Abstract
Objective: This study aims to explore the clinical characteristics of ABCE1 in esophageal cancers and its roles in the proliferation, invasiveness, migration and apoptosis of the esophageal carcinoma Eca109 cell line. Methods: The expression of ABCE1 and its target protein-RNase L, were first studied in tumor tissues of esophageal carcinoma and adjacent non-tumor tissues. The siRNA green fluorescent protein (GFP) expression vector of ABCE1 was prepared and transfected into the esophageal carcinoma Eca109 cells, then the fluorescence microscope was used to study the transfection efficiency. The MTT assay, cell invasion, the transwell and scratch assay were used to study cell proliferation and migration activity; the apoptosis rate was tested by flow cytometry. Western blot and RT-PCR assay were adopted to measure their silencing efficacy. Results: ABCE1 expression is low in the adjacent non-tumor tissues while the expression is high in the esophageal carcinoma; the expression is reversely proportional to the differentiation degrees. The expression of RNase L was in contrary to ABCE1. After transfected with ABCE1-siRNA, the proliferation, invasiveness and migration capabilities of cells decreased significantly whilst the apoptosis rate enhanced greatly (P<0.01). Meanwhile, the expression of ABCE1 in Eca109 cells was blocked (P<0.01) while the expression of RNase L increased significantly (P<0.01). Conclusion: ABCE1 is closely connected with the pathogenesis and development of esophageal carcinoma, which act through the cellular pathways of 2-5A/RNase L.
Keywords: ABCE1, RNase L, esophageal carcinoma, Eca109 cells, siRNA
Introduction
Esophageal carcinoma is characterized as a common malignant tumor with poor prognosis. The pathogenesis and development of esophageal carcinoma is a complicated process, and in most cases, esophageal carcinoma shows no symptoms until the cancer is in an advanced stage, thus making a cure less likely. The overall survival rate still remains low and less than 20% of diagnosed patients can survive for more than five years [1]. So far, the histopathological progression of esophageal cancer have been thoroughly described, while the molecular underpinnings are less well understood [2]. ATP binding cassette E1 (ABCE1) is a member of the ATP-binding cassette (ABC) transporters superfamily and OABP subfamily, is expressed in the cytoplasm of various tissues and organs in mammals [3,4]. ABCE1 encodes a product of 68 KD protein which is described as Ribonuclease L (RNase L) inhibitor [5-7], and by forming a heterodimer with RNase L, ABCE1 protein inhibits the IFN-dependant 2-5A/RNase L system thus regulates a broad range of biological functions including viral infection, tumor cell proliferation, and antiapoptosis [7-10]. Few studies have reported the characters of ATP-Binding Cassette in the pathogenesis and development of esophageal carcinoma [11,12] while ABCE1 was rarely reported. In the current study, we firstly studied the expression of ABCE1 and RNase L at esophageal carcinoma and the adjacent non-tumor tissues resected from esophageal carcinoma patients. Then we adopt the small interfering RNA (siRNA) to construct the siRNA vector of ABCE1, and transfect it into the Eca109 cell lines of esophageal carcinoma to block the expression of gene ABCE1, thus investigate its role in the pathogenesis, development and treatment of esophageal carcinoma.
Materials and methods
The study was approved and registered by our hospital Ethics Committee in January 2010, the related screening and analysis of resection sample were approved by the Ethics committee of Liaoning Medical University, and written consent for using these samples were signed and agreed by all subjects.
Resection sample collection
The esophageal carcinoma tissue and adjacent non-tumor tissue samples (at least 5 cm away from the edge of cancerous tissue) were collected from the sample preservation center of our hospital. These esophageal carcinoma (and adjacent non-tumor tissue) samples were all resected in the department of thoracic surgery from May 2010 to December 2012.
The inclusion criteria of these sample were: (i) Patient didn’t receive any radiotherapy and chemotherapy treatment before; (ii) Each patient had received a medical examination which includes, but not limited to, cranial CT scan, chest CT scan, abdominal CT scan, MRI and ECT, which could define the TNM stage of the patient clearly; (iii) Patients had received radical surgery with sufficient tissue samples prepared in paraffin blocks for further testing; Patients who had at least two concurrent primary tumors were excluded.
All samples were fixed in 10% formaldehyde and paraffin-embedded. The samples were routinely serial sectioned with thickness of 5 μm, and then immunohistochemically stained. The esophageal carcinomas were confirmed by two senior pathologists and were classified according to the standard of histological classification of WHO in 1999, all cases were phased according to the tumor-node-metastasis (TNM) staging system stipulated by the seventh version of American Joint Committee on Cancer (AJCC) Cancer Staging Manual.
Immunohistochemistry
Sections were deparaffinized and hydrated in a stepwise xylene and graded ethanol, washed with PBS, and recovered through microwave irradiation. A 3% H2O2 solution were added and cultured for 10 minutes, and then washed with PBS. Goat blocking serum was supplied and cultured under room temperature and the diluted primary antibodies were applied (1:100). Sections, after staying overnight in 4°C, were applied PBS washing, then added with secondary antibodies and cultured in 37°C for 20 minutes. The newly-prepared DAB chromogenic reagent was applied, and cultured at 37°C for 5 to 10 minutes. Nuclei were then stained with hematoxylin-eosin (HE).
Expressions of ABCE1 and RNase L protein in tissue sample
Western blot was used to determine the protein level in the cancer tissues. Briefly, frozen tissues were homogenated with lysis buffer, then centrifuged at 4°C for 30 min (12000 r/min). The supernatant was collected; BCA method was used to determine protein concentration. A 10% polyacrylamide gel was prepared to load protein samples, 5% nonfat dry milk was added to block the non-specific antigen. The primary antibodies (Rabbit anti-human ABCE1 polyclonal antibody, and Mouse anti-human RNase L polyclonal antibody, Santa Cruz) and secondary antibodies were applied.
Cell lines
Eca109 cell line was purchased from American Type Culture Collection (ATCC, USA). It was cultured in RPMI-1640 medium supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and maintained in a 5% CO2 humidified atmosphere at 37°C.
Preparation of target vector and transfection
Small interfering RNAs (siRNAs) targeting ABCE1 were chemically synthesized by Takara Biotechnology (Dalian) Co., Ltd. The two siRNA sequences were: (Si-1) F: 5’GATCCGCTACAGCGAGTACGTTTACCTGTGAAGCCACAGATGGGGTAAACGTACTCGCTGTAGCTTTTTTG3’; (Si-1) R: 5’AATTCAAAAAAGCTACAGCGAGTACGTTTACCCCATCTGTGGCTTCACAGGTAAACGTACTCGCTGTAGCG3’; (Si-2) F: 5’GATCCGAGTACGATGATCCTCCTGACTGTGAAGCCACAGATGGGTCAGGAGGATCATCGTACTCTTTTTTG3’; (Si-2) R: 5’AATTCAAAAAAGAGTACGATGATCCTCCTGACCCATCTGTGGCTTCACAGTCAGGAGGATCATCGTACTCG3’; the negative scramble siRNA sequence was (Si-N) F: 5’GATCCGCGAGACCTCAGTATGTTACCTGTGAAGCCACAGATGGGGTAACATACTGAGGTC TCGCTTTTTTG3’ (control group).
The DNA sequence were fully ligated into RNAi-Ready pRNAT-U6.1/Neo-siRNA-Expression Vector (Clontech, Mountain View, CA, USA) at 4°C overnight by an TaKaRa DNA Ligation Kit. There were two targeted recombinant plasmids (ABCE1-SiRNA-1, ABCE1-SiRNA-2), and 1 negative control (ABCE1-SiRNA-N), untreated cells served as the blank control. Lipofectamine 2000 was used in transfection with the concentration of 2×105 cells per well (six-well plate) and 5 μg of recombinant plasmids. The inverted fluorescence microscope was used for observation and taking pictures.
MTT assay
The cells from four groups were loaded in 96-well plate with 4×103 cells per well, cultured with the DMEM medium with 10% FBS, at time points of 24, 48, 72 and 96 hours. The medium was removed from each well and 100 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; 0.5 mg/ml in PBS) was added in the absence of light; formazan crystals were produced over a 4 h incubation period. To dissolve crystals, 150 μl of 0.04 N HCl in isopropanol was added to each well and the optical density at 540 nm was measured on a Tecan Spectrophotometer (Tecan SPECTRAFluor, Tecan, Männedorf, Switzerland).
Transwell assay
Forty eight hours after the transfection, the full medium was changed into serum-free culture medium. Eight hours later, they were digested to a suspension with a density of 1×104/ml. Cells were seeded into the transwell chamber (Millipore, USA) which membrane was coated by a dilution of Matrigel (50 mg/L). The chamber was placed into a 24 well culture plate, with 500 μl of 1640 medium containing 15% serum added outside of the chamber, and 200 μl cell suspension were added in the chamber. 72 hours later, the cells were stained and placed under the fluorescence microscope for observation.
Scratch assay
The migration capacity of Eca109 cells was performed in a 24-well culture plate. After transfection, a horizontal wound (scratch) was made on the cells with a tiny spear. Cell migration was observed at 24, 48 and 72 hours after the transfection, the scratch spaces were analyzed.
Flow cytometry
Forty eight hours after the transfection, the cells were washed with PBS twice, and then digested and centrifuged. Then apoptosis detection was processed according to the instructions of the Detection Kit (PE, Annexin V Apoptosis I): Cells were resuspended with 1×binding buffer (1×106/ml). A total of 100 μl was drawn, 5 μl of PE Annexin V and 5 μl of 7-AAD were added. Cells were cultured in a rotary system under room temperature for 15 minutes, added with 400 μl of 1×binding buffer, and placed for analysis within one hour.
Knock-down efficiency
Forty eight hours later, western blot and Real time-PCR were used to analyze the knock-down efficiency. The primers of RT-PCR were designed and synthesized by Sangon Biotech company (Shanghai, China), with the sequence of ABCE1-Forward: 5’CCAGGTGAAGTTTTGGGATTAG3’ and ABCE1-Reverse: 5’AGGTTTGATGATGGCTTTTAGG3’. After total cellular RNA was extracted, the first-strand cDNA was synthesized using a ReverTra Ace reverse transcription kit (Toyobo Co., Ltd., Osaka, Japan). ABCE1 was amplified using the ABI 7500 Real-time PCR System, GAPDH served as internal control. The amplification performed as follows: one cycle of pre-denaturation at 95°C 5 min followed by 40 cycles of denaturing at 95°C for 15 s and extension at 60°C for 1 min. SYBR® Green Real-Time PCR Master Mixes (Life Technologies, USA) were used.
Western blot was used to determine the protein level of ABCE1 in the four groups and the procedure was same as mentioned above.
Statistical analysis
The images are analyzed by Quantity One software. All laboratory data are represented as “Mean ± Standard Deviation (S.D.)”. The chi-square test and single factor analysis of variance (ANOVA) were performed with the SPSS17.0 software, a significant difference was considered for P<0.05.
Results
A total of 134 samples were selected successfully, 105 of them were resected from males and 29 from females, with the average age of 57 years (ranging from 35 to 84). Of them, 22 cases were highly differentiated, 55 cases were moderately differentiated, and 57 cases were differentiated lowly. Thirty eight cases were under stage III (stage I and stage II), and 96 cases were stage III and stage IV (Table 1).
Table 1.
The Relationship between the positive expression of ABCE1 protein in esophageal squamous cell carcinoma and the patient characteristics
| ABCE1 | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| N (134) | - | + | ++ | X2 | P | |
| Gender | ||||||
| Male | 105 | 17 | 25 | 63 | 0.75 | 0.687 |
| Female | 29 | 5 | 9 | 15 | ||
| Age | ||||||
| ≤60 | 74 | 12 | 22 | 40 | 1.73 | 0.421 |
| >60 | 60 | 10 | 12 | 38 | ||
| Location | ||||||
| Mid-thoracic | 82 | 15 | 24 | 43 | 2.924 | 0.232 |
| Lower-thoracic | 52 | 7 | 10 | 35 | ||
| Degree of Differentiation | ||||||
| High | 22 | 9 | 11 | 2 | ||
| Moderate | 55 | 7 | 14 | 34 | 28.607 | 0.000 |
| Low | 57 | 6 | 9 | 42 | ||
| TNM Stages | ||||||
| I+II Stages | 38 | 18 | 13 | 7 | 47.006 | 0.000 |
| III Stage | 96 | 4 | 21 | 71 | ||
ABCE1 is highly expressed in low differentiated esophageal carcinoma tissue
Immunohistochemistry showed the ABCE1 protein was mainly expressed in cytoplasm as yellow or brown-color staining. Its expression was significantly higher in low differentiated cells compared with highly differentiated cells. A few of expressions existed in the cytoplasm of stratum basale of the normal esophageal mucosa cells, yet the expressions decreased in the enveloping layer cells. There are 83.6% (112 cases of 134) positive expressions of ABCE1 protein in esophageal carcinoma tissues while the rate of positive expression was significantly low in adjacent non-tumor tissues (7.46%, 10 of 134, P<0.05). The expression of ABCE1 in the esophageal carcinoma tissues had no correlation with the age, gender, and cancerous location of the patients (showed in Table 1), yet there was a statistical differences between the differentiation of esophageal carcinoma cells (X2=28.607, P=0.000), and the degree of differentiation is negatively correlated with the expression of ABCE1 (Figure 1). There is also a statistical differences in the expression of ABCE1 with the TNM stages in esophageal carcinoma (X2=47.006, P=0.000), and the expression is positively correlated with the TNM stages.
Figure 1.
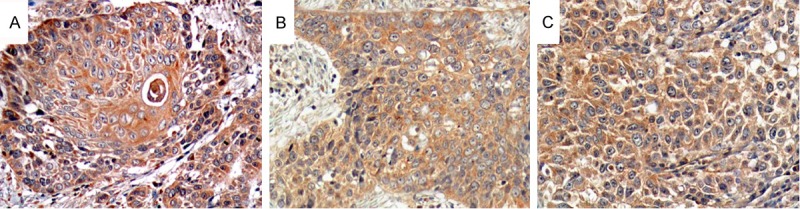
The immunohistochemistry of ABCE1 in esophageal carcinoma tissues, the degree of differentiation is negatively correlated with the expression of ABCE1. A: Highly differentiated tissues; B: Moderate differentiated tissues; C: Lowly differentiated tissues.
Expression of ABCE1 is low in the adjacent non-tumor tissues while high in esophageal carcinoma tissues
The western blot result (Figure 2) showed ABCE1 expressed lowly in adjacent non-tumor tissues of esophageal carcinoma, and expressed highly in esophageal carcinoma tissues. And the lower the degree of differentiation is, the higher the expression. On the contrary, RNase L expressed highly in adjacent non-tumor tissues of esophageal carcinoma, and the higher the degree of differentiation is, the lower the expression is.
Figure 2.
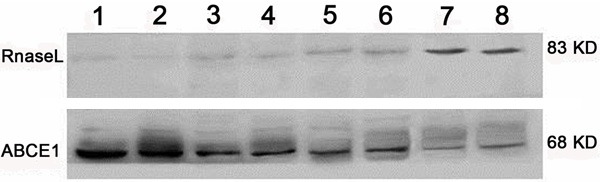
The expression of ABCE1 protein and RNase L protein in the esophageal carcinoma and its adjacent non-tumortissues. Sample 1 and 2 are low differentiated, 3 and 4 are moderate differentiated, 5 and 6 are of high differentiated, and 7 and 8 are the adjacent non-tumor tissues.
Fluorescence microscope observation
As shown in Figure 3, under fluorescence microscope observation, the transfection efficiency of the three groups’ transfected plasmid (ABCE1-SiRNA-1, ABCE1-SiRNA-2 and ABCE1-SiRNA-N) were satisfactory with all exceeded 80%.
Figure 3.
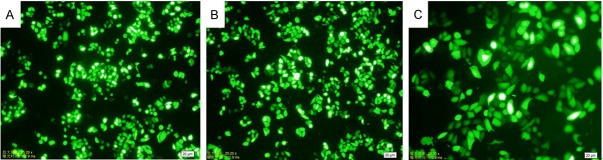
The expression of green fluorescent protein (GFP) in ECA109 cells after transfected 48 hours. A: ABCE1-SiRNA-1 group (×100); B: ABCE1-SiRNA-2 group (×100); C: ABCE1-SiRNA-N group (×200).
Decreasing of proliferation and migration ability after transfection
MTT test showed the proliferation of cells in ABCE1-SiRNA-N and blank control group increased almost 4 folds from 24 to 96 hours after transfection (Figure 4A), while the proliferation in ABCE1-SiRNA-1 and ABCE1-SiRNA-2 were relatively slow, which increased only 1 fold in 72 hours. Compared with negative control, the proliferation were retarded significantly (P<0.01).
Figure 4.
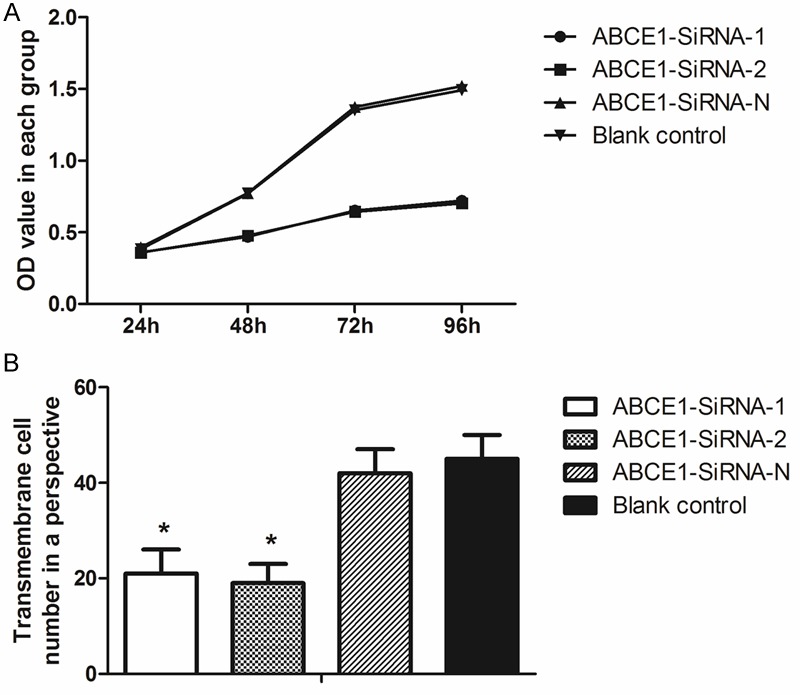
Proliferation and migration ability were decreased after transfection. A: Proliferation tested by MTT assay; B: Column diagram of transmembrane cell count comparison.
The migration ability test of transfected cell was performed 72 hours after the transfection. The transmembrane cell numbers in ABCE1-SiRNA-1 and ABCE1-SiRNA-2 were only half of the numbers in ABCE1-SiRNA-N and blank control group (Figure 4B); this indicated that the migration ability was inhibited significantly in ABCE1-SiRNA-1 and ABCE1-SiRNA-2 groups. The scratch width measure results supported the results of the above-mention test: the reducing space in ABCE1-SiRNA-1 and ABCE1-SiRNA-2 from 0 to 72 hours after the transfection were far below those measured from ABCE1-SiRNA-N and blank control group (data showed in Table 2).
Table 2.
The width of the scratch in each time period of each group of cells (mean±SD, N=5)
| ABCE1-SiRNA-1 | ABCE1-SiRNA-2 | ABCE1-SiRNA-N | Blank control | |
|---|---|---|---|---|
| 0h (mm) | 33.42±1.36 | 32.95±2.12 | 34.21±1.69 | 33.78±1.91 |
| 24h (mm) | 27.89±2.17 | 27.34±1.57 | 26.77±1.45 | 26.06±1.09 |
| 48h (mm) | 23.75±1.38* | 22.16±2.29* | 15.33±1.19 | 14.02±1.73 |
| 72h (mm) | 18.27±1.76* | 17.96±1.97* | 8.21±1.28 | 7.07±2.47 |
P<0.01, compared with group D.
Apoptosis rate increased after transfection
Flow cytometry analysis showed the apoptosis rate increased significantly (P<0.01) in ABCE1-SiRNA-1 and ABCE1-SiRNA-2 transfected cells, compared to negative control; While there is no significant change in the apoptotic rate between cells transfected with ABCE1-SiRNA-N (P>0.05) and blank control (Table 3).
Table 3.
Analysis of the rate of apoptosis of each group of cells
| Group | Apoptosis rate (%) |
|---|---|
| ABCE1-SiRNA-1 | 19.81±1.21* |
| ABCE1-SiRNA-2 | 22.03±1.99* |
| ABCE1-SiRNA-N | 6.01±0.57 |
| Blank control | 5.88±1.75 |
P<0.01, compared with ABCE1-SiRNA-N group.
Decreased expression of ABCE1 in transfected Eca109 cells
Compared with ABCE1-SiRNA-N group and the blank control group, Real time PCR result showed that after the transfection, the RNA expression of ABCE1-SiRNA-1 and ABCE1-SiRNA-2 significantly decreased (Figure 5A), the relative expression of ABCE1 (compared with GAPDH ) were 0.241±0.049, 0.253±0.072, 0.698±0.031, and 0.674±0.009, in ABCE1-SiRNA-1, ABCE1-SiRNA-2, ABCE1-SiRNA-N and blank control group respectively.
Figure 5.
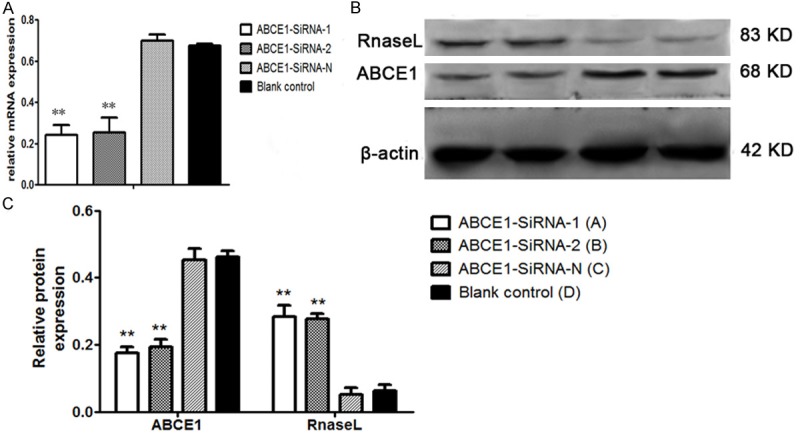
Expression of ABCE1 decreased while RNase L increased after transfection, β-actin served as control. A: Real time-PCR result; B: Western blot result; C: The expression displayed by column chart. **, P<0.01, compared with blank control.
Western blot result (Figure 5B) showed that the expression of ABCE1 protein was inhibited significantly in ABCE1-SiRNA-1 and ABCE1-SiRNA-2 group compared with blank control (P<0.05, Figure 5C), while the expression of ABCE1-SiRNA-N group almost remained unchanged; on the contrary, the expression of RNase L increased significantly in ABCE1-SiRNA-1 and ABCE1-SiRNA-2 group compared with blank control (P<0.05), and the expression in ABCE1-SiRNA-N group was same with blank control.
Discussion
Recent research have indicated that gene ABCE1 has an over-expression in lung adenocarcinoma [13], colorectal carcinoma, rectal carcinoma and prostatic carcinoma [10]. After the targeted silencing of gene ABCE1, there is a distinct change in the proliferation and apoptosis of tumor cells [13-15]. Some findings suggest that the mutation of gene ABCE1 could lead to the changing of pathogenesis, development and metastasis of genetic prostatic carcinoma [16,17].
So far, there are few reports about the role of gene ABCE1 in the esophageal carcinoma. In this report, we observed that ABCE1 was highly expressed in the esophageal cancer patients, compared to adjacent non-tumor tissues. Importantly, we observed that the expression of ABCE1 is closely related to cancer cell differentiation and TNM stages, which suggested that ABCE1 might contribute to invasion and metastasis of esophageal cancer. Based on that, we depleted it in Eca109 cells, which showed high level of ABCE1 expression and strong invasive ability, and confirmed that ABCE1 plays a major role in controlling the cell invasion, migration, and motility. All of our results are in consistence, suggesting targeting ABCE1 could be a strategy for the development of esophageal tumor therapy.
ABCE1 belongs to the gene subfamily of ATP-binding cassette transporters which express in the cytoplasm and the karyotheca. Compiling experimental evidences have showed that ABCE1 protein is the inhibitor the Ribonuclease L (RNase L) [5-7]. Studies have showed that RNase L could specifically bind with and degrade RNA within cells, thus prevent the bio-synthesis of protein at the translation level, and leading to the apoptosis of cells [7-9]. ABCE1 play an important role by forming a heterodimer with RNase L in inhibiting this pathway during the apoptotic process of cells [9,10].
The 2’-5’ oligoadenylate (2-5A)/RNase L system is one of the principle enzymatic pathways induced by interferon (IFN). RNase L mediates cell apoptosis and plays a significant role by regulating RNA expression. Increased 2-5A level within the cells plays an up-regulating role to the RNase L activity, whilst down-regulates the ABCE1 protein [18,19].
The discovery of 21-23 nucleotide RNA duplexes, called small interference RNA (siRNA) may well be one of the transforming events in biology in the past decade, which has become a powerful tool for studies on gene function, carcinoma and viral disease therapy [20,21]. Zheng et al. constructed an siRNA-expression vector for targeting ABCE1 gene in lung carcinoma cells 95-D and NCI-H446 with a satisfactory transfection efficiency of 42.7% [22]. Enlightened by his work, we studied the expression difference of ABCE1 and RNase L in esophageal carcinoma tissues and their adjacent non-tumor tissues. In our study, over 80% of plasmids in ABCE1-SiRNA-1, ABCE1-SiRNA-2 and ABCE1-SiRNA-N group were transfected successfully, while the proliferation, migration and apoptosis rate in the ABCE1-SiRNA-N group have almost showed no difference with the blank control group. These results indicate that the safety and efficacy of interfering vector system is reliable in the cell level, and ABCE1 is an ideal target for esophageal carcinoma.
Our previous investigation also finds out that the siRNA expression vector transfected with gene ABCE1 has an impact on the proliferation, invasiveness, migration and apoptosis of the tumor cells of human small-cell lung cancer [15]. This suggests that gene ABCE1 might serve as the candidate for the pathogenesis and metastasis of tumors. At present a wide array of genes which could possibly affect the pathogenesis and development of esophageal carcinoma have been investigated, targeted RNAi of CXCR4, MMP-2, XIAP, MTA1 and surviving have been reported for the esophageal carcinoma in vitro or in vivo with s down-regulation ranged from 20%-80% [23-25]. Yet few reports research the role of gene ABCE1 to the esophageal carcinoma. Our study demonstrates that ABCE1 could be a future target in the treatment of esophageal carcinoma.
In summary, our study showed that ABCE1 has a high expression in the esophageal carcinoma tissues, and the lower the degree of differentiation is, the higher the expression. The transfection vector system we constructed could inhibit the expression of ABCE1 successfully; the in vivo toxicity and efficiency will be the focus in our future research.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Zhu L, Yan W, Rodriguez-Canales J, Rosenberg AM, Hu N, Goldstein AM, Taylor PR, Erickson HS, Emmert-Buck MR, Tangrea MA. MicroRNA analysis of microdissected normal squamous esophageal epithelium and tumor cells. Am J Cancer Res. 2011;1:574–584. [PMC free article] [PubMed] [Google Scholar]
- 3.Karcher A, Buttner K, Martens B, Jansen RP, Hopfner KP. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure. 2005;13:649–659. doi: 10.1016/j.str.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Braz AS, Finnegan J, Waterhouse P, Margis R. A plant orthologue of RNase L inhibitor (RLI) is induced in plants showing RNA interference. J Mol Evol. 2004;59:20–30. doi: 10.1007/s00239-004-2600-4. [DOI] [PubMed] [Google Scholar]
- 5.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allikmets R, Gerrard B, Hutchinson A, Dean M. Characterization of the human ABC superfamily: isolation and mapping of 21 new genes using the expressed sequence tags database. Hum Mol Genet. 1996;5:1649–1655. doi: 10.1093/hmg/5.10.1649. [DOI] [PubMed] [Google Scholar]
- 7.Diriong S, Salehzada T, Bisbal C, Martinand C, Taviaux S. Localization of the ribonuclease L inhibitor gene (RNS4I), a new member of the interferon-regulated 2-5A pathway, to 4q31 by fluorescence in situ hybridization. Genomics. 1996;32:488–490. doi: 10.1006/geno.1996.0151. [DOI] [PubMed] [Google Scholar]
- 8.van der Deen M, de Vries EG, Timens W, Scheper RJ, Timmer-Bosscha H, Postma DS. ATP-binding cassette (ABC) transporters in normal and pathological lung. Respir Res. 2005;6:59. doi: 10.1186/1465-9921-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodnina MV. Protein synthesis meets ABC ATPases: new roles for Rli1/ABCE1. EMBO Rep. 2010;11:143–144. doi: 10.1038/embor.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian Y, Han X, Tian DL. The biological regulation of ABCE1. IUBMB Life. 2012;64:795–800. doi: 10.1002/iub.1071. [DOI] [PubMed] [Google Scholar]
- 11.Huang L, Lu Q, Han Y, Li Z, Zhang Z, Li X. ABCG2/V-ATPase was associated with the drug resistance and tumor metastasis of esophageal squamous cancer cells. Diagn Pathol. 2012;7:180. doi: 10.1186/1746-1596-7-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsunoda S, Okumura T, Ito T, Kondo K, Ortiz C, Tanaka E, Watanabe G, Itami A, Sakai Y, Shimada Y. ABCG2 expression is an independent unfavorable prognostic factor in esophageal squamous cell carcinoma. Oncology. 2006;71:251–258. doi: 10.1159/000106787. [DOI] [PubMed] [Google Scholar]
- 13.Ren Y, Li Y, Tian D. Role of the ABCE1 gene in human lung adenocarcinoma. Oncol Rep. 2012;27:965–970. doi: 10.3892/or.2012.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu DZ, Tian DL, Ren Y. [Expression and clinical significance of ABCE1 in human lung adenocarcinoma] . Zhonghua Zhong Liu Za Zhi. 2008;30:296–297. [PubMed] [Google Scholar]
- 15.Huang B, Gao Y, Tian D, Zheng M. A small interfering ABCE1-targeting RNA inhibits the proliferation and invasiveness of small cell lung cancer. Int J Mol Med. 2010;25:687–693. [PubMed] [Google Scholar]
- 16.Xiang Y, Wang Z, Murakami J, Plummer S, Klein EA, Carpten JD, Trent JM, Isaacs WB, Casey G, Silverman RH. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2’,5’-oligoadenylates. Cancer Res. 2003;63:6795–6801. [PubMed] [Google Scholar]
- 17.Silverman RH. Implications for RNase L in prostate cancer biology. Biochemistry. 2003;42:1805–1812. doi: 10.1021/bi027147i. [DOI] [PubMed] [Google Scholar]
- 18.Iordanov MS, Paranjape JM, Zhou A, Wong J, Williams BR, Meurs EF, Silverman RH, Magun BE. Activation of p38 mitogen-activated protein kinase and c-Jun NH(2)-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol Cell Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 20.Xia L, Guan W, Wang D, Zhang YS, Zeng LL, Li ZP, Wang G, Yang ZZ. Killing effect of Ad5/F35-APE1 siRNA recombinant adenovirus in combination with hematoporphrphyrin derivative-mediated photodynamic therapy on human nonsmall cell lung cancer. Biomed Res Int. 2013;2013:957913. doi: 10.1155/2013/957913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi X, Zhao G, Zhang H, Guan D, Meng R, Zhang Y, Yang Q, Jia H, Dou K, Liu C, Que F, Yin JQ. MITF-siRNA formulation is a safe and effective therapy for human melasma. Mol Ther. 2011;19:362–371. doi: 10.1038/mt.2010.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng MG, Gao Y, Huang B, Tian DL, Yang CL. Suppression of ABCE1 Leads to Decreased Cell Proliferation and Increased Apoptosis in 95-D and NCI-H446 Lung Carcinoma Cells. Progress in Biochemistry and Biophysics. 2009;36:1475–1482. [Google Scholar]
- 23.Wang T, Mi Y, Pian L, Gao P, Xu H, Zheng Y, Xuan X. RNAi targeting CXCR4 inhibits proliferation and invasion of esophageal carcinoma cells. Diagn Pathol. 2013;8:104. doi: 10.1186/1746-1596-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Shen YG, Xu YJ, Shi ZL, Han HL, Sun DQ, Zhang X. Effects of RNAi-mediated matrix metalloproteinase-2 gene silencing on the invasiveness and adhesion of esophageal carcinoma cells, KYSE150. Dig Dis Sci. 2012;57:32–37. doi: 10.1007/s10620-011-1864-y. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Ding F, Luo A, Chen A, Yu Z, Ren S, Liu Z, Zhang L. XIAP is highly expressed in esophageal cancer and its downregulation by RNAi sensitizes esophageal carcinoma cell lines to chemotherapeutics. Cancer Biol Ther. 2007;6:973–980. doi: 10.4161/cbt.6.6.4195. [DOI] [PubMed] [Google Scholar]


