Abstract
Chronic lymphocytic leukemia (CLL) is a common leukemia in adults, but its pathogenesis is still poorly understood. Interleukin-9 (IL-9) is initially described as a growth factor secreted by helper T cells. Recently, the oncogenic activities of IL-9 were reported in some leukemia but not chronic lymphocytic leukemia (CLL). The purpose of the present study is to investigate the expression of IL-9 from patients with CLL and to evaluate its correlation with clinical characteristics. Serum and peripheral blood mononuclear cells (PBMCs) from patients with CLL were analyzed using ELISA, RT-PCR, and western blot. ELISA analysis indicated IL-9 could be detected in 20 of 47 sera from CLL patients while none serum sample from healthy volunteers contained detectable levels of IL-9. There was a higher expression of IL-9 within PBMCs from patients with CLL compared with B cells of healthy blood donors using RT-PCR and western blot. The upregulated IL-9 was correlated to the clinical staging, ZAP-70 expression, β2 microglobulin expression and IgVH status of CLL patients (P<0.05). Our findings suggest that overexpression of IL-9 may contribute to the pathogenesis of CLL and is associated with some adverse prognostic parameters.
Keywords: IL-9, chronic lymphocytic leukemia, prognosis
Introduction
B-cell CLL continues to be a more common leukemia with no obvious curative approaches [1,2]. CLL is characterized by a dynamic imbalance between the proliferation and apoptosis of leukemia cells and by the accumulation of neoplastic B lymphocytes coexpressing CD5 and CD19 antigens [3-6]. Nevertheless, the pathogenesis of CLL is still poorly understood.
IL-9 is a member of the common γ-chain family of cytokines, using this receptor in combination with the cytokine-specific receptor IL-9 receptor-α (IL-9Rα) [7]. Due to its pleiotropic functions on mast cells, IL-9 has long been recognized as an important regulator of allergic inflammation [8]. But in recent years, a resurgence of interest in IL-9 has been spurred due to an expanded identification of its receptor on various immune cells [9]. A series of observations have pointed to this cytokine as a factor promoting oncogenesis, especially lymphomagenesis [10,11]. The dysregulated expression of IL-9 can be detected in biopsies and serums from patients with Hodgkin’s disease (HD), anaplastic large cell lymphomas (ALCL) [12] as well as nasal natural killer (NK)/T-cell lymphoma [13,14]. Our previous study also demonstrated that there was an elevated serum level of IL-9 in B-cell NHL patients including some DLBCL cases. The present study is aimed to investigate the expression of IL-9 in CLL patients and to illuminate its role in the pathogenesis of CLL.
Materials and methods
Patients and samples
Blood sample from 47 patients with CLL were taken at diagnosis at Shandong Provincial Hospital between January 2010 and December 2011 who met the diagnostic criteria for CLL, while 10 healthy volunteers served as normal control. Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood obtained from 20 CLL patients. All patients were untreated and their lymphocytes exceeded 90%. PBMCs of 10 healthy blood donors were isolated by density gradient centrifugation and subjected to a preliminary phenotypic characterization. When residual non-B cells exceeded 10%, B cells were enriched by negative selection with antibody-coated magnetic beads to obtain a more than 97% pure CD19+ B-cell population. The protocol was approved by the Shandong Provincial Hospital Ethics Committee and written informed consent was obtained from all participants involved in this study.
ELISA for IL-9
Serum samples from 47 CLL patients and 10 healthy volunteers were collected and frozen at -80°C. IL-9 levels in sera were quantified using human ELISA kit (eBioscience) according to the manufacturer’s instructions. The sensitivity limit for quantitative determination was 1pg/ml.
Reverse transcription-polymerase chain reaction (PCR)
Total RNA was extracted from PBMCs of CLL patients using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Then reverse transcription reaction was conducted by means of PrimeScript reverse transcription (RT) reagent Kit (TaKaRa, Dalian, China). The reaction was incubated at 37°C for 15 minutes, and 85°C for 5 seconds. Amplification reactions were performed using SYBR Premix Ex Taq (Perfect Real Time) (TaKaRa, Dalian, China) on ABI 7500 Real-Time quantitative PCR System(Applied Biosystems, Foster City, Ca, USA) with cycling as follows: an initial cycle for 30 s at 95°C, followed by 40 biphasic cycles of 5 seconds at 95°C, 20 seconds at 60°C; Melt Curve Analysis 95°C 0 s, 65°C, 15 s; 95°C, 0 s, 0.1°C/s. PCR products were confirmed as a single product at the desired size on agarose gels and visualized by ethidium bromide staining. Specific primers for RT-PCR were obtained from Biosune (Shanghai, China), and the primer sequences are listed in Table 1. Expression data were normalized to the geometric mean of housekeeping gene β-actin to control the variability in expression levels and analyzed using the 2-ΔΔCT method.
Table 1.
the primer sequences
| Gene Name | Sequence |
|---|---|
| IL-9 | 5‘-CTCTGTTTGGGCATTCCCTCT-3’ |
| 5‘-GGGTATCTTGTTTGCATGGTGG-3’ | |
| β-actin | 5‘-CATTAAGGAGAAGCTGTGCT-3’ |
| 5‘-GTTGAAGGTAGTTTCGTGGA-3’ |
Western blot analysis
Total protein was extracted from PBMCs of CLL patients and B-cells of healthy samples using RIPA and 1% PMSF (Shenergy Biocolor, China). For cytoplasmic and nuclear extracts, cells were washed with phosphate-buffered saline (PBS) and were lysed in NEPER extraction reagent (Pierce) according to the manufacturer’s protocol. The protein concentration of the samples was determined by the BCA assay (Shenergy Biocolor). Cell lysate was then electrophoresed on 10% SDS-polyacrylamide gels and transferred onto nitrocellulose membranes. After the membranes were blocked with 5% skim milk in Tris-saline buffer with 0.1% Tween-20 (TBST), they were subsequently probed with primary antibodies at 4°C overnight. After washing with TBST, secondary antibody conjugated with the horseradish peroxidase (Zhongshan Goldenbridge Biotechnology Company, China) was added to the membranes. Antibodies used in this study included GAPDH Antibody (1:800). All other antibodies were from Abcam. Western blot results were analyzed using the Image J software (NIH, USA) and GraphPad Prime software.
Statistical analysis
All statistical analyses were performed using the SPSS version 17.0 for Windows. The numerical data were statistically analyzed by the 2-tailed Student’s t-test. Chi-square analysis and Fisher’s exact test were used to analyze the relationship between the positive IL-9 expression and prognostic factors. P<0.05 was considered to indicate a statistically significant difference.
Results
Upregulation of IL-9 in sera from CLL patients
To determine the possible involvement of IL-9 in the CLL, we measured serum levels of IL-9 in patients with CLL and healthy controls using ELISA (Figure 1). IL-9 could be detected in 20 of 47 sera from CLL patients while none serum sample from healthy volunteers contained detectable levels of IL-9. The data shows that serum IL-9 levels in patients with CLL were obviously higher than healthy controls (P=0.0083).
Figure 1.
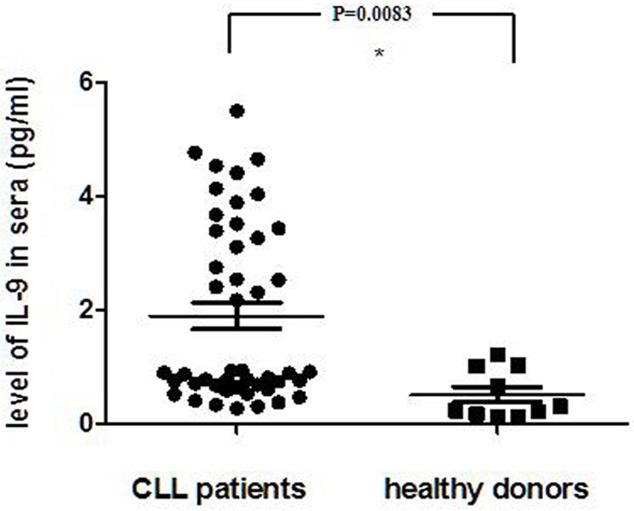
Serum level of IL-9 was detected by ELISA in serum from 47 CLL patients and 10 healthy controls. The data analyzed by 2-tailed Student’s t-test *P=0.0083.
Overexpression of IL-9 mRNA and protein in CLL patients
To confirm these data, the expressions of IL-9 mRNA and protein were determined in 20 CLL patients using RT-PCR and western blot, respectively. As illustrated in Figure 2, there is a higher expression of IL-9 mRNA in CLL patients in contrast to that in the healthy donors. In addition, the analysis on the gray scale of the Western blot bands indicated that the total protein levels of IL-9 in patients were significantly upregulated (p<0.05, Figure 3).
Figure 2.
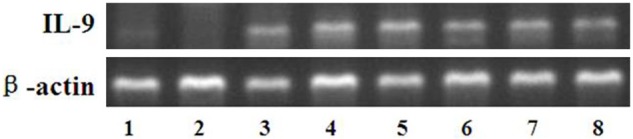
Expression of IL-9 mRNA in CLL patients (lanes 3-8) and healthy donors (lanes 1-2).
Figure 3.
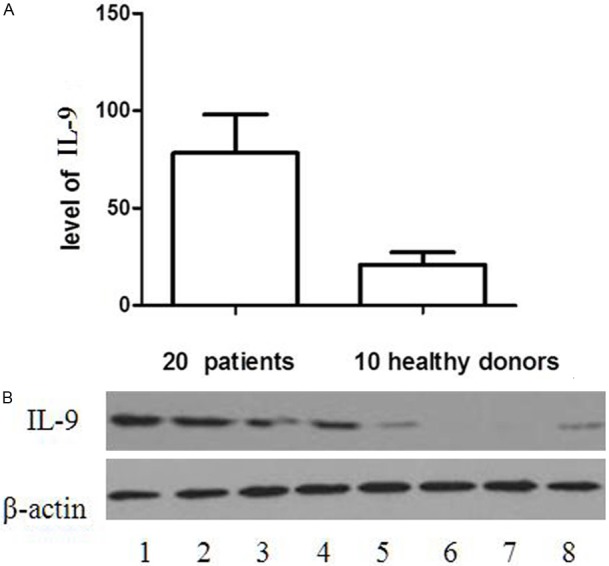
The analysis on the gray scale of the Western blot bands indicated that the total protein levels of IL-9 in CLL patients were significantly upregulated (p=0.003). A: Expression levels were normalized with β-actin. B: Expression of total IL-9 protein in CLL patients (lanes 1-4) and healthy donors (lanes 5-8).
The correlation between positive IL-9 expression and clinical characteristics
Statistical analyses were done to examine the correlation between the serum IL-9 levels by ELISA analysis and the clinical features of CLL. As shown in Table 2, there was no correlation between IL-9 expression and patient age (P=0.6580), gender (P=0.4063). Nevertheless, the expression of IL-9 was strongly correlated to ZAP-70 expression (P=0.0272), β2 microglobulin expression (P=0.0101) and IgVH status (P=0.0320). Together, these results indicated that the overexpression of IL-9 was associated with CLL clinical progression.
Table 2.
the correlation between IL-9 expression and patient characteristics
| Characteristic | IL-9 Expression | P value | |
|---|---|---|---|
|
|
|||
| Positive (%) | Negative (%) | ||
| Sex | |||
| Male | 14 (70.0) | 19 (70.4) | 0.6580 |
| Female | 6 (30.0) | 8 (29.6) | |
| Age (yr) | |||
| <60 | 7 (35.0) | 12 (44.4) | 0.4063 |
| ≥60 | 13 (65.0) | 15 (55.6) | |
| ZAP-70 expression | |||
| Positive | 4 | 0 | 0.0272* |
| Negative | 16 | 27 | |
| IgVH status | |||
| Unmutated (≥98% homology) | 14 | 26 | |
| Mutated (<98% homology) | 6 | 1 | 0.0320* |
| β2 microglobulin expression | |||
| No more than 4 mg/L | 15 | 27 | |
| more than 4 mg/L | 5 | 0 | 0.0101* |
| Ria classification | |||
| I | 1 | 6 | |
| II | 3 | 8 | |
| III | 5 | 11 | |
| IV | 11 | 2 | 0.0035 |
| Binet classification | |||
| A | 3 | 13 | |
| B | 6 | 12 | |
| C | 11 | 2 | 0.0010 |
Fisher’s exact test.
Expression of serum IL-9 correlates with Ria and Binet classification in CLL
Ria and Binet classification, the important prognostic indicators, were also associated with IL-9 expression. Chi-square analysis indicated that there was a strong association between IL-9 overexpression and Ria & Binet classification (P<0.05, Table 2).
Discussion
Interleukin-9 is able to stimulate proliferation of lymphoma cells [15] and protect them from dexamethasone (DEX)-induced apoptosis [16]. The pro-proliferative and anti-apoptotic effect of IL-9 were mainly dependent on its high-affinity binding with IL-9R. This receptor-ligand interaction activates the Janus kinase-signal transducer and activator of transcription (JAK/STAT) pathway and subsequently regulates its downstream apoptosis proteins [15,17]. Extensive studies have confirmed the oncogenic activities of IL-9 in lymphoma. Our previous study has demonstrated that there is an elevated IL-9R level in DLBCL patients [18,19]. Although abnormal expression of IL-9 has been observed in hematological malignancies, this study is the first report on the pathological significance of IL-9 in CLL.
IL-9 is a member of the common γ-chain family of cytokines, using this receptor in combination with the cytokine-specific receptor IL-9 receptor-α (IL-9Rα) [7]. Besides its role during immune responses, its growth factor and antiapoptotic activities on multiple transformed cells suggest a potential role in hematological malignancies. It can induce the proliferation of various lymphoid and hemopoietic cells. The dysregulated expression of IL-9 has been detected in biopsies or serum of patients with some hematological malignancies, such as ATL, HD, ALCL and NKT-cell lymphoma, which provides clinical evidence for its possible involvement in pathogenesis of hematological malignancies [20-22]. However, the pathogenic role of IL-9 in CLL has not been reported before. Our previous study has indicated that IL-9 participated in pathogenesis of B-cell NHL through up-regulation of immunosuppression mediated by Treg cells and mast cells. Based upon the previous study, we strive to prove that IL-9 directly take part in the development of CLL. Our findings suggest that the overexpression of IL-9 is associated with the pathogenesis of CLL. We demonstrated that IL-9 was markedly overexpressed in CLL patients compared with their counterparts.
In addition, we observed that IL-9 overexpression was associated with clinical characteristics in CLL patients. The expression of IL-9 was strongly correlated to the clinical staging of patients with CLL (P<0.05), ZAP-70 expression (P=0.0272), β2 microglobulin expression (P=0.0101) and IgVH status (P=0.0320). These factors are important adverse prognostic indicators of CLL. The correlation between IL-9 upregulation and patient characteristics provided direct clinical evidence for the contribution of IL-9 to the pathogenesis of CLL.
In summary, the results clearly indicate that IL-9 is markedly overexpressed in CLL patients compared to their counterparts. The expression intensity of IL-9 is correlated with adverse prognostic indicators of CLL in pathological sections. Our findings suggest that the signal mediated by IL-9 contributes to the pathogenesis of CLL. It helps us to get a deeper understanding about the molecular mechanism of CLL and could be served as a potentially therapeutic target for CLL patients in the future.
Acknowledgements
This study was partly supported by: National Natural Science Foundation (No. 81270598), Natural Science Foundations of Shandong Province (No. Y2007C053, No. 2009ZRB14176 and No. ZR2012HZ003), Technology Development Projects of Shandong Province (No. 2007GG10, and No. 2010GSF10250), Program of Shandong Medical Leading Talent, and Taishan Scholar Foundation of Shandong Province.
Disclosure of conflict of interest
The authors declare that they have no competing interests.
References
- 1.Negro R, Gobessi S, Longo PG, He Y, Zhang ZY, Laurenti L, Efremov DG. Overexpression of the autoimmunity-associated phosphatase PTPN-22 promotes survival of antigen-stimulated CLL cells by selectively activating AKT. Blood. 2012;119:6278–6287. doi: 10.1182/blood-2012-01-403162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu K, Wang X. Therapeutic advancement of chronic lymphocytic leukemia. J Hematol Oncol. 2012;5:55. doi: 10.1186/1756-8722-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fecteau JF, Messmer D, Zhang S, Cui B, Chen L, Kipps TJ. Impact of oxygen concentration on growth of mesenchymal stromal cells from the marrow of patients with chronic lymphocytic leukemia. Blood. 2013;121:971–974. doi: 10.1182/blood-2012-08-447813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawstron AC, Böttcher S, Letestu R, Villamor N, Fazi C, Kartsios H, de Tute RM, Shingles J, Ritgen M, Moreno C, Lin K, Pettitt AR, Kneba M, Montserrat E, Cymbalista F, Hallek M, Hillmen P, Ghia P. Improving efficiency and sensitivity: European Research Initiative in CLL (ERIC) update on the international harmonised approach for flow cytometric residual disease monitoring in CLL. Leukemia. 2013;27:142–149. doi: 10.1038/leu.2012.216. [DOI] [PubMed] [Google Scholar]
- 5.Samuel S, Beljanski V, Van Grevenynghe J, Richards S, Ben Yebdri F, He Z, Nichols C, Belgnaoui SM, Steel C, Goulet ML, Shamy A, Brown D, Abesada G, Haddad EK, Hiscott J. BCL-2 Inhibitors Sensitize Therapy-resistant Chronic Lymphocytic Leukemia Cells to VSV Oncolysis. Mol Ther. 2013;21:1413–1423. doi: 10.1038/mt.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang C, Zhuang Y, Wang L, Fan L, Wu YJ, Zhang R, Zou ZJ, Zhang LN, Yang S, Xu W, Li JY. High levels of CD20 expression predict good prognosis in chronic lymphocytic leukemia. Cancer Sci. 2013;104:996–1001. doi: 10.1111/cas.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornakova T, Staerk J, Royer Y, Flex E, Tartaglia M, Constantinescu SN, Knoops L, Renauld JC. Acute lymphoblastic leukemia-associated JAK1 mutants activate the Janus kinase/STAT pathway viainterleukin-9 receptor alpha homodimers. J Biol Chem. 2009;284:6773–6781. doi: 10.1074/jbc.M807531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiener Z, Falus A, Toth S. IL-9 increases the expression of several cytokines in activated mast cells, while the IL-9-induced IL-9 production is inhibited in mast cells of histamine-free transgenic mice. Cytokine. 2004;26:122–130. doi: 10.1016/j.cyto.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Noelle RJ, Nowak EC. Cellular sources and immune functions of interleukin-9. Nat Rev Immunol. 2010;10:683–687. doi: 10.1038/nri2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bittner C, Merz H, Krokowski M, Briese J, Wiedemann GJ, Feller AC. [New immunotherapeutic approaches for the treatment of anaplastic large cell lymphoma in a mouse model] . Verh Dtsch Ges Pathol. 2000;84:187–198. [PubMed] [Google Scholar]
- 11.Renauld JC, van der Lugt N, Vink A, van Roon M, Godfraind C, Warnier G, Merz H, Feller A, Berns A, Van Snick J. Thymic lymphomas in interleukin 9 transgenic mice. Oncogene. 1994;9:1327–1332. [PubMed] [Google Scholar]
- 12.Merz H, Houssiau FA, Orscheschek K, Renauld JC, Fliedner A, Herin M, Noel H, Kadin M, Mueller-Hermelink HK, Van Snick J, et al. Interleukin-9 expression in human malignant lymphomas: unique association with Hodgkin’s disease and large cell anaplastic lymphoma. Blood. 1991;78:1311–1317. [PubMed] [Google Scholar]
- 13.Nagato T, Kobayashi H, Kishibe K, Takahara M, Ogino T, Ishii H, Oikawa K, Aoki N, Sato K, Kimura S, Shimizu N, Tateno M, Harabuchi Y. Expression of interleukin-9 in nasal natural killer/T-cell lymphoma cell lines and patients. Clin Cancer Res. 2005;11:8250–8257. doi: 10.1158/1078-0432.CCR-05-1426. [DOI] [PubMed] [Google Scholar]
- 14.Fischer M, Bijman M, Molin D, Cormont F, Uyttenhove C, van Snick J, Sundstrom C, Enblad G, Nilsson G. Increased serum levels of interleukin-9 correlate to negative prognostic factors in Hodgkin’s lymphoma. Leukemia. 2003;17:2513–2516. doi: 10.1038/sj.leu.2403123. [DOI] [PubMed] [Google Scholar]
- 15.Jaffe ES, Harris NL, Stein H, Isaacson PG. Classification of lymphoid neoplasms: the microscope as a tool for disease discovery. Blood. 2008;112:4384–4399. doi: 10.1182/blood-2008-07-077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu L, Lai R, Lin Q, Lau E, Thomazy DM, Calame D, Ford RJ, Kwak LW, Kirken RA, Amin HM. Autocrine release of interleukin-9 promotes Jak3-dependent survival of ALK+ anaplastic large-cell lymphoma cells. Blood. 2006;108:2407–2415. doi: 10.1182/blood-2006-04-020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renauld JC, Vink A, Louahed J, Van Snick J. Interleukin-9 is a major anti-apoptotic factor for thymic lymphomas. Blood. 1995;85:1300–1305. [PubMed] [Google Scholar]
- 18.Feng LL, Gao JM, Li PP, Wang X. IL-9 contributes to immunosuppression mediated by regulatory T cells and mast cells in B-cell non-hodgkin’s lymphoma. J Clin Immunol. 2011;31:1084–1094. doi: 10.1007/s10875-011-9584-9. [DOI] [PubMed] [Google Scholar]
- 19.Lv X, Feng L, Fang X, Jiang Y, Wang X. Overexpression of IL-9 receptor in diffuse large B-cell lymphoma. Int J Clin Exp Pathol. 2013;6:911–916. [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Petrus M, Bryant BR, Phuc Nguyen V, Stamer M, Goldman CK, Bamford R, Morris JC, Janik JE, Waldmann TA. Induction of the IL-9 gene by HTLV-I Tax stimulates the spontaneous proliferation of primary adult T-cell leukemia cells by a paracrine mechanism. Blood. 2008;111:5163–5172. doi: 10.1182/blood-2007-09-113654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsushita K, Arima N, Ohtsubo H, Fujiwara H, Hidaka S, Fukumori J, Tanaka H. Frequent expression of interleukin-9 mRNA and infrequent involvement of interleukin-9 in proliferation of primary adult T-cell leukemia cells and HTLV-I infected T-cell lines. Leuk Res. 1997;21:211–216. doi: 10.1016/s0145-2126(96)00109-9. [DOI] [PubMed] [Google Scholar]
- 22.Ju W, Zhang M, Jiang JK, Thomas CJ, Oh U, Bryant BR, Chen J, Sato N, Tagaya Y, Morris JC, Janik JE, Jacobson S, Waldmann TA. CP-690,550, a therapeutic agent, inhibits cytokine-mediated Jak3 activation and proliferation of T cells from patients with ATL and HAM/TSP. Blood. 2011;117:1938–1946. doi: 10.1182/blood-2010-09-305425. [DOI] [PMC free article] [PubMed] [Google Scholar]


