Abstract
Mammalian genomes include a considerable number of endogenous retroviruses (ERVs), relics of ancestral infectious retroviruses, whose proviruses have invaded the germ-line. The documented ability of infectious retroviruses to cause cancer has greatly contributed to the discovery of ERVs. It also reinforced the concept that ERVs are causative agents of many cancers, a notion that historically has not always stood up to experimental scrutiny. The recent greater appreciation of the complexity of ERV biology and the identification of dedicated host mechanisms controlling ERV activity have revealed novel interactions between ERVs and their hosts with the potential to cause or contribute to disease. In this review, the involvement of ERVs in cancer initiation and progression is discussed, as well as their contribution to our understanding of the process of transformation and to the invention of innovative preventive and therapeutic cancer treatments.
Endogenous retroviruses (ERVs) are remnants of germ-line integrations of exogenous infectious retroviruses (1-4). Once in the germ-line of a species, these proviral DNA copies may further amplify their copy number by reinfection of germ cells (1). ERVs are recognized by their similarity in genomic structure with all retroviruses, typically consisting of a gag, pro, pol and env genes flanked by two long-terminal repeats (LTRs) (Fig. 1). Certain ERVs may also lose the env gene and thus the ability to infect new cells (Fig. 1). These can still propagate efficiently in the germ-line by retrotransposition rather than reinfection (1, 5).
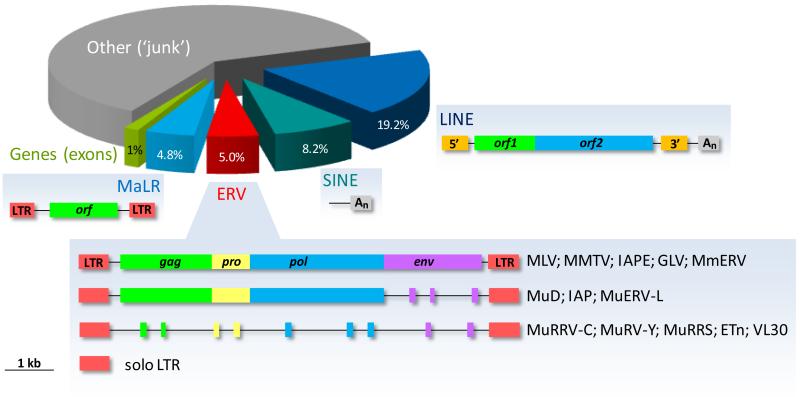
Figure 1. Retroelement composition of the mouse genome.
The proportion of LTR retroelements (MaLRs and ERVs) and non-LTR retrotransposons (LINEs and SINEs) is depicted. The typical genomic structure of distinct ERVs, adopted from (91), is also shown. ORF overlap has been removed for simplicity.
ERVs comprise 5.0% and 4.7% of the mouse and human genomes, respectively (6, 7), and are part of the group of LTR elements, which also contains the mammalian apparent LTR retrotransposons (MaLRs) (Fig. 1). LTR elements belong to a larger family of retroelements that also includes non-LTR retrotransposons (8, 9). The latter comprise a larger part of the genome (27% and 34% of the mouse and human genomes, respectively (6, 7)) and can be distinguished into long and short interspersed nuclear elements (LINEs and SINEs) (Fig. 1).
Ultimately, germ-line integrated ERV sequences degenerate by accumulation of mutations or recombination events, which destroy their ability to produce infectious virus (1, 10). For example, recombination between the two LTRs will excise the whole provirus leaving only a single LTR (referred to as solo LTR, Fig. 1). These make up the majority (90%) of all LTR elements. ERVs that have recently invaded the germ-line are more likely to represent full, sometimes infectious copies and are also more likely to be insertionally polymorphic between individuals. Inversely, older ERVs have sustained significantly more damage to their open reading frames (ORFs), rendering them replication-defective, and have also reached genetic fixation in their host genome (10).
It is important to note that the ability of ERVs to produce infectious virions differs substantially between host species. For example, replication-competent endogenous copies of murine leukemia virus (MLV) or mouse mammary tumor virus (MMTV) can be found in many strains of laboratory mouse (4). Infectious viruses produced by these ERVs can cause new somatic cell infections as well as transmit vertically to progeny as fully exogenous retroviruses. Replication-competent ERVs have also been found in other species of mammal (11). However, no replication-competent human ERV (HERV) has been found to date. HERV ORFs appear to be more defective than murine ERVs and most HERVs are fixed in the human germ-line. One possible exception is the HML2 group of the HERV-K family, which is considered the most recently acquired family (12-14). HERV-K(HML2) proviruses have the most intact ORFs, although none seems to be replication-competent, and at least some of them display insertional polymorphism within the human population (12-14). In addition to ERVs, non-LTR retrotransposons are still active in both humans and mice, where frequent somatic retrotransposition can be demonstrated (8, 13, 15).
Given the ability of ERVs and other retroelements to move in the genome, it is perhaps anticipated that these ‘genomic parasites’ have been incriminated in the pathological processes leading to cancer, which is also viewed as a ‘genomic disease’. Understanding the nature of ERV association with cancer and elucidating the precise mechanisms underlying any such association will be important in reaching a verdict.
ERVs and cancer: a historical perspective
Historically, ERVs have been linked to the development of cancer due to the transforming nature of retroviral infection, a property that was also a major contributor to the discovery of ERVs (11, 16, 17). Furthermore, the study of retroviral infection in animal models has greatly contributed to the delineation of the process of cellular transformation and laid the foundation of modern cancer research (18).
The one animal that supported the study of retroviral infection and discovery of ERVs, as well as their association with cancer, is arguably the mouse (11, 16, 17). Records of spontaneous tumors observed in mice date back to the 1890s and include mice that were caught in the wild or supplied by mouse fanciers, who had been breeding selected phenotypic traits (16). Cancer research scientists both in Europe and North America around that time had been obtaining mice from mouse farms, which often kept family records of the selected breeds (16). This collaboration fertilized the early notion of ‘heritable’ cancer, which was later investigated extensively at the Bussey Institute of Harvard University in Boston, Massachusetts. There, C.C. Little and his mentor W.E. Castle, carried out seminal work on cancer genetics with mice likely originally obtained from a nearby mouse farm run by Miss A. Lathrop, who had also made important observations on the spontaneous incidence of cancers in her mice (19, 20). This work at the Bussey Institute also included the genetics of coat color and transplant rejection and generated the first inbred mouse strains, which proved an invaluable tool not only in the dissection of cancer genetics and transplantation, but also later in all other fields of experimental biology (20). Little’s work also led to the discovery by J.J. Bittner at the Jackson Laboratory in Bar Harbor, Maine, of the ‘milk factor’, a vertically-transmitted extra-chromosomal agent causing mammary cancers in mice (16). This, of course, was MMTV, the prototypic beta-retrovirus responsible for the majority of mammary tumors in mice, and its transmission via milk seemed to argue against Mendelian cancer inheritance. However, several years later, the Dutch investigators P. Bentvelzen and J.H. Daams discovered germ-line integrated copies of MMTV proviruses, which were indeed inherited as Mendelian traits in the GR mouse strain, and which encoded fully-infectious MMTV (16, 17).
Also notable are the independent parallel discoveries of endogenous copies of MLV in inbred mouse strains and of ALV (avian leukosis virus) in domestic fowl. The latter work by R. Weiss in London, England, was one of the earliest suggestions of the existence of ERVs, and is reviewed elsewhere (11, 17). The leukemogenic potential of these ERVs also reinforced the association with cancer. However, at least for the laboratory mouse, ERVs appear to have far-reaching effects. Firstly, many murine ERVs are still very much active and thus a significant source of mutation of the murine germ-line. It is estimated that 1 in 10 spontaneous phenotypes that have been described in mice are caused by insertional mutagenesis by an ERV (21, 22). Secondly, active ERV retrotransposition in the mouse germ-line and intensive inbreeding involved in the establishment of laboratory mice have resulted in extensive insertional ERV polymorphisms among mouse strains (4). Insertional ERV polymorphism is a considerable part of all structural variants among mouse strains identified by genome sequence comparisons, and will undoubtedly affect the phenotypic outcome of a given single-gene mutation in distinct inbred mouse strains (23).
Similarly to the first inbred mouse strain, DBA, bred for and named after its coat color, which is the result of a germ-line integration of an MLV provirus (24), selection of certain phenotypic characteristics by early mouse fanciers and experimental biologists may have inadvertently contributed to the high ERV activity in laboratory mice, rendered polymorphic by subsequent inbreeding. Although this situation is ideal for the study of ERVs in particular, it is something that should be considered in the interpretation of studies involving mice.
Potential mechanistic links between ERVs and cancer
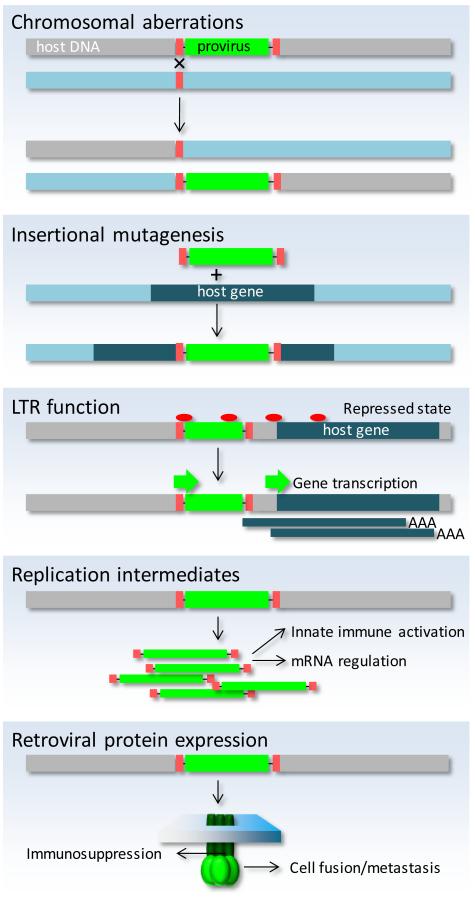
With the possible exception of rare acutely oncogenic variants, exogenous infectious retroviruses cause cancer principally by insertional mutagenesis, a process that has led to the identification of numerous cellular oncogenes or tumor-suppressor genes (16, 18). This well-established transforming potential of exogenous retroviruses is also shared by at least those ERVs that retain infectivity, such as the infectious copies of MMTV or MLV in certain mouse strains (3, 4, 16, 18). However, the only contribution of the germ-line copy of these infectious retroviruses is transmission to progeny, which in the case of both MMTV and MLV can also be achieved by vertical transmission via milk. Thus, the germ-line copies of infectious ERVs may not directly participate in the process of cellular transformation. Instead, it is new somatic integrations as a result of cellular infection with virions encoded by infectious ERVs that mediate the mutagenic effect. However, in contrast to mechanisms necessitating somatic infections by infectious retroviruses, the sheer number and repetitive nature of ERVs in the genome and consequently their presence in every cell may trigger additional pathogenic mechanisms that do not require the production of infectious virions (25, 26), and these are discussed separately (Fig. 2).
Figure 2. Potential mechanisms of ERV-mediated transformation.
Non-allelic homologous recombination between ERVs creates chromosomal rearrangements. New ERV integrations may disrupt a host gene. ERV transcriptional derepression can activate downstream genes. Accumulated incomplete ERV replication intermediates activate innate immunity or disregulate non-coding RNA networks. Certain retroviral proteins (e.g. envelope) mediate immunosuppression and may also be involved in cellular fusion.
Transformation by insertional mutagenesis
The process of transformation by endogenous copies of infectious MMTV and MLV almost invariably involves insertional activation of oncogenes, such as Wnt1 or Notch1 among many others, or disruption of tumor-suppressor genes, and has been extensively described in the mouse (27). However, the search for infectious retroviruses, endogenous or otherwise, that may be causing cancer in humans through a similar mechanism, was not free from controversy (28). The strong link between MMTV infection and mammary tumor in mice inspired an exhaustive, but as yet unsuccessful quest for a retrovirus causing human breast cancer. The lack of convincing evidence, despite major efforts in identifying human tumor viruses, led scientists referring to the elusive target as ‘rumor viruses’ (28). Although the discovery of HTLV-1 (human T-lymphotropic virus 1) unequivocally proved the existence of a tumor-inducing exogenous human retrovirus, no HERV provirus with the potential to produce an infectious retrovirus has been discovered to date. One of the most convincing evidence for infectious HERVs would be finding new somatic HERV integrations, and genome sequencing and analysis tools are sufficiently developed today to provide a certain degree of validity even of negative findings. In this regard, data from cancer genome sequencing identified over 180 LINE-1 somatic integrations in human cancer cells (29). In contrast, the same data identified only a single somatic integration of a small HERV fragment, which was likely the result of a microhomology-mediated DNA repair mechanism, than genuine retrotransposition (29).
Nevertheless, it is conceptually possible that infectious retroviruses derived from HERV precursors do arise spontaneously. These may not cause generalized infection within an individual or even transmission between individuals and would therefore elude detection. Indeed, the HERV-K(HML2) family comprises insertionally polymorphic proviruses with various degrees of inactivation, which collectively, but not individually contain the complete information for a replication-competent recombinant retrovirus (30, 31). Recombination between defective HERV-K(HML2) proviruses has been shown to occur naturally (14, 32), and may be facilitated by exogenous retroviruses, as in the case of HIV-1 infection (33). Although such recombination events restoring ERV infectivity would be exceptionally rare, recent mouse studies have shown that the probability of infectious MLVs arising from defective ERV precursors can be significantly elevated when innate and adaptive immune responses are defective (34, 35). This process ultimately leads to development of leukemias in mice, and additionally seems to be controlled by environmental parameters, such as the intestinal microbiota (34). It is, therefore, possible that infectious HERVs do arise and contribute to cancer development in specific situations involving immunodeficiency.
Tumor-promoting retroviral proteins
Even though most, if not all, ERVs have accumulated replication-inactivating mutations, many still contain intact ORFs that produce retroviral proteins (36, 37). These ERV-derived proteins may serve as a source of antigen for the immune system and, for certain ERVs, also a source of superantigen, with examples including MMTV in mice (38) and HERV-K18 in humans (39). ERV-encoded proteins are unlikely to be distinguished by the immune system from all other host proteins and are thus expected to contribute to T or B cell antigen-receptor repertoire selection processes, as well as peripheral regulatory mechanisms, as part of ‘self’ (40, 41). Nevertheless, immune reactivity to ERV products can often occur spontaneously in infection or cancer and is considered the driving force of several autoimmune disorders in mice (42). Immune reactivity to ERV products can also be experimentally induced in mice and non-human primates (43, 44), suggesting that immunological tolerance to ERV-derived proteins is not complete. ERV proteins may also retain biological function, which has been proposed to contribute to cellular transformation (45). Indeed, expression of HERV-encoded proteins has been detected, by antibody straining, in a variety of human cancers (25). Although certain HERV proteins can also be expressed in healthy, non-transformed tissue, some show exclusive expression in tumors (25, 28). However, direct mechanisms of contribution to cancer development have been proposed for only few of these proteins.
Examples include two accessory proteins, Rec and Np9, produced as alternative splicing products of the HERV-K(HML2) env gene. These proteins are frequently found in transformed cells from testicular germ cell tumors, but not in healthy cells (25). Rec, in particular, interacts with the cellular transcription factor PLZF (promyelocytic leukemia zinc-finger protein), and its overexpression in mice can induce early seminomas (46).
The envelope proteins of several ERVs may also contribute to the development of cancer through their fusogenic or immunosuppressive properties. Both properties of expressed retroviral envelope proteins can also serve physiological processes in the host (3). For example, products of the host syncytin genes, which are derived from retroviral env genes, are essential for placentation, by mediating cell fusion of syncytial cell layers, and for maternal tolerance of the fetus, by immunosuppression (47). Expression of Syncytins, which is normally restricted to the placenta, has been found in human cancer and it is hypothesized that Syncytin-mediated cell fusion participates in transformation or metastasis (48, 49).
Seminal work by T. Heidmann has further defined the immunosuppressive domain (ISD) within the transmembrane subunit of the envelope protein of several retroviruses, as well as its potential role in tumor growth (50). Overexpression of Moloney MLV transmembrane subunit in murine tumor cell lines led to tumor growth in recipient mice that would otherwise immunologically reject them (51). Equally, knockdown of env transcripts in B16 melanoma and Neuro-2a neuroblastoma cell lines, both of which produce infectious MLVs derived from ERV precursors, rendered them susceptible to immune rejection in vivo (52, 53). Thus, ERV-encoded ISDs may contribute to cancer development by establishing the immunosuppression necessary for tumor growth and, in addition to Syncytins, the env of HERV-E (54) and HERV-H (55) has also been shown to possess immunosuppressive potential.
ERV effects on genome function
For every ERV with the ability to produce proteins or virions, there are many more that are transcriptionally silenced or may only produce non-coding RNA transcripts. Both non-coding RNA transcription from ERVs and their repetitive presence in the genome as DNA elements have enormous potential to affect genome function (56). Although some of the complex effects of ERVs on genome function benefit the host and have been co-opted during evolution, others may be pathogenic (56, 57).
The cell possesses an array of enzymes involved in DNA metabolism and some of these are considered essential for preventing accumulation of nucleic acid intermediates from incomplete ERV replication cycles. These include TREX1, a single-stranded DNA exonuclease, and SAMHD1, an enzyme involved in the regulation of cytosolic dNTPs (58) that may also exert nuclease activity (59, 60). These enzymes interfere with the replication of exogenous retroviruses, and may have similar roles against ERVs (59, 60). This potential role against ERVs is thought to be at the center of the pathogenic mechanism initiated when these pathways are not operative. Indeed, mutations in the genes encoding these enzymes lead to autoimmune or autoinflammatory human conditions, associated with elevated production of type I IFNs (61). Notably, SAMHD1 mutations have also been identified in a variety of human cancers (62-64), suggesting that SAMHD1 is a tumor-suppressor gene, although the precise mechanism connecting SAMHD1 mutations with cancer development is not currently clear.
In addition to the effect of generic ERV transcripts and replication intermediates, non-coding RNAs of certain ERVs can have more direct effects on genome function. Repression of several proto-oncogenes, as well as other cellular genes, relies on the pre-mRNA splicing factor PSF (polypyrimidine tract-binding protein (PTB)-associated splicing factor) (65). In turn, PSF activity is negatively regulated by biding to non-coding RNAs, one of which is transcribed from a HERV-K11 provirus (66), thus potentially linking HERV transcription with proto-oncogene activation.
ERVs additionally influence genome function without producing an RNA transcript (67-70). They provide promoter activity, numerous binding sites for transcription or other DNA binding factors, or homology sites for DNA recombination. Indeed, ERV LTRs are perfect docking sites for DNA binding factors, which are critically involved in cancer development (2). One such factor is p53, an important tumor-suppressor maintaining genome integrity and preventing mutation, often referred to as the ‘the guardian of the genome’ (71). One in three of all p53 binding sites identified in a human cancer cell line were in fact within the LTRs of two HERV families, which, when bound by p53, could activate transcription of downstream genes (72). Thus, ERV LTRs may also participate in the tumor-suppressing p53 response.
However, ERV LTRs may also promote cellular transformation by in cis activation of downstream oncogenes. Transcriptional derepression of the CSF1R gene, encoding colony-stimulating factor-1 receptor, by a demethylated MaLR LTR acting as an alternative promoter has been linked with survival of cancer B cells in Hodgkin’s lymphoma (73). This study revealed how loss of epigenetic silencing of retroelement LTRs directly contributes to tumor growth.
Lastly, the repetitive nature of ERVs provides an ideal substrate for non-allelic homologous DNA recombination (2, 32, 56), that results in a variety of chromosomal rearrangements. Recombination between ERVs is responsible for notable germ-line genomic rearrangements leading to known human genetic disorders (32). Such recombination events can also occur in somatic cells, resulting in chromosomal translocations, which in turn underpin cellular transformation. Examples include recurrent chromosomal translocations in human prostate cancer, which create fusions between a HERV-K provirus and an otherwise dormant oncogene of the ETS (E26 transformation-specific) family (74). Further study of chromosomal translocations in cancer is expected to add to the list of ERVs driving these translocations.
Elevated ERV activity in cancers: harmful cause or useful consequence?
It is evident that ERVs do possess properties that might cause or contribute to cancer development and the activity of many ERVs is frequently elevated in transformed cells. There are numerous examples of ERV protein expression or virion production in mouse and human cancer cells (75, 76). However, both ERV proteins and virions can be seen also in either certain healthy tissues, such as the placenta (77-79), or in infection and other non-neoplastic diseases. Therefore, elevated ERV activity, even if restricted to cancer cells, should not be taken to signify causality. Rather, most of this activity is likely to represent lack of ERV regulation under these conditions.
ERV activity in a cell is regulated both by cell-intrinsic factors and external signals (1, 3, 34, 80). Epigenetic silencing seems to be a major way, in which the cell prevents transcription of repetitive elements, including ERVs (1, 3). Global DNA hypomethylation, leading to epigenetic derepression is a hallmark of cellular transformation and a major contributor to oncogene activation. In the altered epigenetic landscape of transformed cells, ERV derepression should be expected as a consequence (81).
Nevertheless, causal or otherwise, characteristic patterns of ERV expression are often seen in various tumors (25), offering a unique approach to immunotherapy, trials of which have already been contemplated (44). Largely tumor-restricted expression has been proposed for several ERVs, including HERV-K(HML6) in melanomas, HERV-K(HML2) in germ-cell carcinomas, HERV-E in renal cell carcinomas, or even Syncytin-1 in diverse cancers (82, 83). HERV expression in some of these cases has also been demonstrated to lead to immune reactivity against HERV-derived epitopes (84, 85). Of note, hematopoietic stem cell transplantation in patients with renal cell carcinoma resulted in tumor regression likely due to a graft-versus-tumor effect. In such patients, anti-tumor CTLs were found to be targeting a HERV-E-encoded epitope (85), demonstrating the potential for anti-ERV immune responses in tumor regression.
Tumor immunotherapy by anti-ERV CTL responses can also be experimentally demonstrated in mice. CTL epitopes have been described in EL-4 lymphoma cells and C26 colon carcinoma encoded by an endogenous MMTV and MLV, respectively (86, 87). Immune priming with a peptide from the envelope protein of Emv2, an endogenous defective MLV in C57BL/6 mice, leads to eradication of syngeneic B16 melanoma cells, which also express the same MLV envelope (43). B16 cells are also known to produce recombinant MLVs, which have restored Emv2 infectivity and have re-infected these cells (88). Infectious MLVs produced by tumor cells and spreading in the host, will inevitable complicate the interpretation of immunological studies, if tumor transplantation is also accompanied by retroviral infection of the host. It would appear that many if not most of the transplantable tumor cells carry infectious retroviruses. Some are the result of recombination between defective endogenous precursors, as in B16 and Neuro-2a cells, which are likely linked to the original transformation event (53, 88, 89). Others may be unwittingly derived from MLV-induced tumors. Also, the controversy surrounding XMRV (xenotropic murine leukemia virus-related virus), a recombinant contaminant virus that arose when human prostate cancer cells were serially propagated in mice (90), offers a cautionary tale. Although the overall impact of production of infectious retroviruses by transplantable tumors in mouse studies, and their translatability to human cancer, is difficult to establish without further investigation, the association further supports the possibility of cancer immunotherapy targeted against ERVs.
Conclusions
The tumorigenic mechanisms employed by exogenous retroviruses, namely insertional mutagenesis and acquisition of oncogenes, helped pave the way to understanding cellular transformation events. This increased understanding of the complex processes of transformation no longer required infection with retroviruses, as similar genomic insults are generated by a variety of other mechanism. However, this increased understanding of genome structure and function has also highlighted the potential importance of ERVs, the ultimate genomic parasites, although obtaining compelling evidence remains a challenge. Just as their predecessors, exogenous retroviruses, have done in the past, ERVs may illuminate additional mechanistic details of cancer initiation and progression, as well as offer new prospects for cancer immunotherapy.
Acknowledgements
I wish to thank Jonathan Stoye at NIMR for helpful discussion and critical reading of the manuscript.
This work was supported by the UK Medical Research Council (U117581330).
Abbreviations
- ERV
endogenous retrovirus
- HERV
human endogenous retrovirus
- ISD
immunosuppressive domain
- LINE
long interspersed nuclear element
- LTR
long-terminal repeat
- MaLR
mammalian apparent LTR retrotransposon
- MLV
murine leukemia virus
- MMTV
mouse mammary tumor virus
- ORF
open reading frame
- SINE
short interspersed nuclear element
Footnotes
Disclosures The author has no financial conflicts of interest.
References
- 1.Dewannieux M, Heidmann T. Endogenous retroviruses: acquisition, amplification and taming of genome invaders. Curr. Opin. Virol. 2013 doi: 10.1016/j.coviro.2013.08.005. doi: 10.1016/j.coviro.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 3.Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012;10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 4.Stocking C, Kozak CA. Murine endogenous retroviruses. Cell. Mol. Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magiorkinis G, Gifford RJ, Katzourakis A, De Ranter J, Belshaw R. Env-less endogenous retroviruses are genomic superspreaders. Proc. Natl. Acad. Sci. USA. 2012;109:7385–7390. doi: 10.1073/pnas.1200913109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 8.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramerov DA, Vassetzky NS. SINEs. Wiley. Interdiscip. Rev. RNA. 2011;2:772–786. doi: 10.1002/wrna.91. [DOI] [PubMed] [Google Scholar]
- 10.Blomberg J, Benachenhou F, Blikstad V, Sperber G, Mayer J. Classification and nomenclature of endogenous retroviral sequences (ERVs): problems and recommendations. Gene. 2009;448:115–123. doi: 10.1016/j.gene.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Weiss RA. On the concept and elucidation of endogenous retroviruses. Philos. Trans. R. Soc. Lond. , B, Biol. Sci. 2013;368:20120494. doi: 10.1098/rstb.2012.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 2005;79:12507–12514. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 16.Cardiff RD, Kenney N. A compendium of the mouse mammary tumor biologist: from the initial observations in the house mouse to the development of genetically engineered mice. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss RA. The discovery of endogenous retroviruses. Retrovirology. 2006;3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimkin MB. A. E. C. Lathrop (1868-1918): Mouse Woman of Granby. Cancer Res. 1975;35:1597–1598. [PubMed] [Google Scholar]
- 20.Crow JF. C. C. Little, cancer and inbred mice. Genetics. 2002;161:1357–1361. doi: 10.1093/genetics/161.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoye JP, Fenner S, Greenoak GE, Moran C, Coffin JM. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988;54:383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- 23.Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, Nellaker C, Goodstadt L, Nicod J, Bhomra A, et al. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477:326–329. doi: 10.1038/nature10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins NA, Copeland NG, Taylor BA, Lee BK. Dilute (d) coat colour mutation of DBA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature. 1981;293:370–374. doi: 10.1038/293370a0. [DOI] [PubMed] [Google Scholar]
- 25.Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. Endogenous retroviruses and cancer. Cell. Mol. Life Sci. 2008;65:3366–3382. doi: 10.1007/s00018-008-8496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young GR, Stoye JP, Kassiotis G. Are human endogenous retroviruses pathogenic? An approach to testing the hypothesis. Bioessays. 2013;35:794–803. doi: 10.1002/bies.201300049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan H, Johnson C. Insertional oncogenesis by non-acute retroviruses: implications for gene therapy. Viruses. 2011;3:398–422. doi: 10.3390/v3040398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voisset C, Weiss RA, Griffiths DJ. Human RNA “rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol. Mol. Biol. Rev. 2008;72:157–196. doi: 10.1128/MMBR.00033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, III, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes JF, Coffin JM. Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics. 2005;171:1183–1194. doi: 10.1534/genetics.105.043976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, et al. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J. Virol. 2012;86:262–276. doi: 10.1128/JVI.00602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu P, Lubben W, Slomka H, Gebler J, Konert M, Cai C, Neubrandt L, Prazeres da Costa O, Paul S, Dehnert S, et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 2012;37:867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 36.Benit L, Dessen P, Heidmann T. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 2001;75:11709–11719. doi: 10.1128/JVI.75.23.11709-11719.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villesen P, Aagaard L, Wiuf C, Pedersen FS. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross SR. Mouse mammary tumor virus molecular biology and oncogenesis. Viruses. 2010;2:2000–2012. doi: 10.3390/v2092000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tai AK, Lin M, Chang F, Chen G, Hsiao F, Sutkowski N, Huber BT. Murine Vbeta3+ and Vbeta7+ T cell subsets are specific targets for the HERV-K18 Env superantigen. J. Immunol. 2006;177:3178–3184. doi: 10.4049/jimmunol.177.5.3178. [DOI] [PubMed] [Google Scholar]
- 40.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat. Immunol. 2009;10:1162–1169. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young GR, Ploquin MJ, Eksmond U, Wadwa M, Stoye JP, Kassiotis G. Negative selection by an endogenous retrovirus promotes a higher-avidity CD4+ T cell response to retroviral infection. PLoS Pathog. 2012;8:e1002709. doi: 10.1371/journal.ppat.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baudino L, Yoshinobu K, Morito N, Santiago-Raber ML, Izui S. Role of endogenous retroviruses in murine SLE. Autoimmun. Rev. 2010;10:27–34. doi: 10.1016/j.autrev.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Kershaw MH, Hsu C, Mondesire W, Parker LL, Wang G, Overwijk WW, Lapointe R, Yang JC, Wang RF, Restifo NP, et al. Immunization against Endogenous Retroviral Tumor-associated Antigens. Cancer Res. 2001;61:7920–7924. [PMC free article] [PubMed] [Google Scholar]
- 44.Sacha JB, Kim IJ, Chen L, Ullah JH, Goodwin DA, Simmons HA, Schenkman DI, von Pelchrzim F, Gifford RJ, Nimityongskul FA, et al. Vaccination with cancer- and HIV infection-associated endogenous retrotransposable elements is safe and immunogenic. J. Immunol. 2012;189:1467–1479. doi: 10.4049/jimmunol.1200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oricchio E, Sciamanna I, Beraldi R, Tolstonog GV, Schumann GG, Spadafora C. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007;26:4226–4233. doi: 10.1038/sj.onc.1210214. [DOI] [PubMed] [Google Scholar]
- 46.Galli UM, Sauter M, Lecher B, Maurer S, Herbst H, Roemer K, Mueller-Lantzsch N. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene. 2005;24:3223–3228. doi: 10.1038/sj.onc.1208543. [DOI] [PubMed] [Google Scholar]
- 47.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Duelli D, Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–448. doi: 10.1016/s1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 49.Bjerregaard B, Holck S, Christensen IJ, Larsson LI. Syncytin is involved in breast cancer-endothelial cell fusions. Cell. Mol. Life Sci. 2006;63:1906–1911. doi: 10.1007/s00018-006-6201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. Lond. , B, Biol. Sci. 2013;368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mangeney M, Heidmann T. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc. Natl. Acad. Sci. USA. 1998;95:14920–14925. doi: 10.1073/pnas.95.25.14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mangeney M, Pothlichet J, Renard M, Ducos B, Heidmann T. Endogenous retrovirus expression is required for murine melanoma tumor growth in vivo. Cancer Res. 2005;65:2588–2591. doi: 10.1158/0008-5472.CAN-04-4231. [DOI] [PubMed] [Google Scholar]
- 53.Pothlichet J, Heidmann T, Mangeney M. A recombinant endogenous retrovirus amplified in a mouse neuroblastoma is involved in tumor growth in vivo. Int. J. Cancer. 2006;119:815–822. doi: 10.1002/ijc.21935. [DOI] [PubMed] [Google Scholar]
- 54.Naito T, Ogasawara H, Kaneko H, Hishikawa T, Sekigawa I, Hashimoto H, Maruyama N. Immune abnormalities induced by human endogenous retroviral peptides: with reference to the pathogenesis of systemic lupus erythematosus. J. Clin. Immunol. 2003;23:371–376. doi: 10.1023/a:1025369500466. [DOI] [PubMed] [Google Scholar]
- 55.Mangeney M, de PN, Thomas G, Heidmann T. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J. Gen. Virol. 2001;82:2515–2518. doi: 10.1099/0022-1317-82-10-2515. [DOI] [PubMed] [Google Scholar]
- 56.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 57.Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 58.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 59.Luban J. Innate immune sensing of HIV-1 by dendritic cells. Cell Host Microbe. 2012;12:408–418. doi: 10.1016/j.chom.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Atianand MK, Fitzgerald KA. Molecular basis of DNA recognition in the immune system. J. Immunol. 2013;190:1911–1918. doi: 10.4049/jimmunol.1203162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 66.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc. Natl. Acad. Sci. USA. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 68.van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 69.Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 70.Conley AB, Piriyapongsa J, Jordan IK. Retroviral promoters in the human genome. Bioinformatics. 2008;24:1563–1567. doi: 10.1093/bioinformatics/btn243. [DOI] [PubMed] [Google Scholar]
- 71.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 72.Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. USA. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Kochert K, Bouhlel MA, Richter J, Soler E, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 2010;16:571–9. 1. doi: 10.1038/nm.2129. [DOI] [PubMed] [Google Scholar]
- 74.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 75.Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 76.Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, Fodinger D, Seppele H, Schanab O, Magin-Lachmann C, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003;63:8735–8741. [PubMed] [Google Scholar]
- 77.Dirksen ER, Levy JA. Virus-like particles in placentas from normal individuals and patients with systemic lupus erythematosus. J. Natl. Cancer Inst. 1977;59:1187–1192. doi: 10.1093/jnci/59.4.1187. [DOI] [PubMed] [Google Scholar]
- 78.Kalter SS, Helmke RJ, Heberling RL, Panigel M, Fowler AK, Strickland JE, Hellman A. Brief communication: C-type particles in normal human placentas. J. Natl. Cancer Inst. 1973;50:1081–1084. doi: 10.1093/jnci/50.4.1081. [DOI] [PubMed] [Google Scholar]
- 79.Vernon ML, McMahon JM, Hackett JJ. Additional evidence of type-C particles in human placentas. J. Natl. Cancer Inst. 1974;52:987–989. doi: 10.1093/jnci/52.3.987. [DOI] [PubMed] [Google Scholar]
- 80.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, Sasaki C, Costa J, Lizardi PM. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009;448:151–167. doi: 10.1016/j.gene.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cegolon L, Salata C, Weiderpass E, Vineis P, Palu G, Mastrangelo G. Human endogenous retroviruses and cancer prevention: evidence and prospects. BMC Cancer. 2013;13:4. doi: 10.1186/1471-2407-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katoh I, Kurata SI. Association of Endogenous Retroviruses and Long Terminal Repeats with Human Disorders. Front. Oncol. 2013;3:234. doi: 10.3389/fonc.2013.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62:5510–5516. [PubMed] [Google Scholar]
- 85.Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy JP, Hanada K, Mena O, Kurlander R, Tawab A, et al. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J. Clin. Invest. 2008;118:1099–1109. doi: 10.1172/JCI34409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Malarkannan S, Serwold T, Nguyen V, Sherman LA, Shastri N. The mouse mammary tumor virus env gene is the source of a CD8+ T-cell-stimulating peptide presented by a major histocompatibility complex class I molecule in a murine thymoma. Proc. Natl. Acad. Sci. USA. 1996;93:13991–13996. doi: 10.1073/pnas.93.24.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang AY, Gulden PH, Woods AS, Thomas MC, Tong CD, Wang W, Engelhard VH, Pasternack G, Cotter R, Hunt D, et al. The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc. Natl. Acad. Sci. USA. 1996;93:9730–9735. doi: 10.1073/pnas.93.18.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li M, Huang X, Zhu Z, Gorelik E. Sequence and Insertion Sites of Murine Melanoma-Associated Retrovirus. J. Virol. 1999;73:9178–9186. doi: 10.1128/jvi.73.11.9178-9186.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pothlichet J, Mangeney M, Heidmann T. Mobility and integration sites of a murine C57BL/6 melanoma endogenous retrovirus involved in tumor progression in vivo. Int. J. Cancer. 2006;119:1869–1877. doi: 10.1002/ijc.22066. [DOI] [PubMed] [Google Scholar]
- 90.Delviks-Frankenberry K, Cingoz O, Coffin JM, Pathak VK. Recombinant origin, contamination, and de-discovery of XMRV. Curr. Opin. Virol. 2012;2:499–507. doi: 10.1016/j.coviro.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dewannieux M, Ribet D, Heidmann T. Risks linked to endogenous retroviruses for vaccine production: a general overview. Biologicals. 2010;38:366–370. doi: 10.1016/j.biologicals.2010.01.006. [DOI] [PubMed] [Google Scholar]