SUMMARY
The complex, interconnected architecture of cell-signaling networks makes it challenging to disentangle how cells process extracellular information to make decisions. We have developed an optogenetic approach to selectively activate isolated intracellular signaling nodes with light and use this method to follow the flow of information from the signaling protein Ras. By measuring dose and frequency responses in single cells, we characterize the precision, timing, and efficiency with which signals are transmitted from Ras to Erk. Moreover, we elucidate how a single pathway can specify distinct physiological outcomes: by combining distinct temporal patterns of stimulation with proteomic profiling, we identify signaling programs that differentially respond to Ras dynamics, including a paracrine circuit that activates STAT3 only after persistent (>1 hr) Ras activation. Optogenetic stimulation provides a powerful tool for analyzing the intrinsic transmission properties of pathway modules and identifying how they dynamically encode distinct outcomes.
INTRODUCTION
The signaling networks that cells use to respond to extracellular stimuli have complex branched and feedback architectures. Thus, it is challenging to disentangle how information flows through such networks to encode precise responses. Ideally, we would like to be able to reach into a cellular network and selectively activate isolated nodes to observe how perturbations are propagated through the system (Figure 1A). Optogenetic perturbation has emerged as a powerful approach to interrogate complex neuronal circuitry: light-gated channels allow activation of individual neurons within a complex network (Boyden et al., 2005) and have been used to elucidate subcircuits responsible for behaviors such as movement (Gradinaru et al., 2007), sensing (Li et al., 2011), or memory (Liu et al., 2012).
Figure 1. Cellular Optogenetics: Approaches for Dissecting Complex Signaling Networks.
(A) Optogenetic inputs can be used to stimulate a single intracellular node to isolate subnetworks within the full physiological signaling response network.
(B) By applying time-varying light inputs, it is possible to dissect how dynamics are transmitted and drive specific responses.
(C) These approaches can be used to understand how a shared signaling node can yield distinct responses when reused in multiple physiological pathways. Specificity can be encoded by distinct combinations of activated pathways (left panel) or by the dynamics of activation (e.g., duration or amplitude) of a single pathway (right panel).
(D) Direct optogenetic activation of shared nodes provides a powerful approach to dissect these mechanisms of encoding.
Can parallel tools be used to analyze cell-signaling networks? We and others have recently developed cellular optogenetic tools that can be used to control the activity of isolated signaling proteins within living cells (Kennedy et al., 2010; Levskaya et al., 2009; Strickland et al., 2012; Wu et al., 2009). Here we explore how these tools can be used to track information flow through cellular signaling networks. Our approach harnesses the phyto-chrome B (Phy)-PIF light-gated protein interaction system from plants (Levskaya et al., 2009). The Phy-PIF interaction can be controlled by stimulation with red light (650nm—ON) and infrared light (750 nm—OFF) and switches between states in a matter of seconds. When the Phy-PIF module is linked to signaling proteins whose activity is controlled by recruitment, we can use light to activate signaling with complex time-variant patterns (Figure 1B). This optogenetic strategy is related to activation via chemical dimerizer modules (Spencer et al., 1993) but allows more flexible and precise spatial and temporal control of activity.
We apply this optogenetic approach to study signal transmission by the Ras/Erk mitogen-activated protein kinase (MAPK) cascade. The Ras/Erk cascade is a shared signaling module that is activated by many extracellular signals and can lead to diverse outcomes, including cell proliferation, differentiation, or arrest (Bishop et al., 1994; Meloche and Pouysségur, 2007). The functional plasticity of key shared signaling modules such as Ras/Erk presents a conundrum: when a shared internal signaling node is activated, how does the cell know which response to initiate?
Two mechanisms have been proposed to resolve this paradox (Figure 1C). First, signaling information can be combinatorially encoded: two different external stimuli that activate the same internal signaling pathway may also induce other stimulus-specific pathways, and these distinct combinations may encode a specific downstream response (Barber et al., 2007; Koronakis et al., 2011; Prehoda et al., 2000). Alternatively, signaling information can be dynamically encoded: two different extracellular stimuli might lead to activation of the same internal node, but with different temporal patterns of activation, and these differences in dynamics could be decoded by downstream modules to yield distinct responses (Purvis and Lahav, 2013). Discriminating between these models is challenging because most network perturbations that block specific pathways (e.g., small-molecule inhibition; small interfering RNA [siRNA]) also perturb pathway dynamics. Here we show that optogenetic stimulation provides a general technique to directly manipulate the dynamics of a single pathway to assess its response (Figure 1D).
We use light-controlled Ras to interrogate signaling at two levels. First, by coupling this tool to a live-cell reporter of the downstream MAPK Erk, we quantitatively characterize the intrinsic signal-transmission properties of the Ras/Erk module. Measuring dose-response curves from individual cells reveals that Ras-to-Erk signaling can accurately transmit quantitative information about stimulus level. Measuring frequency-response curves shows that this pathway is a high-bandwidth transmission device—although it rejects transient input fluctuations of less than 4 min, it efficiently transmits input signals across the broad range of timescales from 4 min to multiple hours. This high-bandwidth behavior is ideally suited for a shared internal node that is reused to transmit multiple responses, often encoded with different intrinsic dynamics, and may be a common feature of many multi-use signaling pathways.
Second, we take advantage of the high temporal resolution of optogenetic control to stimulate cells with systematic temporal patterns of Ras/Erk activity. We then use an array-based proteomics approach to screen for downstream signaling outputs that are selectively activated by specific Ras dynamic profiles. We identify outputs activated by transient Ras activity (20 min), along with others that require sustained Ras activity (>1 hr). This approach identifies a dynamically regulated Erk target, STAT3, which is activated by a paracrine signaling circuit that requires persistent (>1 hr) Erk activation.
These results suggest a modular organization of signaling networks in which different parts of the network perform different signal-processing functions: shared pathways like Ras/Erk are optimized to efficiently transmit diverse signals across a wide range of timescales, whereas downstream effector modules filter and decode stimulus-specific dynamic information. This optogenetic approach provides a powerful general method to characterize the transmission properties of key internal signaling nodes and how differences in their dynamics are decoded.
RESULTS
Engineering Optogenetic Control of Ras
Many signaling proteins depend on proper localization for activity. Thus, a general strategy for engineering light control is to use the Phy-PIF system to colocalize an upstream activator with its downstream effector. We previously engineered control over Rho-family GTPases by recruiting the appropriate upstream guanine nucleotide exchange factor (GEF) to the plasma membrane using the Phy-PIF module (Levskaya et al., 2009).
To engineer light activation of Ras, we screened several RasGEF domains and identified the catalytic segment of the protein SOS (referred to as SOScat) as a domain whose ability to activate Ras is highly dependent on recruitment to the plasma membrane(Gureasko et al., 2008).We constructed a cytoplasmic PIF-SOScat fusion (tagged with YFP on its N terminus) and a membrane-localized PhyB (PhyB-mCherry-CAAX) and refer to this pair of constructs as the opto-SOS system (Figure 2A).
Figure 2. Opto-SOS: Engineering a Light-Gated Switch to Drive Ras Activation.
(A) Light drives the heterodimerization of membrane-localized Phy with a cytoplasmic PIF-tagged SOScat construct, leading to Ras activation and nuclear translocation of BFP-Erk2.
(B) Representative fluorescent images of YFP-PIF-SOScat (upper panels) and BFP-Erk (lower panels) from an NIH 3T3 opto-SOS cell under inactivating and activating light, showing light-dependent cytoplasmic depletion of SOScat and nuclear Erk accumulation.
(C) The fold-change in nuclear Erk intensity in response to activating (650 nm) and inactivating (750 nm) light inputs (n = 3 cells). Still images from (B) are taken at starred time points. Mean + SEM. (D) NIH 3T3 opto-SOS cells proliferate after stimulation with either 100 ng/µl PDGF or red light, measured by an increase in S-G2 DNA content after serum starvation (histograms, shaded area). (E) PC12 opto-SOS cells differentiate after stimulation with either 100 ng/µl NGF or red light, measured by neurite outgrowth after 24 hr.
(F) Growth factor activates both Ras/Erk and PI3K/Akt signaling; optogenetic inputs could be selective for Ras/Erk.
(G) Time course of phospho-Erk and phospho-Akt western blots of NIH 3T3 opto-SOS cells stimulated by either 100 ng/µl PDGF or red light; opto-SOS activation is specific for Erk but not Akt signaling.
See also Figure S1.
We used fluorescently tagged Erk2 (BFP-Erk) as an Erk activation reporter—upon phosphorylation, Erk is translocated to the nucleus (Burack and Shaw, 2005; Cohen-Saidon et al., 2009; Shankaran et al., 2009). Paired with the opto-SOS system, this reporter enables simultaneous manipulation and monitoring of Erk activity in living cells (Figure 2A).
When opto-SOS and the BFP-Erk were expressed in NIH 3T3 cells and the cells were treated with activating red light (650 nm), we observed recruitment of YFP-PIF-SOScat to the plasma membrane (and, conversely, depletion from the cytoplasm; Figures S2A–S2C available online), coupled with nuclear translocation of Erk (Figure 2B). The magnitude of Erk translocation was comparable to that induced by platelet-derived growth factor (PDGF; Figure S2E) and was dependent on addition of phycocyanobilin (PCB), the chromophore required for PhyB activity. Subsequent exposure of the cells to inactivating light (infrared—750 nm) reversed Erk nuclear localization within minutes (Figure 2C). We verified that light levels required for activation are not phototoxic to the cell (Figures S1J–S1L).
Optogenetically Activated Ras Leads to Hallmark Biochemical and Physiological Responses
Also, we used anti-phospho-Erk western blots to biochemically assay Erk activation. Red-light stimulation of NIH 3T3 and PC12 cells expressing opto-SOS resulted in high levels of Erk phosphorylation, comparable to those observed with PDGF or nerve growth factor (NGF) stimulation (Figures 2G and S1). Erk phosphorylation persisted as long as input light was present but was reversed within minutes in response to inactivating light (Figure S1B). The opto-Sos and Erk reporter constructs had no effect on the normal response to PDGF or NGF (Figure S1D).
Importantly, optogenetic Ras stimulation selectively activates the Erk MAPK cascade. For example, although growth factors activate both Ras/Erk and PI3K/Akt signaling (Chen et al., 2012), Akt phosphorylation was undetectable in light-stimulated opto-SOS cells (NIH 3T3 and PC12) (Figures 2F, 2G, and S1). Although many prior studies indicate that PI3K is downstream from Ras, these results suggest that Ras activity alone is not sufficient for PI3K activation (Mendoza et al., 2011).
Optogenetically activated Ras is sufficient to drive key physiological and morphological aspects of cell-fate decisions that are normally driven by growth factor stimulation (Bishop et al., 1994; Chambard et al., 2007). For example, PDGF causes serum-starved, G1-arrested NIH 3T3 cells to proliferate, and NIH 3T3 cells expressing opto-SOS also re-enter the cell cycle when stimulated by light (16 hr) (Figure 2D). Similarly, NGF stimulation causes neurite outgrowth and differentiation in PC12 cells. PC12 cells expressing opto-SOS show neurite outgrowth with 24 hr light stimulation (Figure 2E).
Light Control Enables Measurement of Single-Cell Dose-Response Curves of the Isolated Ras/Erk Module
We first used this optogenetic system to ask how precisely steady-state signals can be transmitted through the Ras/Erk pathway. Many analyses suggest that cell-signaling circuits are noisy systems, with limited capacity to display analog sensitivity—the ability to reliably distinguish subtle differences in input levels, rather than simply turning ON and OFF. Directly measuring the dose-response precision of a pathway is difficult because most such experiments are done in bulk and cannot separate noise that is due cell-to-cell variability from noise that is intrinsic to each individual cell.
Using both live-cell stimulation and readout makes it possible to precisely measure dose-response curves in individual cells (Figure 3A). We can simultaneously measure the activity of nodes directly above and below the Ras/Erk MAPK module, i.e., the amount of SOScat recruited to the membrane (the light-induced INPUT) and the degree of nuclear transport of Erk (OUTPUT) (Figure 3A). This approach gives us the unprecedented ability to quantitatively analyze input/output relationships in the isolated Ras/Erk module, free from the complexities of combinatorial, feedback, and feedforward linkages outside this core module (Figures 3A and 3B).
Figure 3. Single-Cell SOS-to-Erk Dose Responses Show that Stimulus Levels Are Precisely Transmitted by Individual Cells.
(A) Both light-gated input (membrane localization of PIF-YFP-SOScat) and output (nuclear BFP-Erk fold-change) can be measured in live cells over time, revealing full dose-response relationships in each cell.
(B) All cells in a field of light-stimulated NIH 3T3 opto-SOS cells exhibit membrane SOScat translocation (upper panels) and nuclear Erk translocation (lower panels).
(C) Single-cell dose-response curves from four representative cells. Each point shows the membrane SOScat and nuclear Erk nuclear fold-change induced by a defined red/infrared light ratio (mean + SD).
(D) The SOScat/Erk activity induced in each cell by light across a population of 25 single cells. Envelope shows the variability of single-cell fits across the population.
(E) SOScat-to-Erk dose responses for two representative cells, where envelopes show the confidence in dose-response curve fit for each cell, overlaid on the cell-to-cell variability from (D) (dotted lines).
We measured Ras/Erk dose-response curves by applying different ratios of 650/750 nm (light to a field of ~25 cells) and quantifying both SOScat membrane recruitment (INPUT) and Erk nuclear localization (OUTPUT) in each cell. Treating SOScat recruitment as the INPUT (instead of light intensity) makes the dose measurements independent of variability in opto-SOS expression. We ensured that nuclear Erk levels reached steady state by applying each light dose for 10 min (in control experiments, membrane SOScat levels reach steady state within 1 min, and nuclear Erk reaches steady state in ~6 min; Figure S1). To assess the reliability of the dose response of each cell, we applied different light doses in a random order, returning to each dose multiple times during the experiment (Figures 3C and S2; Movie S1). Notably, the stable steady-state responses we observe when optogenetically stimulating the isolated Ras/Erk module qualitatively differ from the pulses of Erk nuclear localization observed with extracellular epidermal growth factor (EGF) stimulation (Albeck et al., 2013; Cohen-Saidon et al., 2009). These differences suggest that the network links that regulate pulsatile Erk activation originate outside of the core Ras/Erk module.
Optogenetic dose-response analysis reveals significant differences between population level and individual cell responses (Figures 3D and 3E). Across the population of ~25 cells, a clear saturatable response was observed, but there was a very high level of variability (Figure 3D). When analyzed at the single-cell level, however, the SOScat-to-Erk dose response showed much higher precision (Figure 3E), tracing out dose-response curves (with a Hill coefficient ~2) that could be reproducibly repeated over many hours (Figure S2I; Movie S2). These responses did not show any memory—regardless of past history, a given level of SOScat recruitment led to a reproducible level of intermediate nuclear Erk for each cell (Figures S2E–S2G). Nevertheless, the maximum Erk amplitude and the input switching threshold varied between individual cells (Figure 3C), explaining the higher population-level variability. To ensure that the differences between cells did not arise because of differential expression levels of the opto-SOS system, we repeated these dose-response measurements in clonally derived 3T3 cell lines with stable optogenetic component expression. Even among these identical cells, we observed high cell-to-cell dose-response variability (Figure S2J).
These results suggest that individual cells vary from one another, most likely because of variability in the expression level of various relevant molecular components. However, each individual cell has a very consistent response that gives it the capability to precisely sense and discriminate subtle input differences. Thus, our findings suggest that cells can reliably measure stimulus level more precisely than has been suggested by recent population-level analyses (Cheong et al., 2011).
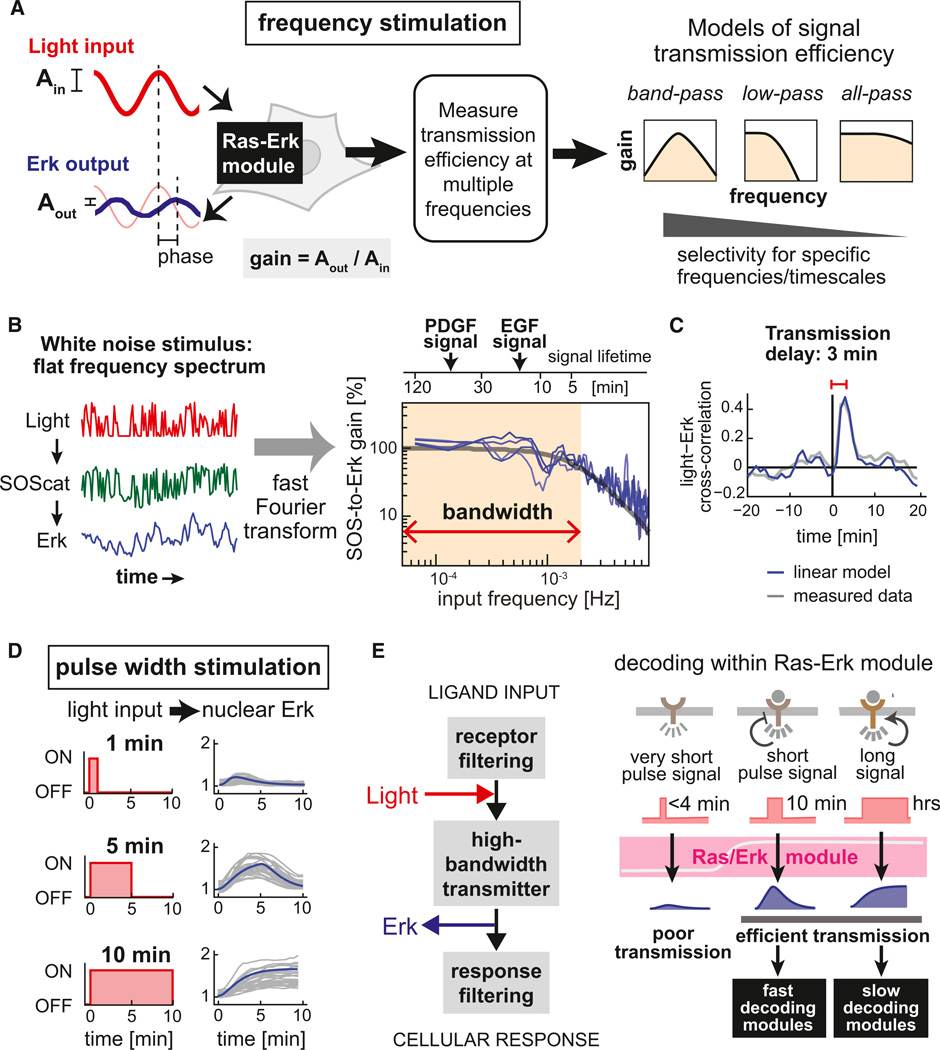
Frequency-Response Analysis Shows that the Ras/Erk Module Is a High-Bandwidth, Low-Pass Filter
Optogenetic control over the Ras/Erk pathway allows us to address a fundamental question in cell signaling: how does a signaling pathway filter or transmit dynamic input signals? This is a crucial question for the Ras/Erk module, which responds over a very wide range of physiologically relevant timescales (Santos et al., 2007; Sasagawa et al., 2005). Thus, we performed the first quantitative comparison of the Ras/Erk module’s signal transmission across the range of relevant timescales (spanning 1 min to multiple hours), using the engineering technique of frequency-response analysis (Oppenheim et al., 1997).
Frequency-response analysis is typically performed by stimulating a complex system with an input at a specific frequency and measuring the system’s resulting response (Figure 4A, left panel). Two parameters, the ratio of output to input amplitude (“gain”) and the delay in the output oscillation relative to the input (“phase”), characterize this response at each frequency. By performing this measurement at many frequencies, a full dynamic response can be obtained (Figure 4A).
Figure 4. The Ras/Erk Module Is a High-Bandwidth, Low-Pass Filter, Faithfully Transmitting Dynamic Signals from 4 min to 2 hr.
(A) By applying oscillating light inputs and measuring responses, a pathway’s gain (output versus input amplitude) and phase shift (delay in peak response) can be obtained at each frequency. Possible frequency-response behaviors range from the narrow response of a band-pass filter to broad all-pass transmission.
(B) Left panel: Measuring the response to a light input containing multiple frequencies can efficiently reconstruct entire frequency responses from individual cells. Right panel: The frequency responses of five cells (blue curves) match a linear second-order low-pass filter (gray line). Upper timeline shows activation pulse timescales that correspond to stimulus frequencies shown on the x axis; typical Erk-response lifetimes (for PDGF and EGF stimulation) are shown.
(C) The mean cross-correlation between light input and nuclear Erk fold-change from five cells (gray line) and the predicted low-pass filter response of the linear frequency response model from (B) (blue line) show a 3 min delay of Erk activation following light input.
(D) Single-cell pulse responses from 25 cells (gray lines), shown with predictions of the linear frequency response model from (B) (blue lines).
(E) A model of signal transmission by the Ras/Erk module. Very short input stimuli are filtered and ignored by the Ras/Erk module, whereas inputs from minutes to hours are faithfully and efficiently transmitted by the MAPK cascade. The distinct patterns of Erk activity are then likely decoded by downstream dynamic filtering modules (black boxes).
See also Figure S3.
Signaling systems can potentially behave as one of several different types of dynamic filters (Figure 4A): (1) band-pass modules, which respond best to a specific input frequency or pulse length; (2) low-pass filters, which efficiently transmit low-frequency, persistent signals but suppress high-frequency (short duration) noisy signals above a cutoff frequency; and (3) high-bandwidth, all-pass modules, which faithfully transmit inputs with a wide range of different dynamic properties. (A fourth class of behavior, high-pass filters, are possible in electronics but mechanistically improbable in biochemical reaction systems because the finite reaction time of such systems will intrinsically filter high-frequency signals; see Samoilov et al., 2002.) These three filter types are increasingly permissive, transmitting an increasingly broad range of signals to downstream modules.
Physiological signaling systems that use dynamics to encode specific outcomes are expected to display band-pass behavior, yielding maximal gain at frequencies corresponding to the natural timescale of response (Mettetal et al., 2008). We thus hypothesized that the Ras/Erk pathway, typically associated with specific dynamic responses, might perform band-pass filtering, selectively transmitting a certain input timescale at the expense of others like a radio tuned to a specific frequency.
To experimentally measure frequency responses in single cells, we utilized an efficient Fourier transform-based technique that is prevalent in engineering but previously inaccessible in biology. Instead of stimulating cells with one frequency at a time, we applied a fluctuating input that simultaneously contains information at many frequencies. We then measured each cell’s response over time and used the Fourier transform to mathematically separate the contributions at each frequency. This approach requires both fast-timescale stimulation and readout, which our optogenetic system provides. After measuring membrane fluctuations in SOScat recruitment and Erk nuclear translocation in each cell once per minute for ~3 hr (Figure 4B), we computed the Ras/Erk pathway’s gain (Figure 4B) and phase shift (Figure S3F) as a function of input frequency. Similar results are obtained when we use the lower-throughput approach of stimulating cells with individual frequency sinusoidal inputs (Figures S3E–S3G).
The measured frequency-response curves provide a number of insights into the Ras/Erk pathway’s fundamental signal transmission properties. The frequency-response curve (Figure 4B) is largely flat across frequencies that correspond to input timescales ranging from 4 min to 2 hr. This timescale range also encompasses the timescales on which Erk responses have been measured: EGF generates single or periodic 15 min pulses (Cohen-Saidon et al., 2009), and PDGF drives >1 hr sustained Erk activation (Murphy et al., 2002).
At the same time, we also observe that for sufficiently high-frequency stimulation—short input pulses of less than 4 min—the Erk transmission efficiency drops dramatically (Figure 4B). In this respect, the Ras/Erk module is a low-pass filter that suppresses transmission of input pulses of less than 4 min. This critical timescale marking the boundary of signal transmission and suppression is highly reproducible between cells. It also represents a property intrinsic to the Ras/Erk module, not our optogenetic input, as prior measurements of PIF membrane translocation indicate high-fidelity signal transmission (i.e., a flat frequency response) in this range (Toettcher et al., 2011). We conclude that the Ras/Erk module is a high-bandwidth, low-pass filter: it efficiently transmits signals from input stimuli over a broad timescale range (4 min to at least 2 hr), but it also filters transient minutes-scale inputs (Figure 4B).
By measuring the cross-correlation between our fluctuating input and Erk output, we also extracted the transmission delay (Figure 4C): it takes 3 min to transmit the signal from SOScat to Erk nuclear translocation. This delay, and the signal dissipation that occurs during this time, is likely correlated with the setpoint of the low-pass filtering threshold. A system capable of rejecting inputs shorter than a cutoff time must (1) delay its response at least as long as the cutoff time, and (2) dissipate inputs of this timescale so as not to initiate a response (Weinberger and Shenk, 2007) (Figures S3K–S3M).
To test the predictive power of the measured Ras/Erk frequency-response profile, we used a simple linear model to predict responses to single stimulus pulses of variable duration. Our model, a second-order linear low-pass filter with a single cutoff frequency of 2 mHz, closely matches both the experimentally measured gain and phase shift across all frequencies tested (Figures 4B and S3). Overall, short (1 min) input pulses led to attenuated Erk responses, whereas longer (5 and 10 min) pulses drove high-amplitude Erk output (Figure 4D).
Taken together, our observations suggest a model for dynamic transmission through the Ras/Erk pathway (Figure 4E). Very transient input fluctuations are suppressed, generating little or no Erk nuclear accumulation. Such filtering may prevent responses to spurious stochastic events in the cell. However, inputs persisting longer than a few minutes, likely corresponding to genuine receptor activation events, are efficiently transmitted to Erk activation with equal amplitude, including those in the range of canonical “ transient” (minutes) and “sustained” (hours) Erk activation observed for natural growth factors.
In summary, the Ras/Erk module appears to act as a wide signal transmission pipe that can efficiently transmit a broad range of signals (except the most transient), behavior that may be exactly what is required for a module that is reused by physiologically distinct responses operating with diverse dynamics (Figure 4E). Nonetheless, because physiological responses that incorporate the Ras/Erk module often show specific dynamic encoding, we hypothesized that there must exist signal-processing modules downstream from Ras/Erk that can decode the different dynamical signals that are efficiently passed through this high-bandwidth module.
An Array-Based Proteomic Screen Identifies Downstream Modules that Decode Transient versus Sustained Ras/Erk Dynamics
The broad transmission capability of the Ras/Erk modules suggests that a significant amount of dynamic signal decoding may occur downstream. To test this hypothesis, we used our optogenetic stimulation system to screen for downstream modules that selectively respond to specific Ras/Erk dynamics. Although prior studies have profiled cellular responses to PDGF and EGF, which respectively drive sustained and transient Ras/Erk module dynamics, these growth factors activate multiple pathways and differ from one another in many more ways than just their dynamic responses. In contrast, optogenetic stimulation allows one to directly vary Ras/Erk dynamics in an otherwise identical cellular context.
To broadly search for dynamics-dependent downstream modules, we took advantage of reverse-phase protein arrays (RPPA), a high-throughput proteomic technique for measuring 180 phospho- and total protein levels (Amit et al., 2007; Tibes et al., 2006). NIH 3T3 cells expressing opto-SOS were stimulated with either a constant 120 min light stimulus (sustained) or a 20 min light pulse (transient). Lysates were collected at 0, 15, 30, 60, and 120 min after initiation of stimulation. As a control, we also characterized NIH 3T3 cells stimulated with PDGF (Figure 5A). Of the 180 antibody probes tested, 27 showed significant changes in response to PDGF or optogenetic Ras activation (Figures 5B, 5C, and S4A; full RPPA data set is available upon request). The 27 hits clustered into 3 classes of responders, which we termed classes I–III (Figure 5B).
Figure 5. A Proteomic Screen Identifies Proteins downstream of Ras/Erk Signaling that Decode Dynamic Inputs.
(A) Downstream proteins are measured in response to 100 ng/µl PDGF or transient (20 min) and sustained (2 hr) light inputs by reverse phase protein array (RPPA).
(B and C) Scatter plots showing PDGF-responsive proteins (B; black circles) and light-responsive proteins (C; red circles) as well as the rest of the 180 protein RPPA panel (gray circles). Black diagonal lines indicate identical responses between unstimulated and stimulated conditions.
(D–F) Twenty-seven proteins show a clear response to either light or PDGF. The responses of class I (D), II (E), and III (F) nodes are shown after PDGF, constant light, and pulsed light. Upper panels: RPPA results for each responding protein are feature normalized (i.e., mean-subtracted and divided by SD) and shown in false color. Lower panels show relative protein levels over time, normalized for protein loading, for three representative proteins in each class.
See also Figure S4.
Class I responding proteins (Figure 5B) showed a strong response to PDGF but were not activated by light. Some of these outputs may be completely independent of Ras/Erk signaling, representing a clear distinct branch of signaling from the PDGF receptor. Others may require Ras/Erk signaling, but Ras stimulation alone is insufficient for their activation. These outputs are therefore good candidates for divergent or combinatorial control (Figure 1A). This class includes activated forms of receptor tyrosine kinases (EGFR and HER2; these antibodies may also cross-react with PDGFR), the tyrosine kinase Src, the MAPK Jnk, and the canonical PI3K target Akt (Figures 2F and 2G). PI3K/Akt signaling, previously linked to Ras stimulation in response to growth factor, is a likely candidate for combinatorial control (Fivaz et al., 2008).
Class II responding proteins are potently activated by both PDGF and light and track both transient and sustained light input dynamics (Figure 5B, middle panel). These 12 proteins are members of “fast” response modules, responding quickly to increases and decreases in light-stimulated Ras. Members of this class include the canonical MAPK cascade members (phosphorylated Raf, Mek, and Erk), the Erk target P90RSK, and the mTOR pathway’s upstream signaling kinase GSK3. We found that PKCβ quickly responds to light-activated Ras, reaching levels comparable to those seen after PDGF treatment, an unexpected result because PLCγ/PKC signaling is typically thought to be induced by receptor-level activation, not Ras (we hypothesized that PKCβ would be a member of class I). Thus, this optogenetic approach can reveal crosstalk between pathways downstream of canonical receptor-level control.
Finally, class III responding proteins are activated by PDGF but were only triggered by sustained optogenetic Ras activity, ignoring a transient light pulse (class III; Figure 5B). These seven proteins are members of response modules capable of differentiating sustained from transient Ras/Erk pathway inputs. This class includes components involved in mTORC1/2 signal transduction (Rictor, p70S6K, S6, and NDRG1) (Garcίa-Martίnez and Alessi, 2008), as well as the epithelial-mesenchymal transition transcriptional repressor SNAIL. Perhaps the most striking response was the phosphorylation of STAT3, which only increased after more than 1 hr of sustained light input, rising to a level comparable to that induced by PDGF by 2 hr.
Previous work identified one class III-like module—the c-Fos transcription factor subunit—that selectively responds to sustained rather than transient Erk activation (Murphy et al., 2002). c-Fos was not a member of the RPPA antibody panel. We therefore used western blotting to directly probe for total and phosphorylated c-Fos in both NIH 3T3 and PC12 opto-SOS cell lysates. We observed that phospho-c-Fos (and to a lesser extent total c-Fos) selectively responded to sustained but not transient light stimulation, consistent with a class III response (Figures S4B and S4C).
An Erk-to-STAT3 Paracrine Signaling Circuit Filters Dynamic Input Signals
We have shown that light activation of the Ras/Erk module drives two classes of responses: fast modules capable of tracking the dynamics of input stimuli, and slow modules that filter transient activation signals, responding only to sustained pathway activation. To gain more insight into how this dynamics-dependent filtering is implemented, we focused on the phosphorylation of STAT3 that occurs after sustained but not transient optogenetic Ras stimulation.
This finding was particularly interesting because it could not be easily explained by known signaling linkages. STAT3 activation typically proceeds downstream of two input pathways: the Janus kinase (JAK) signaling pathway that is activated by interleukin-6 (IL-6) family cytokines and the Src pathway that is activated by stimulation of RTKs such as PDGFR (Turkson et al., 1998). Our RPPA results exclude Src activation as a plausible mechanism: as a class I node, Src is not activated by optogenetic stimulation of Ras. We hypothesized that there may be an uncharacterized linkage between Ras activation and the autocrine/paracrine release of IL-6 cytokine family ligands (Park et al., 2003) (Figure 6A). Such an extracellular cytokine link might be responsible for STAT3 activation in response to light.
Figure 6. The Erk-to-STAT3 Circuit Is a Paracrine Persistence Detector.
(A) Proposed model whereby Ras-stimulated cell activates JAK/STAT signaling in neighboring cells by cytokine secretion.
(B) Immunofluorescence images of mixed WT and opto-SOS NIH 3T3 cells: untreated (left), stimulated with the STAT3 activator IL-6 (middle), or stimulated with light (right). Membrane Phy-mCherry-CAAX expression (red) and nuclear phospho-STAT3 (green) are shown.
(C) These data suggest a model whereby light-stimulated opto-SOScells secrete aSTAT3-activating ligand but are themselves insensitive toSTAT3-activating ligand.
(D) An experimental test for persistence detection: measuring whether a 2 hr continuous light input is sensed differently than two 1 hr light inputs.
(E) Immunofluorescence images of mixed opto-SOS and WT NIH 3T3 cells stimulated by the light inputs in (D). Fluorescent channels are as in (B).
(F) Quantification of nuclear pSTAT3 (mean + SEM) from opto-SOS and WT cells in (E). Dotted line shows signal corresponding to low STAT3 response in light-treated opto-SOS cells.
See also Figures S5 and S6.
To test whether STAT3 activation was due to the release of an extracellular factor, we performed optogenetic stimulation experiments on a two-cell system—we cocultured 3T3 cells containing opto-Sos with wild-type (WT) 3T3 cells that could not respond to red light. In this system, the WT 3T3 cells function as “receiver” cells that could only respond to biochemical signals (such as a secreted cytokine) and not optical ones. We used immunofluorescence to monitor STAT3 activation in each cell type. Opto-SOS cells are identified in the coculture by their dual fluorescent expression of Phy-mCherry-CAAX and TagBFP-PIF-SOScat. In a control experiment, when stimulated with recombinant IL-6, both WT and opto-SOS cells induced high levels of nuclear phospho-STAT3 (Figure 6B). As predicted by our paracrine signaling model, we found that light induces STAT3 phosphorylation in WT 3T3 cells only when they are cocultured with opto-SOS 3T3 cells, which presumably are secreting an extracellular factor (Figures 6B and S5A–S5C). Further investigation indicated that this paracrine signaling is transcription and Erk dependent (Figures S6A and S6B). The paracrine signal acts through IL-6 family receptors, as a neutralizing antibody to the GP130 receptor subunit blocked the effect (Figure S5D). Additional cytokine array studies suggest that the cytokine mediating the effect is not IL-6 but the related cytokine leukemia inhibitory factor (LIF) (Figures S5E–S5G).
Surprisingly, although our results were consistent with paracrine signaling between opto-SOS and WT 3T3 cells, we did not observe autocrine activation: opto-SOS cells did not phosphorylate STAT3 in response to light (Figures 6B and 6C). This observation is not due solely to the expression of the opto-SOS genetic constructs, as both WT and opto-SOS cells respond to exogenous IL-6 family cytokines (Figure 6B). However, this sensitivity is decreased in opto-SOS cells previously stimulated with light (Figures 6C–6E). Our data support a more complex model of cell-cell communication whereby light-activated Ras/Erk signaling initiates a paracrine circuit by secretion of a STAT3-activating ligand, while simultaneously intracellularly inhibiting the cell’s own autocrine STAT3 response (Figure 6C).
In light of this observation, it is perhaps surprising that STAT3 was identified by our RPPA proteomic screen from a homotypic population of NIH 3T3 opto-SOS cells. However, we found that expression levels of both Phy and PIF components are variable in opto-SOS cells (Figures S2B and S3A), and nuclear phospho-STAT3 is very strongly induced by light-induced paracrine signaling (Figures 6F and 6G). It is likely that a small fraction of the opto-SOS population exhibits low Phy/PIF expression and is unresponsive to light and is therefore capable of STAT3 activation. Even a small population of these strongly activated cells would lead to a detectable RPPA signal when measured in a bulk population.
How does this paracrine-signaling circuit between Erk and STAT3 decode transient versus sustained dynamic inputs? The simplest explanation is that synthesizing and secreting an extra-cellular factor is a slow process, requiring 1–2 hr to reach sufficient levels to activate nearby cells. Alternatively, we reasoned that the Erk-to-STAT3 circuit might be a bona fide persistence detector (Mangan et al., 2003; Murphy et al., 2002), selectively responding to a sustained 2 hr light input but filtering out transient inputs even when their combined duration is 2 hr. To discriminate these models, we stimulated cells with inputs that each consisted of 2 hr of total activation time, either delivered in a continuous pulse or as two 1 hr pulses of activating (650 nm) light separated by 1 hr of inactivating (750 nm) light (Figure 6D).
Strikingly, in coculture experiments, we found that sequential 1 hr light inputs did not result in any measurable STAT3 activation, whereas a single 2 hr pulse robustly activated STAT3 (Figures 6E and 6F), despite the fact that the same total duration of light was delivered in both cases. This behavior is consistent with a persistence detector. This result was not due merely to a short cytokine lifetime after secretion, as a 2 hr light input triggered STAT3 activation that was still sustained 2 hr after inactivation and incubation in the dark (Figures S6H and S6I). Taken together, our results suggest that sophisticated dynamically gated circuits can control cytokine release. These findings demonstrate the power of using time-varying optogenetic inputs for uncovering new signal-processing relationships, even between canonical pathways.
DISCUSSION
Optogenetic Switches as Tools to Sculpt Dynamics of Intracellular Pathways
A logical approach to understand how dynamics encode information in cells is to systematically change the dynamic activation profile of a regulatory node of interest and to determine how the outputs change as a function of these dynamics. How can we control dynamics of key nodes? There have been some notable successes in the past few years in trying to “sculpt” dynamical profiles of signaling pathways to determine their functional significance. Applying time-varying chemoattractant or salt inputs has helped elucidate the systems-level organization of bacterial chemotaxis (Shimizu et al., 2010) and the yeast osmolarity response (Hersen et al., 2008; Mettetal et al., 2008; Muzzey et al., 2009), respectively. More recently, delivering stress inputs combined with tailored time courses of small-molecule inhibitors has shed light on the regulation of stress-responsive transcription in yeast (Hao et al., 2013) and how mammalian cells regulate cell fate using p53 dynamics (Purvis et al., 2012).
Yet such approaches can be difficult to generalize. For example, one cannot simply pump growth factors in and out of culture because of their tight binding (the half-life can range from 10 min for PDGF-BB to ~10 hr for VEGF) (Rusnati et al., 2009) and the complications of processes like receptor internalization, which are autonomously driven and have complex intrinsic dynamics. In addition, the combination of natural ligands and time-variant drugs is challenging and can still involve complex fan-out signaling to multiple pathways beyond a single module of interest. Most of these approaches involve using external stimuli and thus do not allow the study of isolated internal signaling modules. Chemical dimerizers provide an alternative approach to directly control internal signaling nodes, but these are constrained in temporal flexibility—both in their speed of reversibility and their ability to be toggled on and off in multiple cycles (Komatsu et al., 2010). In short, optogenetic control of individual nodes provides one of the most generalizable and precise ways to control intracellular signals and analyze how their dynamic activity is functionally interpreted.
Precision Sensing at the Single-Cell Level
It has been suggested that high noise in signaling systems can limit the cell’s ability to detect and discriminate multiple stimulus levels, and that cells primarily make binary decisions using coarse-grained information (Cheong et al., 2011). Our single-cell dose-response measurements suggest an alternative interpretation—although cells in a population may not show coherent dose responses, each individual cell can display precise analog sensitivity. In our probing of Ras/Erk signaling, we find that despite significant variability between cells, each individual cell’s dose response is highly precise and stable over many hours (Figures 3C–3E). If, in individual cells, the sensitivity of downstream responses were matched to the parameters of the Ras/Erk response, then even a noisy cell population could differentially respond to subtle differences in intermediate levels of stimuli.
The Ras/Erk Module Is a High-Bandwidth Channel for Efficiently Transmitting Diverse Dynamic Signals
If cells dynamically encode information, then in general, we expect that frequency analysis of physiological response pathways would show band-pass behavior—a specific response would be maximal when the cell is stimulated by a single optimal frequency. Indeed, frequency-response analysis of the bacterial chemotaxis (Shimizu et al., 2010) and yeast osmolarity response pathways (Hersen et al., 2008; Mettetal et al., 2008) show that these physiological response pathways exhibit band-pass filtering, preferentially transmitting stimuli at a timescale that is precisely tuned to the cell’s physiological response. In both cases, band-pass filtering behavior arises from adaptation mediated by negative feedback.
But what happens when one analyzes the frequency-response behavior of isolated submodules within a more complex physiological response network? Here we analyze the frequency response of the isolated Ras/Erk module and find that it is not a band-pass filter but rather is a high-bandwidth, low-pass filter. Input signals with pulse widths ranging from 4 min to at least 2 hr are faithfully transmitted with equal amplitude (high bandwidth), whereas inputs of a few minutes or shorter are dramatically attenuated (low-pass filter).
Why does the Ras/Erk module show this distinct high-bandwidth signal transmission behavior, compared to intact physiological response pathways that show narrow bandwidth transmission? We postulate that it may be advantageous for intracellular modules that are shared by many physiological pathways (and across different cell types) to be capable of responding across a large range of timescales, transducing dynamic information about a broad range of response programs. Indeed, we find that at its core, the Ras/Erk pathway is a wide “pipe” capable of efficiently transmitting a diverse range of dynamic signals from the plasma membrane to the nucleus (Figure 7A).
Figure 7. Dynamic Signal Processing downstream of the Ras/Erk Module.
(A) A broad range of dynamics are transmitted through the Ras/MAPK module to Erk activation (representative timecourses shown in blue) and are differentially sensed by “fast” and “slow” downstream decoding modules (representative timecourses shown in green).
(B) Diverse responses identified by our array-based proteomics screen. Shown are pathway combinations activated by PDGF but not Ras (brown nodes), “fast” responding modules (red nodes), and “slow” responding modules (blue nodes) across a number of canonical signaling pathways. Gold arrows represent connections for which optogenetic Ras/Erk signaling is sufficient for activation, and gray nodes/arrows are not directly measured by our technique.
A corollary of this model is that there may be an overall hierarchical organization to how cellular pathways dynamically process signals (Figure 4E). Upstream components, such as receptors, may encode information dynamically by converting distinct extracellular chemical or physical inputs into intracellular signals with specific intrinsic frequencies (e.g., through feedback control of receptor signaling duration). Such information is then efficiently transmitted, and potentially amplified, through shared intracellular nodes that are agnostic about frequency (e.g., the Ras/Erk module). Finally, the information that is transmitted faithfully by such a shared high-bandwidth module can then be physiologically interpreted by dynamic decoding modules that are either downstream or parallel to the core transmission module. This type of modular, hierarchical organization may be linked to the process of how pathways and their schemes for information encoding may have evolved.
A Proteomic Screen Identifies Dynamics-Dependent Downstream Decoding Modules
We were able to identify dynamically sensitive modules downstream from the Ras/Erk pathway by coupling variable optogenetic Ras activation with a proteomic screen (Figure 7B). We found some “FAST” responses that quickly tracked input dynamics, exhibiting activation after both transient and sustained Ras activation (e.g., P90RSK, PLC/PKC signaling). We also found “SLOW” responses that filtered out and ignored transient Ras/Erk pathway activity and instead responded exclusively to sustained inputs (e.g., mTOR pathway members, SNAIL, STAT3).
Measuring downstream responses to optogenetic control of Ras/Erk signaling led to three surprising observations. First, it enabled identification of novel Ras effector programs (e.g., PKCβ activity) that were previously ascribed to other signaling pathways. Second, some pathways thought to be acting immediately downstream of Ras were not engaged after light-dependent activation of endogenous Ras (e.g., PI3K/Akt), suggesting that after growth factor treatment, additional, combinatorial cues may be required for their activation (Mendoza et al., 2011). Third, it revealed that several downstream signaling pathways have intrinsic dynamic dependencies, selectively responding to sustained but not transient Ras activation. These dynamic dependencies are difficult to extract by treatment with normal extracellular stimuli because of their complex, multibranched signaling.
We show that this analysis can be used to uncover extracellular as well as intracellular signaling connections: mixing light-responsive and WT 3T3 cells revealed an Erk-to-STAT3 paracrine signaling circuit. This circuit has two key properties: (1) it is preferentially activated in cells that did not previously activate Ras/Erk signaling, and (2) it ensures that only sustained Erk activation drives cytokine release. We imagine that this peculiar dual role, combining self-inhibition with transactivation, might play an important spatial role in signaling.
The cytokine-mediated activation of STAT3 is a “ persistence detector,” preferentially responding to input signals that do not undergo a transient drop in intensity (Figures 6D–6F, S6H, and S6I). This observation indicates that the Erk-to-STAT3 module is capable of sophisticated signal processing and is not merely a slow transcriptional response accumulating extracellular cytokine over time (which would be insensitive to a transient drop in input). Network motifs such as the coherent feedforward loop (Mangan and Alon, 2003; Murphy et al., 2002), known to be capable of persistence detection, may play a role in this process.
This optogenetic approach should, in principle, be applicable to a wide range of signaling nodes, and it would be extremely useful to use such an approach to catalog the basis set of responses triggered by a set of the most commonly used signaling nodes. This basis set of responses might provide the elemental building blocks that can be used to disentangle and understand more complex signaling behaviors.
EXPERIMENTAL PROCEDURES
Plasmids
Fragments encoding key domains were amplified from plasmids using PCR and ligated into a pHR lentiviral backbone using Gibson assembly (Gibson et al., 2009).
Cell Culture
NIH 3T3 cell lines were generated and cultured as described in Toettcher et al. (2011). PC12 cell lines were cultured as described in Santos et al. (2007).
Microscopy
Bright-field and confocal microscopy was performed on a Nikon Eclipse Ti microscope at 37° and in 5% CO2, as described in detail in the Extended Experimental Procedures.
Cell Lysate Collection, RPPA, and Western Blots
For RPPA and western blots, cell lysates were collected as described in Davies et al. (2012). For RPPA, lysates were processed and analyzed by the MD Anderson Cancer Center RPPA Core Facility. For western blots, lysates were loaded onto 4%–12% Bis-Tris gels, transferred to nitrocellulose, probed with primary and secondary antibodies, and imaged using Li-Cor Odyssey imaging system.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde, permeabilized in ice-cold 90% methanol, blocked, and probed with primary and secondary antibodies before immediate imaging by confocal microscopy.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. El-Samad, D. Sivak, and members of the Lim and Weiner labs. This work was supported by a Cancer Research Institute Postdoctoral Fellowship (J.E.T.); NIH grant GM096164 (W.A.L. and O.D.W.); the Howard Hughes Medical Institute and NIH grants GM55040, GM62583, PN2EY016546, and P50GM081879 (W.A.L.); and NIH grant GM084040 (O.D.W.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information included Extended Experimental Procedures, six figures, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.11.004.
REFERENCES
- Albeck JG, Mills GB, Brugge JS. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell. 2013;49:249–261. doi: 10.1016/j.molcel.2012.11.002. Published online December 6, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- Barber MA, Donald S, Thelen S, Anderson KE, Thelen M, Welch HC. Membrane translocation of P-Rex1 is mediated by G protein betagamma subunits and phosphoinositide 3-kinase. J. Biol. Chem. 2007;282:29967–29976. doi: 10.1074/jbc.M701877200. [DOI] [PubMed] [Google Scholar]
- Bishop JM, Capobianco AJ, Doyle HJ, Finney RE, McMahon M, Robbins SM, Samuels ML, Vetter M. Proto-oncogenes and plasticity in cell signaling. Cold Spring Harb. Symp. Quant. Biol. 1994;59:165–171. doi: 10.1101/sqb.1994.059.01.020. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Burack WR, Shaw AS. Live Cell Imaging of ERK and MEK: simple binding equilibrium explains the regulated nucleocytoplasmic distribution of ERK. J. Biol. Chem. 2005;280:3832–3837. doi: 10.1074/jbc.M410031200. [DOI] [PubMed] [Google Scholar]
- Chambard JC, Lefloch R, Pouysségur J, Lenormand P. ERK implication in cell cycle regulation. Biochim. Biophys. Acta. 2007;1773:1299–1310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Chen JY, Lin JR, Cimprich KA, Meyer T. A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision. Mol. Cell. 2012;45:196–209. doi: 10.1016/j.molcel.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong R, Rhee A, Wang CJ, Nemenman I, Levchenko A. Information transduction capacity of noisy biochemical signaling networks. Science. 2011;334:354–358. doi: 10.1126/science.1204553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Saidon C, Cohen AA, Sigal A, Liron Y, Alon U. Dynamics and variability of ERK2 response to EGF in individual living cells. Mol. Cell. 2009;36:885–893. doi: 10.1016/j.molcel.2009.11.025. [DOI] [PubMed] [Google Scholar]
- Davies MA, Fox PS, Papadopoulos NE, Bedikian AY, Hwu WJ, Lazar AJ, Prieto VG, Culotta KS, Madden TL, Xu Q, et al. Phase I study of the combination of sorafenib and temsirolimus in patients with meta-static melanoma. Clin. Cancer Res. 2012;18:1120–1128. doi: 10.1158/1078-0432.CCR-11-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivaz M, Bandara S, Inoue T, Meyer T. Robust neuronal symmetry breaking by Ras-triggered local positive feedback. Curr. Biol. 2008;18:44–50. doi: 10.1016/j.cub.2007.11.051. [DOI] [PubMed] [Google Scholar]
- García-Martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem. J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, Schneider MB, Deisseroth K. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J. Neurosci. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gureasko J, Galush WJ, Boykevisch S, Sondermann H, Bar-Sagi D, Groves JT, Kuriyan J. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 2008;15:452–461. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Budnik BA, Gunawardena J, O’Shea EK. Tunable signal processing through modular control of transcription factor translocation. Science. 2013;339:460–464. doi: 10.1126/science.1227299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersen P, McClean MN, Mahadevan L, Ramanathan S. Signal processing by the HOG MAP kinase pathway. Proc. Natl. Acad. Sci. USA. 2008;105:7165–7170. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat. Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V, Hume PJ, Humphreys D, Liu T, H rning O, Jensen ON, McGhie EJ. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. USA. 2011;108:14449–14454. doi: 10.1073/pnas.1107666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kang L, Piggott BJ, Feng Z, Xu XZ. The neural circuits and sensory channels mediating harsh touch sensation in Caenorhabditis elegans. Nat Commun. 2011;2:315. doi: 10.1038/ncomms1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Meloche S, Pouysségur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettetal JT, Muzzey D, Gómez-Uribe C, van Oudenaarden A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science. 2008;319:482–484. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LO, Smith S, Chen R-H, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- Muzzey D, Gómez-Uribe CA, Mettetal JT, van Oudenaarden A. A systems-level analysis of perfect adaptation in yeast osmoregulation. Cell. 2009;138:160–171. doi: 10.1016/j.cell.2009.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim AV, Willsky AS, Nawab SH. Signals & Systems. Second Edition. Upper Saddle River, NJ: Prentice Hall; 1997. [Google Scholar]
- Park JI, Strock CJ, Ball DW, Nelkin BD. The Ras/Raf/MEK/extracellular signal-regulated kinase pathway induces autocrine-paracrine growth inhibition via the leukemia inhibitory factor/JAK/STAT pathway. Mol. Cell. Biol. 2003;23:543–554. doi: 10.1128/MCB.23.2.543-554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnati M, Bugatti A, Mitola S, Leali D, Bergese P, Depero LE, Presta M. Exploiting surface plasmon resonance (SPR) technology for the identification of fibroblast growth factor-2 (FGF2) antagonists endowed with antiangiogenic activity. Sensors (Basel) 2009;9:6471–6503. doi: 10.3390/s90806471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoilov M, Arkin A, Ross J. Signal processing by simple chemical systems. J. Phys. Chem. A. 2002;106:10205–10221. [Google Scholar]
- Santos SDM, Verveer PJ, Bastiaens PIH. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat. Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- Sasagawa S, Ozaki Y, Fujita K, Kuroda S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat. Cell Biol. 2005;7:365–373. doi: 10.1038/ncb1233. [DOI] [PubMed] [Google Scholar]
- Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, Wiley HS. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol. Syst. Biol. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu TS, Tu Y, Berg HC. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol. Syst. Biol. 2010;6:382. doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- Toettcher JE, Gong D, Lim WA, Weiner OD. Light-based feedback for controlling intracellular signaling dynamics. Nat. Methods. 2011;8:837–839. doi: 10.1038/nmeth.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol. Cell. Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger LS, Shenk T. An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer. PLoS Biol. 2007;5:e9. doi: 10.1371/journal.pbio.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. Genetically-encoded photoactivatable Rac reveals spatiotemporal coordination of Rac and Rho during cell motility. Nature. 2009;461:104–110. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.