we could possibly have identified more unique (and overlapping) genes for each depot. The latter option is still available to the scientific community by using the open access, GEO-submitted, raw datasets of our microarrays.
The ratios of SAT/EAT, VAT/EAT, and SAT/VAT also identified several genes that were up- or down-regulated. SELE (E-Selectin), for instance, displayed the highest expression specifically in SAT when compared to EAT or VAT, which is consistent with previous studies.28 ITLN1 (intelectin or omentin) displayed the highest expression in VAT, also consistent with previous studies.29 EGFL6 (epidermal growth factor-like domain multiple-6) was expressed at higher levels in SAT, as previously reported30, when compared to EAT. In contrast, IL6 (interleukin 6) was significantly higher in both SAT and VAT compared to EAT suggesting that IL6 mediated metabolic effects may originate with both SAT and VAT. Despite this similarity, SAT and VAT may function substantially different from each other for a number of metabolic activities.31 Taking together the single gene and ratios analysis, we observed substantial overlaps in gene expression levels between depots suggesting the presence of shared functions. The relatively small number of uniquely expressed genes in each depot suggests the presence of a small degree of specialization by each depot. The functional impact of these genes, however, may be of particular importance although that cannot be determined from the present type of analysis.
A second level analysis was performed to determine biological networks and pathways of functional significance. The network identified as most significantly enriched in SAT, relative to VAT and the second most relative to EAT, was the Connective Tissue Disorders, Dermatological Disease network. Given the anatomic proximity of SAT to the skin, this pathway highlights a potentially important role for SAT in dermatological diseases. Many skin disorders are immunologically mediated and the ability of SAT to produce such factors could implicate it in the pathogenesis of this group of disorders. Despite this seemingly obvious observation, more experimental data would be required to support this possibility.
In VAT, the network identified as most significantly enriched was Cardiovascular System Development. This is also not surprising given the known role for VAT in metabolic syndrome which is a significant cardiovascular risk factor. In the SAT/VAT pairing, genes formed networks representative again of dermatological disease, in addition to networks indicating the presence of inflammation, and cellular growth and movement. In the VAT/EAT pairing, the majority of networks were indicative of dysregulation of cell cycle and signaling as found in cancer biology. The SAT/VAT pairing, although similar to the SAT/EAT pathway for dermatological disease pathway enrichment, again strongly influenced by genes more differentially regulated in SAT, was different from both SAT/EAT and VAT/EAT in the networks that were representative of presence of developmental, skeletal, and muscular system disorders, as well as cancer. Taken together, these networks reflect similarities between SAT and VAT because, when paired with EAT, both displayed networks that are reflective of the presence of inflammation. Similar data of microarrays using SAT and VAT adipose from obese patients, have been reported by others.24 The top pathways were also indicative of presence of inflammation and immune response to inflammation in SAT and VAT, relative to EAT, which is consistent with previous findings showing that adult adipose tissue-derived stem cells are highly enriched in immune-related genes.32
In addition, we performed correlation analyses of the qPCR data from all 10 patients with metabolic parameters such as fasting glucose levels, HDL, LDL, total cholesterol, triglycerides, systolic blood pressure, diastolic blood pressure, and BMI. We found strong correlations between some of these clinical features with gene expression in VAT or EAT. Interestingly, the correlations found were only for VAT and EAT, but not for SAT, raising the possibility for significant roles by these two adipose tissues in metabolic functions. However, following Bonferroni corrections for multiple testing, and due to the small sample size for this type of analysis, none of the correlations remained statistically significant. Future studies using larger sample sizes would be needed to replicate these initial results.
We also examined the expression levels of a set of genes selected because of their roles in adipocyte development as well as in the pathophysiology of metabolic diseases. Specifically, attenuation of FGF1, FGF2, FGF4, FGF5, FGF7 and FGF10 in mouse beta cells has been associated with the development of diabetes33, while, FGF21 has been linked with both obesity and diabetes, via a mechanism that regulates PGC-1α and the browning of murine white adipose tissue34,35 and possibly via Irisin36. Moreover, the enterokine FGF19 has been linked with the resolution of diabetes in animal models37, while, the murine adipose-derived FGF21 has been proposed to exert autocrine/paracrine effects on UCP1 in part by enhancing murine adipose tissue PGC-1α protein levels.38 Our data indicate that neither FGF19 nor FGF21 are expressed in the three white adipose depots tested here, suggesting that FGF19/21 are unlikely to exert autocrine effects in adult human white adipose located in the region of the waist. FNDC5 (the precursor of Irisin), on the other hand, was robustly expressed in SAT, VAT, and EAT, while, PGC-1α was also expressed in these depots but at lower levels. This raises the possibility for PGC-1α and FNCD5/Irisin driving the browning of human white adipose via an autocrine mechanism.
In conclusion, gene expression profiling of SAT, VAT, and EAT revealed substantial sharing of RNAs, suggesting that their functional differences may lie with differences in the levels of expression. Several uniquely expressed genes and corresponding networks also suggest that SAT may have unique roles in dermatologic conditions and that VAT and EAT may play roles in cardiovascular disorders and cancer. This microarray approach may help elucidate further functions of human white adipose tissue that is located in the region of the human waist.
Supplementary Material
Acknowledgments
This research was supported by research funds from the Geisinger Clinic and the National Institute of Health grants DK072488 (GSG, CDS, GA) and DK088231 (GSG) and DK091601 (GSG). We would like to thank Dr. Charles Schworer and Ms. Alicia Golden for help with the microarray experiments and the data analyses.
Financial support-grants: This research was supported by research funds from the Geisinger Clinic and the National Institute of Health grants DK072488 (GSG, CDS, GA) and DK088231 (GSG) and DK091601 (GSG).
Footnotes
The authors have no conflict of interest to disclose.
Supplementary information is available at the journal’s website.
REFERENCES
- 1.Armani A, Mammi C, Marzolla V, Calanchini M, Antelmi A, Rosano GM, et al. Cellular models for understanding adipogenesis, adipose dysfunction, and obesity. J Cell Biochem. 2010;110:564–572. doi: 10.1002/jcb.22598. [DOI] [PubMed] [Google Scholar]
- 2.Haas B, Schlinkert P, Mayer P, Eckstein N. Targeting adipose tissue. Diabetol Metab Syndr. 2012;4:43. doi: 10.1186/1758-5996-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urs S, Smith C, Campbell B, Saxton AM, Taylor J, Zhang B, et al. Gene expression profiling in human preadipocytes and adipocytes by microarray analysis. J Nutr. 2004;134:762–770. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre AM, Laville M, Vega N, Riou JP, van Gaal L, Auwerx J, et al. Depot-specific differences in adipose tissue gene expression in lean and obese subjects. Diabetes. 1998;47:98–103. doi: 10.2337/diab.47.1.98. [DOI] [PubMed] [Google Scholar]
- 5.Modesitt SC, Hsu JY, Chowbina SR, Lawrence RT, Hoehn KL. Not all fat is equal: differential gene expression and potential therapeutic targets in subcutaneous adipose, visceral adipose, and endometrium of obese women with and without endometrial cancer. Int J Gynecol Cancer. 2012;22:732–741. doi: 10.1097/IGC.0b013e3182510496. [DOI] [PubMed] [Google Scholar]
- 6.Gil A, Olza J, Gil-Campos M, Gomez-Llorente C, Aguilera CM. Is adipose tissue metabolically different at different sites? Int J Pediatr Obes. 2011;6(Suppl 1):13–20. doi: 10.3109/17477166.2011.604326. [DOI] [PubMed] [Google Scholar]
- 7.Billon N, Dani C. Developmental origins of the adipocyte lineage: new insights from genetics and genomics studies. Stem Cell Rev. 2012;8:55–66. doi: 10.1007/s12015-011-9242-x. [DOI] [PubMed] [Google Scholar]
- 8.Fried SK, Kral JG. Sex differences in regional distribution of fat cell size and lipoprotein lipase activity in morbidly obese patients. Int J Obes. 1987;11:129–140. [PubMed] [Google Scholar]
- 9.Ktotkiewski M, Sjostrom L, Bjorntorp P, Smith U. Regional adipose tissue cellularity in relation to metabolism in young and middle-aged women. Metabolism. 1975;24:703–710. doi: 10.1016/0026-0495(75)90038-4. [DOI] [PubMed] [Google Scholar]
- 10.Busetto L, Digito M, Dalla Monta P, Carraro R, Enzi G. Omental and epigastric adipose tissue lipolytic activity in human obesity. Effect of abdominal fat distribution and relationship with hyperinsulinemia. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme. 1993;25:365–371. doi: 10.1055/s-2007-1002121. [DOI] [PubMed] [Google Scholar]
- 11.Paula HA, Ribeiro Rde C, Rosado LE, Abranches MV, Franceschini Sdo C. Classic anthropometric and body composition indicators can predict risk of metabolic syndrome in elderly. Ann Nutr Metab. 2012;60:264–271. doi: 10.1159/000337943. [DOI] [PubMed] [Google Scholar]
- 12.Poliakova N, Despres JP, Bergeron J, Almeras N, Tremblay A, Poirier P. Influence of obesity indices, metabolic parameters and age on cardiac autonomic function in abdominally obese men. Metabolism. 2012;61:1270–1279. doi: 10.1016/j.metabol.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Min JL, Nicholson G, Halgrimsdottir I, Almstrup K, Petri A, Barrett A, et al. Coexpression network analysis in abdominal and gluteal adipose tissue reveals regulatory genetic loci for metabolic syndrome and related phenotypes. PLoS Genet. 2012;8:e1002505. doi: 10.1371/journal.pgen.1002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janiszewski PM, Ross R, Despres JP, Lemieux I, Orlando G, Carli F, et al. Hypertriglyceridemia and waist circumference predict cardiovascular risk among HIV patients: a cross-sectional study. PloS one. 2011;6:e25032. doi: 10.1371/journal.pone.0025032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mutch DM, Temanni MR, Henegar C, Combes F, Pelloux V, Holst C, et al. Adipose gene expression prior to weight loss can differentiate and weakly predict dietary responders. PloS one. 2007;2:e1344. doi: 10.1371/journal.pone.0001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Ambrosi J, Catalan V, Diez-Caballero A, Martinez-Cruz LA, Gil MJ, Garcia-Foncillas J, et al. Gene expression profile of omental adipose tissue in human obesity. FASEB J. 2004;18:215–227. doi: 10.1096/fj.03-0591fje. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly DP. Medicine. Irisin, light my fire. Science. 2012;336:42–43. doi: 10.1126/science.1221688. [DOI] [PubMed] [Google Scholar]
- 19.Matzko ME, Argyropoulos G, Wood GC, Chu X, McCarter RJ, Still CD, et al. Association of ghrelin receptor promoter polymorphisms with weight loss following Roux-en-Y gastric bypass surgery. Obesity surgery. 2012;22:783–790. doi: 10.1007/s11695-012-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Still CD, Wood GC, Chu X, Erdman R, Manney CH, Benotti PN, et al. High allelic burden of four obesity SNPs is associated with poorer weight loss outcomes following gastric bypass surgery. Obesity. 2011;19:1676–1683. doi: 10.1038/oby.2011.3. [DOI] [PubMed] [Google Scholar]
- 21.Wood GC, Chu X, Manney C, Strodel W, Petrick A, Gabrielsen J, et al. An electronic health record-enabled obesity database. BMC medical informatics and decision making. 2012;12:45. doi: 10.1186/1472-6947-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Than NG, Romero R, Tarca AL, Draghici S, Erez O, Chaiworapongsa T, et al. Mitochondrial manganese superoxide dismutase mRNA expression in human chorioamniotic membranes and its association with labor, inflammation, and infection. J Matern Fetal Neonatal Med. 2009;22:1000–1013. doi: 10.3109/14767050903019676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lillvis JH, Erdman R, Schworer CM, Golden A, Derr K, Gatalica Z, et al. Regional expression of HOXA4 along the aorta and its potential role in human abdominal aortic aneurysms. BMC Physiol. 2011;11:9. doi: 10.1186/1472-6793-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korsic M, Gotovac K, Nikolac M, Dusek T, Skegro M, Muck-Seler D, et al. Gene expression in visceral and subcutaneous adipose tissue in overweight women. Front Biosci (Elite Ed) 2012;4:2834–2844. doi: 10.2741/e587. [DOI] [PubMed] [Google Scholar]
- 25.Karastergiou K, Fried SK, Xie H, Lee MJ, Divoux A, Rosencrantz MA, et al. Distinct developmental signatures of human abdominal and gluteal subcutaneous adipose tissue depots. The Journal of clinical endocrinology and metabolism. 2013;98:362–371. doi: 10.1210/jc.2012-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N, et al. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J Exp Med. 2007;204:2603–2614. doi: 10.1084/jem.20070157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwasa H, Kurabayashi M, Nagai R, Nakamura Y, Tanaka T. Genetic variations in five genes involved in the excitement of cardiomyocytes. J Hum Genet. 2001;46:549–552. doi: 10.1007/s100380170039. [DOI] [PubMed] [Google Scholar]
- 28.Bosanska L, Michalsky D, Lacinova Z, Dostalova I, Bartlova M, Haluzikova D, et al. The influence of obesity and different fat depots on adipose tissue gene expression and protein levels of cell adhesion molecules. Physiol Res. 2010;59:79–88. doi: 10.33549/physiolres.931705. [DOI] [PubMed] [Google Scholar]
- 29.Tan BK, Adya R, Randeva HS. Omentin: a novel link between inflammation, diabesity, and cardiovascular disease. Trends Cardiovasc Med. 2010;20:143–148. doi: 10.1016/j.tcm.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Oberauer R, Rist W, Lenter MC, Hamilton BS, Neubauer H. EGFL6 is increasingly expressed in human obesity and promotes proliferation of adipose tissue-derived stromal vascular cells. Mol Cell Biochem. 2010;343:257–269. doi: 10.1007/s11010-010-0521-7. [DOI] [PubMed] [Google Scholar]
- 31.McLaughlin T, Lamendola C, Liu A, Abbasi F. Preferential fat deposition in subcutaneous versus visceral depots is associated with insulin sensitivity. The Journal of clinical endocrinology and metabolism. 2011;96:E1756–E1760. doi: 10.1210/jc.2011-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen BJ, Gilissen C, Roelofs H, Schaap-Oziemlak A, Veltman JA, Raymakers RA, et al. Functional differences between mesenchymal stem cell populations are reflected by their transcriptome. Stem Cells Dev. 2010;19:481–490. doi: 10.1089/scd.2009.0288. [DOI] [PubMed] [Google Scholar]
- 33.Hart AW, Baeza N, Apelqvist A, Edlund H. Attenuation of FGF signalling in mouse beta-cells leads to diabetes. Nature. 2000;408:864–868. doi: 10.1038/35048589. [DOI] [PubMed] [Google Scholar]
- 34.Feingold KR, Grunfeld C, Heuer JG, Gupta A, Cramer M, Zhang T, et al. FGF21 Is Increased by Inflammatory Stimuli and Protects Leptin-Deficient ob/ob Mice from the Toxicity of Sepsis. Endocrinology. 2012;153:2689–2700. doi: 10.1210/en.2011-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flier JS. Hormone Resistance in Diabetes and Obesity: Insulin, Leptin, and FGF21. Yale J Biol Med. 2012;85:405–414. [PMC free article] [PubMed] [Google Scholar]
- 36.Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, et al. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–1624. doi: 10.1126/science.1198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher FM, Kleiner S, Douris N, Fox EC, Mepani RJ, Verdeguer F, et al. FGF21 regulates PGC-1alpha and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012;26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.