Abstract
The development of gastrointestinal diseases has been found to be associated with Helicobacter pylori (H. pylori) infection and various biochemical stresses in stomach and intestine. These stresses, such as oxidative, osmotic and acid stresses, may bring about bi-directional effects on both hosts and H. pylori, leading to changes of protein expression in their proteomes. Therefore, proteins differentially expressed in H. pylori under various stresses not only reflect gastrointestinal environment but also provide useful biomarkers for disease diagnosis and prognosis. In this regard, proteomic technology is an ideal tool to identify potential biomarkers as it can systematically monitor proteins and protein variation on a large scale of cell’s translational landscape, permitting in-depth analyses of host and pathogen interactions. By performing two-dimensional polyacrylamide gel electrophoresis (2-DE) followed by liquid chromatography-nanoESI-mass spectrometry (nanoLC-MS/MS), we have successfully pinpointed alkylhydroperoxide reductase (AhpC), neutrophil-activating protein and non-heme iron-binding ferritin as three prospective biomarkers showing up-regulation in H. pylori under oxidative, osmotic and acid stresses, respectively. Further biochemical characterization reveals that various environmental stresses can induce protein structure change and functional conversion in the identified biomarkers. Especially salient is the antioxidant enzyme AhpC, an abundant antioxidant protein present in H. pylori. It switches from a peroxide reductase of low-molecular-weight (LMW) oligomers to a molecular chaperone of high-molecular-weight (HMW) complexes under oxidative stress. Different seropositivy responses against LMW or HMW AhpC in H. pylori-infected patients faithfully match the disease progression from disease-free healthy persons to patients with gastric ulcer and cancer. These results has established AhpC of H. pylori as a promising diagnostic marker for gastrointestinal maladies, and highlight the utility of clinical proteomics for identifying disease biomarkers that can be uniquely applied to disease-oriented translational medicine.
Keywords: Proteomics, Biomarker, Helicobacter pylori, Gastritis, Gastric ulcer, Gastric cancer, Alkylhydroperoxide reductase, Neutrophil-activating protein, Non-heme iron-binding ferritin
Core tip: This work aims to review the literature on the application of proteomics methodology for identifying proteins differentially expressed in Helicobacter pylori (H. pylori) under different environmental stresses, resulting in the identification of several biomarkers related to gastrointestinal disorders induced by H. pylori infection.
INTRODUCTION
The discovery of Helicobacter pylori (H. pylori), a Gram-negative and microaerophilic bacterium has revolutionized the diagnosis and treatment of gastrointestinal diseases[1,2]. Prolonged infection and inflammation because of persistent H. pylori colonization lead to chronic gastritis and severe gastric pathologies, such as peptic ulcer and gastric cancer[3]. It is worth noting that variable clinical manifestations associated with H. pylori infection are common in the endoscopic examination of patients, about 80%-90% patients being asymptomatic gastritis (GA), 10%-15% gastric or duodenal ulcer (GU) and 1%-2% gastric cancers (GC)[4]. Gastric cancers are generally believed to arise from sites of H. pylori infection accompanied by chronic irritation and inflammation[5-7]. Such inflammatory ulcers of stomach are manifestly the result of severe immune response triggered by the host’s immune system upon H. pylori infection[8,9]. Therefore, to identify the mechanism underlying such pathogenesis and the capricious clinical outcomes in the gastric infections by H. pylori under varied environmental stresses in vivo is crucial to the development of efficient therapeutic approach against H. pylori infection and the associated gastrointestinal maladies[10].
In this review, we have focused on the potential roles of three biomarkers identified in the course of our decade-long research in the pathogenesis of varied gastrointestinal diseases induced by H. pylori infection. The antioxidant enzyme alkylhydroperoxide reductase (AhpC) of H. pylori was found to be expressed with larger amounts in GC strains and exhibited a higher seropositivity for GC patients than gastritis (GA) ones. Based on the significant difference between AhpC in H. pylori isolated from patients with GA, GU and GC, it is conceivable that the antioxidant protein AhpC of H. pylori may be applied as a prognostic or diagnostic protein marker to monitor different stages of tissue damages from H. pylori-infected diseases. The relevance of up-regulated proteins NapA (neutrophil-activating protein) and non-heme iron-binding ferritin (Pfr) under osmotic and acid stresses, respectively is also explored regarding their corresponding regulation of cellular homeostasis. Finally, the potential diagnostic and therapeutic value of these novel biomarkers in H. pylori-related gastroenterological maladies is proposed for their translational medical applications.
ENVIRONMENTAL STRESSES AND GASTROINTESTINAL DISEASES
The variable clinical outcomes of H. pylori infection is suggested to result from different severity and distribution of H. pylori-induced gastric inflammation plus environmental stresses in stomach and intestine[11]. Several studies have also shown the correlation between the protection of host cells against H. pylori infection and stress-induced protein factors[12,13]. Figure 1 depicts H. pylori under various stresses in vivo, such as oxidative, osmotic and acid stresses, which may underlie the pathogenesis of the gastrointestinal infections and lay a firm basis for the development of efficient diagnosis, prognosis and therapeutic managements.
Figure 1.
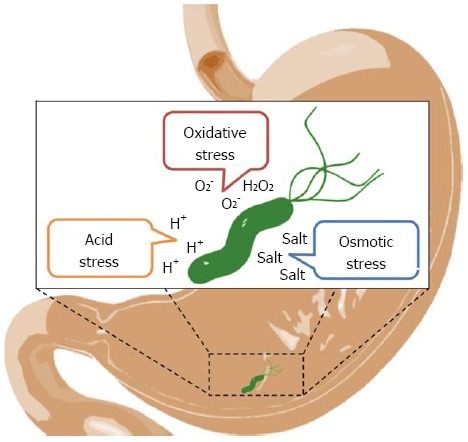
Diagram of Helicobacter pylori exposed to various environmental stresses in stomach and intestine. The development of gastrointestinal diseases is suggested to be associated with Helicobacter pylori (H. pylori) infection and environmental stresses in stomach and intestine. These stresses, such as oxidative, acid and osmotic stresses[33-36], may bring detrimental effects on H. pylori.
Oxidative stress
Oxidative stress was suggested to be associated with chronic inflammation and malignant transformation. It was also reported to be the causative factor for most cancers, including gastric cancer[14]. Nevertheless, the pathogenic agent inducing the gastric inflammation was not known until the discovery of H. pylori[15]. H. pylori infection can activate inflammatory responses and trigger immune system, leading to discharge of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), superoxide anion, and nitric oxide from phagocytes. The excessive ROS production may thereby trigger oxidative stress to cause inflammatory damage to host gastric mucosa, whereas H. pylori will also face the attack of oxygen-related free radicals released from phagocytes (Figure 1)[16]. To survive under such ROS environment generated inside the host, H. pylori has developed some defensive mechanisms to protect against oxidative stress. Therefore, mapping proteome changes of H. pylori in response to oxidative stress can potentially identify indicators or markers for both the inflammation status and the progression of gastrointestinal diseases.
Osmotic stress
High dietary salt intake has been considered as a long-term risk factor for gastric carcinogenesis[17,18]. A link between high salt consumption and gastric cancer has been shown in both animal model and human epidemiologic studies[19-21]. High salt environment is thought to induce disturbance of osmotic homeostasis, which may potentially affect host cells and H. pylori in stomach and intestine (Figure 1)[22]. Although aforementioned evidences suggest high-salt diet may mediate augmentation in the severity of H. pylori-induced disease outcomes, the molecular mechanisms underlying the purported cause-effect correlation between osmotic stress and H. pylori infection still remain poorly understood. Several studies have tried to use genetic approaches for identifying the key regulators in H. pylori in response to high concentration of NaCl. The change of gene-expression levels of two virulent factors, CagA and VacA, were reported by Loh et al[23] and Gancz et al[24], respectively. Both results not only support the synergistic effect between H. pylori infection and diets rich in sodium chloride but point out the potential of investigating whole-proteome change under osmotic stress to explore biomarker discovery.
Acid stress
H. pylori is unique for its ability to adapt to the extreme acidic environment of pH 1-2 in human stomach lumen as evident by the fact that H. pylori infection usually lasts for decades or more (Figure 1)[25,26]. Survival of H. pylori under severe acid shock has been generally proposed to depend on its urease activity, which can release ammonia from urea, thereby neutralize gastric acid in stomach[27,28]. However, other regulatory proteins and molecular mechanisms can also contribute to the acid adaptation of H. pylori in human stomach. To address this, several reports have analyzed differential gene expression in H. pylori under acidic conditions by employing DNA microarray and their findings supported that acidity is a signal modulating the expression of multiple genes related to metal ion homeostasis[29-31]. Therefore, it is of interest and import to know whether proteomics could identify urease and its metal metabolism related proteins as indicators for acid stress in stomach, which may reflect the progression of gastrointestinal diseases.
CLINICAL PROTEOMICS FOR GASTROINTESTINAL DISEASE, BIOMARKER DISCOVERY AND VALIDATION
Proteomics is a robust methodology, which can depict snapshots of protein compositions of particular cells or tissues at defined time points. Clinician scientists have realized its potential and begun to apply innovative proteomic technology to clinical research especially on its application to translational medicine, which generally consists of several phases, that is (1) biomarker discovery; (2) biomarker verification; and finally (3) biomarker validation and clinical trial[32].
To this end, we have set up a proteomics-based platform to systematically identify and validate potential biomarkers in H. pylori for gastrointestinal diseases (Figure 2), which also represents in essence the application of translational medicine from clinics to lab research and then back to clinics. Firstly, H. pylori clinical isolates were obtained from gastric biopsy specimens from patients with different pathological manifestations, such as gastritis, duodenal ulcer, gastric ulcer and gastric cancer. All isolated strains were further confirmed for their urease activity and the helical morphology of these organisms observed by phase-contrast microscopy[33]. Upon exposing to different stress conditions, H. pylori proteomes are compared and analyzed by two-dimensional polyacrylamide gel electrophoresis (2-DE) followed by liquid chromatography-nanoESI-mass spectrometry (nanoLC-MS/MS)[33-36]. By making use of rapidly-evolved powerful bioinformatics analysis programs, highly up- or down-regulated proteins in H. pylori under various stresses could be identified as candidates of potential biomarkers. Further immunoblotting in H. pylori strains from gastrointestinal patients of varied clinical outcomes can then be used to validate whether these candidates are engendered to reflect environmental stresses in stomach and intestine[37].
Figure 2.
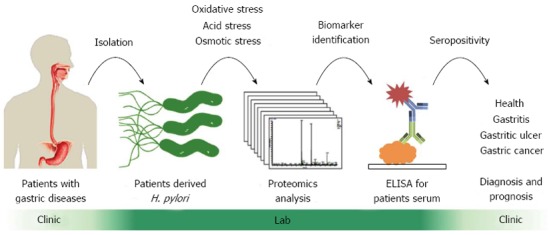
Flow chart of clinical proteomics platform designed for discovery and validation of Helicobacter pylori biomarkers for the diagnosis of gastrointestinal diseases. We have set up a proteomics-based platform[32] to systematically identify and validate potential biomarkers in Helicobacter pylori (H. pylori) for gastrointestinal diseases, which can provide a major methodological support for translational medicine encompassing basic biomedical research and clinical therapeutic medicine. ELISA: Enzyme-linked immunosorbent assay.
To translate the lab findings into clinics, enzyme-linked immunosorbent assay (ELISA) is an ideal tool to employ these H. pylori antigens for the detection of antibodies in sera from patients with different gastric diseases[37]. For large-scale ELISA analysis, H. pylori recombinant antigens are produced by molecular cloning, overexpression in Escherichia coli, and followed by protein purification[37,38]. Recombinant H. pylori antigens are further coated in 96-well plates for screening seropositivities of patient sera, useful for clinical diagnosis and prognosis of disease progression. In summary, our clinical proteomics pipeline provides a rapid and scalable platform for the identification of biomarkers in gastrointestinal diseases and can potentially be applied to other diseases as well, such as cataract and cancer[39,40].
FUNCTIONAL SWITCH OF IDENTIFIED H. PYLORI BIOMARKERS IN RESPONSE TO ENVIRONMENTAL STRESSES
Our clinical proteomics platform has found out several stress-related biomarkers from H. pylori proteome (Table 1). Under oxidative stress induced by H2O2, several antioxidant (alkylhydroperoxide reductase AhpC, catalase KatA, serine protease HtrA) and virulent proteins (cytotoxin-associated protein CagA, vacuolating cytotoxin VacA, adhesin AlpA) are up-regulated in isolated H. pylori from patients with different gastrointestinal diseases[35], revealing part of the mechanism concerning ROS-induced gastric carcinogenesis. In response to osmotic and acid stresses, H. pylori overexpress both iron-binding (NapA, Pfr) and acid-neutralizing proteins (urease subunits UreA, UreB)[34,36], indicating that these two stresses may share similar effects on H. pylori. To further understand the molecular mechanism, AhpC, NapA and Pfr are overexpressed and purified for biochemical characterization with or without stresses. Interestingly, various stresses lead to protein structure change and functional switch of these proteins (Figure 3), indicative of molecular and cellular connotation for H. pylori to adapt to different stress environments.
Table 1.
Proteomic analysis identifies upregulated proteins in Helicobacter pylori under different stresses
| Stress | Source | Disease | Upregulated proteins (function) | Ref. |
| Oxidative stress | Reactive oxygen species (ROS): H2O2, O2- | Gastritis, duodenal ulcer, gastric ulcer, gastric cancer | AhpC, KatA, HtrA (antioxidant) | Huang et al[35] |
| CagA, VacA, AlpA (virulent) | ||||
| Osmotic stress | High salt: NaCl | Gastric cancer | NapA, Pfr (iron-binding) | Liao et al[36] |
| UreA, UreB (acid neutralizing) | ||||
| Acid stress | Gastric acid: H+ | Gastro-esophageal reflux disease, irritable bowel syndrome | Pfr, NapA (iron-binding) | Huang et al[34] |
| UreA, UreB, UreG (acid neutralizing) |
AhpC: Alkylhydroperoxide reductase; NapA: Neutrophil-activating protein; Pfr: Non-heme iron-binding ferritin.
Figure 3.
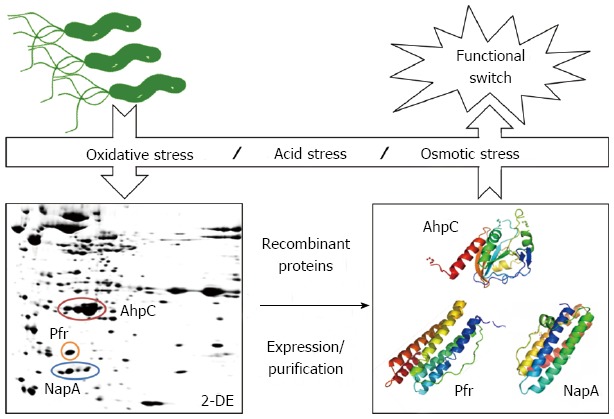
Functional switch of identified Helicobacter pylori biomarkers in response to environmental stresses. Alkylhydroperoxide reductase (AhpC), neutrophil-activating protein (NapA) and non-heme iron-binding ferritin (Pfr) are identified by clinical proteomics as potential biomarkers under different environmental stresses. Further biochemical characterization showed the protein structure change was accompanied by functional switch of these proteins under varied environmental conditions, such as oxidative, acid and osmotic stresses[34,36,38]. The PDB ID of Helicobacter pylori (H. pylori) AhpC, NapA and Pfr are 1ZOF[61], 1JI4[62] and 3EGM[63], respectively.
AhpC and oxidative stress
H. pylori AhpC, an important member of 2-Cys peroxiredoxin (Prx) family[41], is abundantly expressed for balancing the intracellular levels of ROS generated from metabolisms or mild environmental stresses[42]. AhpC cooperates with thioredoxin (Trx) to decompose H2O2 and organic peroxides by its peroxidase activity[43,44]. Previously, we found AhpC also displays dual-activities in vivo, which switches from a peroxide reductase of low-molecular-weight (LMW) oligomers under microaerobic conditions to a molecular chaperone of high-molecular-weight (HMW) complexes in the presence of Trx after long-term (16 h) oxidative stress[38]. In brief, LMW-AhpC efficiently reduces toxic ROS by its peroxide reductase or peroxidase activity through a canonical reaction cycle by coupling reactions involving Trx, thioredoxin reductase (TrxR) and NADPH. Upon facing severe and long-term oxidative stress, LMW-AhpC initially undergoes a peroxidative reaction cycle and becomes peroxidized, converting cysteine at its active site to cysteine sulfinic acid (-SO2H) and sulfonic acid (-SO3H). The oxidized LMW-AhpC would be converted to HMW-AhpC with chaperone activity for prevention of mis- or un-folded proteins from aggregation[45]. In such a scenario, AhpC acts as a potent sensor for ROS and as chaperones for H. pylori to survive and persist in the oxidative environment of human stomachs.
NapA and osmotic stress
NapA, a 17-kDa protein, is known as a major antigen in the human immune response to H. pylori infection, which can stimulate neutrophils to generate oxygen radicals when the microorganism adheres to endothelial cells[46]. In addition, H. pylori NapA is also reported as an iron-binding protein with dodecameric structure[47], indicating that NapA may potentially work with dual functionalities under different stresses. Interestingly, NapA is found not only to be up-regulated in H. pylori under high salt stress but expose its hydrophobic surfaces to exterior environment[36]. We also found the structure of the NapA monomer was not affected by salt, but that the native oligomeric structure of NapA dissociated under high salt concentration, resulting in losing Fe2+-binding ability[36]. The loss in iron binding of NapA under high-salt osmotic stress leads to release of Fe2+ ions, which would react with oxygen and produce H2O2[48]. Elevation of H2O2 concentration would potentially increase the risk of oxidant-related cellular injury and DNA damage in the gastric lumen or mucosa[49]. Therefore, a high dietary salt intake would aggravate the circumstances of H. pylori-associated infections, resulting in inflammation-related gastrointestinal diseases.
Pfr and acid stresses
Unlike animal ferritin as ferroxidase to convert active ferrous ions to the more innocuous ferric ions, ferritin Pfr (a special prokaryote class of ferritin family) of H. pylori was shown to be essential for iron storage in subcellular granules and enabled H. pylori to survive under severe iron starvation. Pfr can also protect H. pylori from acid-amplified iron toxicity[50-52] caused by iron-mediated Fenton reaction[53,54]. Similar to AhpC with dual function, we further found H. pylori ferritin (Pfr) can switch from an iron-storage protein with ferroxidase activity to a DNA-binding/protective function[34]. The conformational change had occurred when ferritin was exposed to acid, leading to exposure of more hydrophobic region to exterior environment at pH 5 than at pH 7. Therefore, the discovery of the new function of ferritin indicates that anti-acid mechanism for the survival of H. pylori may involve several sequential steps. In the acidic environment of the gastric lumen or mucosa, the amplified metal-induced free radicals generated in the presence of ferrous ions and H2O2 may force H. pylori to exert some protection on its own vital DNA, triggering an increasing protein expression level of ferritin and a change in the function from an iron-storage role with ferroxidase activity to that of DNA binding and protection[34,50,54]. This lends support to the supposition that ferritin can act as an indicator or sensor reflecting the acid environment in stomach, and potentially as a biomarker to distinguish duodenal ulcer and gastric cancer due to their differences in acid secretion[55].
H. PYLORI AHPC AS A BIOMARKER TO MONITOR DIFFERENT STAGES OF INFLAMMATION ASSOCIATED GASTROINTESTINAL DISEASES
H. pylori AhpC was identified by our clinical proteomics platform to be a potential biomarker for gastric diseases[35]. As shown in Figure 4, we previously reported that none or low titers of serum antibody against LMW AhpC were detected in gastritis patients with weak inflammation in cases of H. pylori infection[30]. However, higher titers of antibodies against HMW AhpC would be produced in gastric ulcer and cancer patients with more severe inflammation[30]. Therefore, H. pylori AhpC can signal and monitor different stages of inflammation associated with H. pylori infection in gastrointestinal diseases. Also, both HMW AhpC and the serum level of antibody against AhpC are important to predict the risk of gastric cancer among H. pylori-positive patients (Figure 4).
Figure 4.
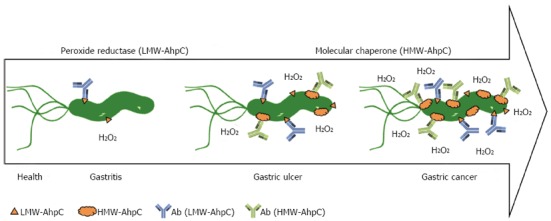
Alkylhydroperoxide reductase of Helicobacter pylori, a biomarker used to monitor different stages of inflammation associated with gastrointestinal diseases. In cases of Helicobacter pylori (H. pylori) infection, none or low titers of serum antibody against low-molecular-weight (LMW)-alkylhydroperoxide reductase (AhpC) were detected in patients of gastritis with weak inflammation. However, higher titers of antibodies against high-molecular-weight (HMW)-AhpC were found in gastric ulcer and cancer patients with severe inflammation. Therefore, H. pylori AhpC can signal and monitor different stages of inflammation associated with H. pylori infection in gastrointestinal diseases[37].
Recently, the use of H. pylori AhpC as a potential vaccine against H. pylori infection[56] or utilization of its monoclonal antibodies for detecting H. pylori[57,58] were reported, again emphasizing its importance during the progression of inflammation-related gastrointestinal diseases. New technologies for producing H. pylori AhpC recombinant proteins, such as SDS-PAGE electroelution-based rapid purification[59] and different prokaryotic expression systems[60], have also been developed. Taking all these findings and technologies into account, H. pylori AhpC is a potential biomarker for gastrointestinal diseases and it may be credible and promising to develop biomarkers kits of H. pylori for clinical application in the future, serving as a non-invasive detection method for diagnosis and prognosis. Most importantly, the successful identification of H. pylori AhpC as a biological marker highlights the utility of clinical proteomics for identifying disease biomarkers that can be translated from lab research into clinics.
ACKNOWLEDGMENTS
We thank the research participation of Dr. Ming-Hong Chuang as a PhD student at the Institute of Biochemical Sciences, National Taiwan University; Dr. Ming-Shiang Wu at the Division of Gastroenterology, Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan; and Dr. Deng-Chyang Wu at the Division of Gastroenterology, Department of Internal Medicine, Chung-Ho Memorial Hospital, Kaohsiung Medical University, Kaohsiung, Taiwan.
Footnotes
Supported by (in part) Kaohsiung Medical University, Academia Sinica, and the National Science Council, Taipei, Taiwan, No. 96-2311-B-037-005-MY3, No.99-2314-B-037-042, and No. 99-2745-B-037-005 to Chiou SH
P- Reviewers: Kozlowski H, Ohshima K S- Editor: Zhai HH L- Editor: A E- Editor: Liu XM
References
- 1.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Hopkins RJ, Girardi LS, Turney EA. Relationship between Helicobacter pylori eradication and reduced duodenal and gastric ulcer recurrence: a review. Gastroenterology. 1996;110:1244–1252. doi: 10.1053/gast.1996.v110.pm8613015. [DOI] [PubMed] [Google Scholar]
- 3.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 4.Wu MS, Chow LP, Lin JT, Chiou SH. Proteomic identification of biomarkers related to Helicobacter pylori-associated gastroduodenal disease: challenges and opportunities. J Gastroenterol Hepatol. 2008;23:1657–1661. doi: 10.1111/j.1440-1746.2008.05659.x. [DOI] [PubMed] [Google Scholar]
- 5.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handa O, Naito Y, Yoshikawa T. Helicobacter pylori: a ROS-inducing bacterial species in the stomach. Inflamm Res. 2010;59:997–1003. doi: 10.1007/s00011-010-0245-x. [DOI] [PubMed] [Google Scholar]
- 8.Amedei A, Cappon A, Codolo G, Cabrelle A, Polenghi A, Benagiano M, Tasca E, Azzurri A, D’Elios MM, Del Prete G, et al. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J Clin Invest. 2006;116:1092–1101. doi: 10.1172/JCI27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki H, Hibi T. Oxidative stress in Helicobacter pylori-associated gastroduodenal disease. J Clin Biochem Nutr. 2006;39:56–63. [Google Scholar]
- 10.Blaser MJ, Berg DE. Helicobacter pylori genetic diversity and risk of human disease. J Clin Invest. 2001;107:767–773. doi: 10.1172/JCI12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pignatelli B, Bancel B, Plummer M, Toyokuni S, Patricot LM, Ohshima H. Helicobacter pylori eradication attenuates oxidative stress in human gastric mucosa. Am J Gastroenterol. 2001;96:1758–1766. doi: 10.1111/j.1572-0241.2001.03869.x. [DOI] [PubMed] [Google Scholar]
- 12.Yeo M, Park HK, Kim DK, Cho SW, Kim YS, Cho SY, Paik YK, Hahm KB. Restoration of heat shock protein70 suppresses gastric mucosal inducible nitric oxide synthase expression induced by Helicobacter pylori. Proteomics. 2004;4:3335–3342. doi: 10.1002/pmic.200400951. [DOI] [PubMed] [Google Scholar]
- 13.Matsukawa Y, Asai Y, Kitamura N, Sawada S, Kurosaka H. Exacerbation of rheumatoid arthritis following Helicobacter pylori eradication: disruption of established oral tolerance against heat shock protein? Med Hypotheses. 2005;64:41–43. doi: 10.1016/j.mehy.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 14.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 15.Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1:1273–1275. [PubMed] [Google Scholar]
- 16.Matysiak-Budnik T, Mégraud F. Helicobacter pylori infection and gastric cancer. Eur J Cancer. 2006;42:708–716. doi: 10.1016/j.ejca.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 18.Wang XQ, Terry PD, Yan H. Review of salt consumption and stomach cancer risk: epidemiological and biological evidence. World J Gastroenterol. 2009;15:2204–2213. doi: 10.3748/wjg.15.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs CS, Mayer RJ. Gastric carcinoma. N Engl J Med. 1995;333:32–41. doi: 10.1056/NEJM199507063330107. [DOI] [PubMed] [Google Scholar]
- 20.Inoue M, Tsugane S. Epidemiology of gastric cancer in Japan. Postgrad Med J. 2005;81:419–424. doi: 10.1136/pgmj.2004.029330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsugane S. Salt, salted food intake, and risk of gastric cancer: epidemiologic evidence. Cancer Sci. 2005;96:1–6. doi: 10.1111/j.1349-7006.2005.00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox JG, Dangler CA, Taylor NS, King A, Koh TJ, Wang TC. High-salt diet induces gastric epithelial hyperplasia and parietal cell loss, and enhances Helicobacter pylori colonization in C57BL/6 mice. Cancer Res. 1999;59:4823–4828. [PubMed] [Google Scholar]
- 23.Loh JT, Torres VJ, Cover TL. Regulation of Helicobacter pylori cagA expression in response to salt. Cancer Res. 2007;67:4709–4715. doi: 10.1158/0008-5472.CAN-06-4746. [DOI] [PubMed] [Google Scholar]
- 24.Gancz H, Jones KR, Merrell DS. Sodium chloride affects Helicobacter pylori growth and gene expression. J Bacteriol. 2008;190:4100–4105. doi: 10.1128/JB.01728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louw JA, Marks IN. Peptic ulcer disease. Curr Opin Gastroenterol. 2004;20:533–537. doi: 10.1097/00001574-200411000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;128:833–848. doi: 10.1053/j.gastro.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bury-Moné S, Skouloubris S, Labigne A, De Reuse H. The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Mol Microbiol. 2001;42:1021–1034. doi: 10.1046/j.1365-2958.2001.02689.x. [DOI] [PubMed] [Google Scholar]
- 28.Hong W, Sano K, Morimatsu S, Scott DR, Weeks DL, Sachs G, Goto T, Mohan S, Harada F, Nakajima N, et al. Medium pH-dependent redistribution of the urease of Helicobacter pylori. J Med Microbiol. 2003;52:211–216. doi: 10.1099/jmm.0.05072-0. [DOI] [PubMed] [Google Scholar]
- 29.McGowan CC, Necheva A, Thompson SA, Cover TL, Blaser MJ. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol Microbiol. 1998;30:19–31. doi: 10.1046/j.1365-2958.1998.t01-1-01079.x. [DOI] [PubMed] [Google Scholar]
- 30.Ang S, Lee CZ, Peck K, Sindici M, Matrubutham U, Gleeson MA, Wang JT. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect Immun. 2001;69:1679–1686. doi: 10.1128/IAI.69.3.1679-1686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allan E, Clayton CL, McLaren A, Wallace DM, Wren BW. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology. 2001;147:2285–2292. doi: 10.1099/00221287-147-8-2285. [DOI] [PubMed] [Google Scholar]
- 32.Chiou SH, Wu CY. Clinical proteomics: current status, challenges, and future perspectives. Kaohsiung J Med Sci. 2011;27:1–14. doi: 10.1016/j.kjms.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Chuang MH, Wu MS, Lin JT, Chiou SH. Proteomic analysis of proteins expressed by Helicobacter pylori under oxidative stress. Proteomics. 2005;5:3895–3901. doi: 10.1002/pmic.200401232. [DOI] [PubMed] [Google Scholar]
- 34.Huang CH, Lee IL, Yeh IJ, Liao JH, Ni CL, Wu SH, Chiou SH. Upregulation of a non-heme iron-containing ferritin with dual ferroxidase and DNA-binding activities in Helicobacter pylori under acid stress. J Biochem. 2010;147:535–543. doi: 10.1093/jb/mvp200. [DOI] [PubMed] [Google Scholar]
- 35.Huang CH, Chiou SH. Proteomic analysis of upregulated proteins in Helicobacter pylori under oxidative stress induced by hydrogen peroxide. Kaohsiung J Med Sci. 2011;27:544–553. doi: 10.1016/j.kjms.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 36.Liao JH, Sun YH, Hsu CH, Lin YC, Wu SH, Kuo CJ, Huang CH, Chiou SH. Up-regulation of neutrophil activating protein in Helicobacter pylori under high-salt stress: structural and phylogenetic comparison with bacterial iron-binding ferritins. Biochimie. 2013;95:1136–1145. doi: 10.1016/j.biochi.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 37.Huang CH, Chuang MH, Lo WL, Wu MS, Wu YH, Wu DC, Chiou SH. Alkylhydroperoxide reductase of Helicobacter pylori as a biomarker for gastric patients with different pathological manifestations. Biochimie. 2011;93:1115–1123. doi: 10.1016/j.biochi.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Chuang MH, Wu MS, Lo WL, Lin JT, Wong CH, Chiou SH. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci USA. 2006;103:2552–2557. doi: 10.1073/pnas.0510770103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang CH, Wang YT, Tsai CF, Chen YJ, Lee JS, Chiou SH. Phosphoproteomics characterization of novel phosphorylated sites of lens proteins from normal and cataractous human eye lenses. Mol Vis. 2011;17:186–198. [PMC free article] [PubMed] [Google Scholar]
- 40.Ho PY, Chueh SC, Chiou SH, Wang SM, Lin WC, Lee IL, Yang HY, Peng HC, Lai MK. ΑB-crystallin in clear cell renal cell carcinoma: tumor progression and prognostic significance. Urol Oncol. 2013;31:1367–1377. doi: 10.1016/j.urolonc.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 42.Olczak AA, Seyler RW, Olson JW, Maier RJ. Association of Helicobacter pylori antioxidant activities with host colonization proficiency. Infect Immun. 2003;71:580–583. doi: 10.1128/IAI.71.1.580-583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker LM, Raudonikiene A, Hoffman PS, Poole LB. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J Bacteriol. 2001;183:1961–1973. doi: 10.1128/JB.183.6.1961-1973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Olczak AA, Walton JP, Maier RJ. Contribution of the Helicobacter pylori thiol peroxidase bacterioferritin comigratory protein to oxidative stress resistance and host colonization. Infect Immun. 2005;73:378–384. doi: 10.1128/IAI.73.1.378-384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang CH, Chuang MH, Wu YH, Chuang WC, Jhuang PJ, Chiou SH. Characterization of site-specific mutants of alkylhydroperoxide reductase with dual functionality from Helicobacter pylori. J Biochem. 2010;147:661–669. doi: 10.1093/jb/mvp209. [DOI] [PubMed] [Google Scholar]
- 46.Satin B, Del Giudice G, Della Bianca V, Dusi S, Laudanna C, Tonello F, Kelleher D, Rappuoli R, Montecucco C, Rossi F. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J Exp Med. 2000;191:1467–1476. doi: 10.1084/jem.191.9.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tonello F, Dundon WG, Satin B, Molinari M, Tognon G, Grandi G, Del Giudice G, Rappuoli R, Montecucco C. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol Microbiol. 1999;34:238–246. doi: 10.1046/j.1365-2958.1999.01584.x. [DOI] [PubMed] [Google Scholar]
- 48.Dominici S, Valentini M, Maellaro E, Del Bello B, Paolicchi A, Lorenzini E, Tongiani R, Comporti M, Pompella A. Redox modulation of cell surface protein thiols in U937 lymphoma cells: the role of gamma-glutamyl transpeptidase-dependent H2O2 production and S-thiolation. Free Radic Biol Med. 1999;27:623–635. doi: 10.1016/s0891-5849(99)00111-2. [DOI] [PubMed] [Google Scholar]
- 49.Gong M, Ling SS, Lui SY, Yeoh KG, Ho B. Helicobacter pylori gamma-glutamyl transpeptidase is a pathogenic factor in the development of peptic ulcer disease. Gastroenterology. 2010;139:564–573. doi: 10.1053/j.gastro.2010.03.050. [DOI] [PubMed] [Google Scholar]
- 50.Frazier BA, Pfeifer JD, Russell DG, Falk P, Olsén AN, Hammar M, Westblom TU, Normark SJ. Paracrystalline inclusions of a novel ferritin containing nonheme iron, produced by the human gastric pathogen Helicobacter pylori: evidence for a third class of ferritins. J Bacteriol. 1993;175:966–972. doi: 10.1128/jb.175.4.966-972.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bereswill S, Waidner U, Odenbreit S, Lichte F, Fassbinder F, Bode G, Kist M. Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology. 1998;144(Pt 9):2505–2516. doi: 10.1099/00221287-144-9-2505. [DOI] [PubMed] [Google Scholar]
- 52.Waidner B, Greiner S, Odenbreit S, Kavermann H, Velayudhan J, Stähler F, Guhl J, Bissé E, van Vliet AH, Andrews SC, et al. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect Immun. 2002;70:3923–3929. doi: 10.1128/IAI.70.7.3923-3929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.French CS. Temperature characteristics for the metabolism of chlorella: III. The catalytic decomposition of hydrogen peroxide by chlorella pyrenoidosa. J Gen Physiol. 1934;18:209–213. doi: 10.1085/jgp.18.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo Y, Henle ES, Linn S. Oxidative damage to DNA constituents by iron-mediated fenton reactions. The deoxycytidine family. J Biol Chem. 1996;271:21167–21176. [PubMed] [Google Scholar]
- 55.Calam J, Baron JH. ABC of the upper gastrointestinal tract: Pathophysiology of duodenal and gastric ulcer and gastric cancer. BMJ. 2001;323:980–982. doi: 10.1136/bmj.323.7319.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Riordan AA, Morales VA, Mulligan L, Faheem N, Windle HJ, Kelleher DP. Alkyl hydroperoxide reductase: a candidate Helicobacter pylori vaccine. Vaccine. 2012;30:3876–3884. doi: 10.1016/j.vaccine.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 57.Mohammadian T, Doosti M, Paknejad M, Siavoshi F, Massarrat S, Soukhtanloo M. Production of polyclonal antibody against alkyl hydroperoxide reductase of Helicobacter pylori and its antigenicity. Hybridoma (Larchmt) 2008;27:481–485. doi: 10.1089/hyb.2008.0054. [DOI] [PubMed] [Google Scholar]
- 58.Amini Najafabadi H, Paknejad M, Farshad S, Mohammadian T, Seyyed Ebrahimi SS, Amini Najafabadi A. Immunodot blot assay to detect Helicobacter pylori using monoclonal antibodies against the 26 kDa protein. Hybridoma (Larchmt) 2012;31:403–410. doi: 10.1089/hyb.2012.0066. [DOI] [PubMed] [Google Scholar]
- 59.Mohammadian T, Doosti M, Paknejad M, Siavoshi F, Massarrat S. Preparative SDS-PAGE Electroelution for Rapid Purification of Alkyl Hydroperoxide Reductase from Helicobacter pylori. Iran J Public Health. 2010;39:85–91. [PMC free article] [PubMed] [Google Scholar]
- 60.Mehmood K, Hasan F. Construction and use of a prokaryotic expression system for Helicobacter pylori AhpC. BMC Res Notes. 2012;5:328. doi: 10.1186/1756-0500-5-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papinutto E, Windle HJ, Cendron L, Battistutta R, Kelleher D, Zanotti G. Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter pylori. Biochim Biophys Acta. 2005;1753:240–246. doi: 10.1016/j.bbapap.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 62.Zanotti G, Papinutto E, Dundon W, Battistutta R, Seveso M, Giudice G, Rappuoli R, Montecucco C. Structure of the neutrophil-activating protein from Helicobacter pylori. J Mol Biol. 2002;323:125–130. doi: 10.1016/s0022-2836(02)00879-3. [DOI] [PubMed] [Google Scholar]
- 63.Cho KJ, Shin HJ, Lee JH, Kim KJ, Park SS, Lee Y, Lee C, Park SS, Kim KH. The crystal structure of ferritin from Helicobacter pylori reveals unusual conformational changes for iron uptake. J Mol Biol. 2009;390:83–98. doi: 10.1016/j.jmb.2009.04.078. [DOI] [PubMed] [Google Scholar]


