Abstract
The cell division cycle 20 homolog (CDC20) expression is increased in diverse human cancers and plays a vital role in tumorigenesis and progression. However, the clinical significance of CDC20 expression in gastric cancer (GC) remains largely unknown. The aim of this study was to investigate the clinicopathologic features and prognostic significance of CDC20 in GC. The CDC20 mRNA expression was measured by quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR). Immunohistochemistry (IHC) was used to detect the expression of CDC20 protein in 131 clinicopathologically characterized GC cases. The relationship between CDC20 expression and clinicopathological features was analyzed by appropriate statistics. Kaplan-Meier analysis and Cox proportional hazards regression models were used to investigate the correlation between CDC20 expression and prognosis of GC patients. The relative mRNA expression of CDC20 were significantly higher in GC tumor tissues than in the corresponding noncancerous tissues (P<0.001). Simultaneously, CDC20 protein expression was positively correlated with tumor size (P=0.02), histological grade (P=0.037), lymph node involvement (P=0.009), and TNM stage (P=0.015). Furthermore, Kaplan-Meier analysis indicated that patients with high CDC20 expression had poor overall survival (P<0.001). Multivariate analysis showed that high CDC20 expression was an independent predictor of overall survival. In conclusion, our data indicated that CDC20 upregulation was associated with aggressive progression and poor prognosis in GC. CDC20 was identified for the first time as an independent marker for predicting the clinical outcome of GC patients.
Keywords: CDC20, immunohistochemistry, gastric cancer
Introduction
Gastric cancer (GC) is the fourth most prevalent forms of human cancers and the second leading cause of cancer-related death in the world, especially in East Asian countries. Its incidence rate is 20 per 100,000 annually [1]. Nowadays, gastrectomy remains the mainstay treatment for GC, but the prognosis for advanced stage patients is still very poor. The median survival time for patients with GC is only 6-9 months [2]. In China, the 5-year overall survival rate of patients with GC is only about 40%, although recent advances in chemotherapy and surgical techniques [3]. This is primarily attributed to the following reasons: lack of diagnostic markers for early detection, weak prognostic value of histological indicators, limited efficiency of current treatment for advanced disease and lack of molecular markers utilized for targeted therapy [4-6]. Therefore, it is of great significance to make a better understanding of gastric carcinogenesis and to identify novel molecular markers for the improvement of clinical management of patients with GC.
The cell division cycle 20 homolog (CDC20) is a regulatory protein that is a target molecule in the cell-cycle checkpoint [7]. CDC20 [7] is a component of the mammalian cell cycle mechanism that activates the anaphase-promoting complex (APC). Its expression is essential for cell division, and its protein activity may be controlled by a balance between ubiquitination and de-ubiquitination. APC activation is required for anaphase initiation and mitosis exit. An abnormal level or dysfunction of CDC20 may therefore abolish mitotic arrest and thus promote premature anaphase by deregulating APC activation, resulting in aneuploidy in the daughter cells [8]. Interestingly, some researchers have shown that CDC20 might also play a role in human carcinogenesis [9-13]. Overexpression of CDC20 protein has now been strongly linked to poor prognosis in cancer, such as colorectal cancer [9], pancreatic cancer [10] and non-small cell lung cancer [11]. However, no information is available regarding CDC20 expression in human GC. To explore the vital role of CDC20 in the tumorigenesis and progression of GC, we examined expression patterns of CDC20 in GC tissues, analyzed the relationship between CDC20 expression and clinicopathological factors of GC.
Materials and methods
Patients and specimens
For qRT-PCR analysis, we collected 27 paired fresh GC tumor tissue samples and corresponding noncancerous tissue samples from patients who underwent surgery between October 2011 and April 2012. In addition, a cohort of 131 formalin-fixed, paraffin-embedded tissues of GC diagnosed between 2002 and 2007 at the Department of General Surgery, The Second Affiliated Hospital of Soochow University was retrieved. The cases selected were based on distinctive pathologic diagnosis of GC, undergoing curative resection for tumor without preoperative chemotherapy and radiotherapy, and availability of resection tissue and follow-up data. The histopathological type and the stage of GC were determined according to the criteria of the World Health Organization classification. The 131 patients included 77 males and 54 females aged from 32 to 85 years (median, 75 years). The clinicopathological characteristics of these 131 patients are summarized in Table 1. The patients’ consent was obtained for the use of the tissue samples and records, and the study protocol was approved and permission for use of the clinical data was given by the Institutional Research Ethics Committee of The Second Affiliated Hospital of Soochow University.
Table 1.
Correlation between CDC20 expression and clinicopathological characteristics of GC patients
| Characteristics | Total cases (n=131) | CDC20 protein expression | P value | |
|---|---|---|---|---|
|
|
||||
| High (n=68) | Low (n=63) | |||
| Age (years) | ||||
| <60 | 47 | 26 | 21 | 0.559 |
| ≥60 | 84 | 42 | 42 | |
| Gender | ||||
| Male | 77 | 41 | 36 | 0.714 |
| Female | 54 | 27 | 27 | |
| Tumor size (cm) | ||||
| ≤5 | 59 | 24 | 35 | 0.02 |
| >5 | 72 | 44 | 28 | |
| Histological grade | ||||
| Well differentiated (G1) | 12 | 2 | 10 | 0.037 |
| Moderately differentiated (G2) | 67 | 37 | 30 | |
| Poorly differentiated (G3) | 52 | 29 | 23 | |
| Lymph node involvement | ||||
| 0 | 8 | 0 | 8 | 0.009 |
| 1 | 31 | 16 | 15 | |
| >1 | 92 | 52 | 40 | |
| TNM stage | ||||
| I+II | 35 | 12 | 23 | 0.015 |
| III+IV | 96 | 56 | 40 | |
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis
Total RNA was isolated from the 27 pairs of GC tumor tissue samples and corresponding noncancerous tissue samples using TRIZOL reagent (Invitrogen, Carlsbad, CA). RNA was reverse-transcribed using SuperScript First Strand cDNA System (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
The CDC20 sense primer was 5’-GGCACCAGTG-ATCGACACATTCGCAT-3’, and the antisense primer was 5’-GCCATAGCCTCAGGGTCTCATCTG-CT-3’. For the β-actin gene, the sense primer was 5’-ATAGCACAGCCTGGATAGCAACGTAC-3’, and the antisense primer was 5’-CACCTTCTAC-AATGAGCTGCGTGTG-3’. qRT-PCR was done using SYBR Green PCR master mix (Applied Biosystems) in a total volume of 20 µl on the 7900HT fast Real-time PCR system (Applied Biosystems) as follows: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 60 s. A dissociation procedure was performed to generate a melting curve for confirmation of amplification specificity. β-actin was used as the reference gene. The relative levels of gene expression were represented as ΔCt=Ctgene-Ctreference, and the fold change of gene expression was calculated by the 2-ΔΔCt Method. Experiments were repeated in triplicate.
Immunohistochemistry (IHC)
All specimens were fixed with 4% formaldehyde, dewaxed, embedded and cut into 4 μm serial sections. Briefly, antigen retrieval was carried out in 10 mmol/l citrate buffer (pH 6.0) for 15 minutes at 100°C in a microwave oven. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 10 minutes at room temperature. The sections were then incubated overnight at 4°C with anti-CDC20 antibody (Santa Cruz Biotechnology). After washing with PBS, sections were incubated with secondary antibodies for 30 minutes at 37°C. The sections were then washed three times with PBS and treated with 3, 3’-diaminobenzedine (DAB) for approximately 5 minutes. Finally, the sections were counterstained with hematoxylin, dehydrated, mounted and examined by light microscopy. Negative controls were probed with PBS under the same experimental conditions.
Immunohistochemical staining was assessed by two independent experienced pathologists who were blinded to all clinicopathological features. Five high power fields in each specimen were randomly selected. A modified immunoreactivity score method to evaluate the immunostaining results was performed by multiplying stain intensity by stain area (staining index [SI]). Stain intensity is as follows: no staining (score 0), weak staining (score 1), moderate staining (score 2), or strong staining (score 3). Staining area is as follows: less than 25% (score 1), 25% to 50% (score 2), 50% to 75% (score 3), or more than 75% (score 4) of tumor cells. The expression levels of CDC20 were determined by the SI, which scores as 0, 1, 2, 3, 4, 6, 8, 9, and 12. An optimal cutoff value was identified as follows: the SI score of 4 or greater was used to define tumors as high CDC20 expression and the SI score of 3 or less as low CDC20 expression.
Statistical analysis
The data are expressed as the mean ± SD. The Student’s t test and one-way analysis of variance test were used to compare data between the different groups. The χ2 test was used to assess the correlations between CDC20 protein expression and clinicopathologic factors. Survival curves were calculated by the Kaplan-Meier method, and the significance of intergroup differences in survival was determined by log-rank test. Multivariate survival analysis based on the Cox proportional hazard model was carried out to identify the significant independent prognostic factors. A P value <0.05 was considered statistically significant.
Results
Expression of CDC20 mRNA by qRT-PCR in GC tissues
Our qRT-PCR results showed that CDC20 mRNA expression was upregulated in 21 of the 27 GC samples compared with the paired noncancerous tissues. The mean expression value of CDC20 mRNA in cancer tissues was significantly higher than the value in relevant normal tissues (2.63 ± 0.84 vs 0.98 ± 0.28, P<0.001, Figure 1).
Figure 1.
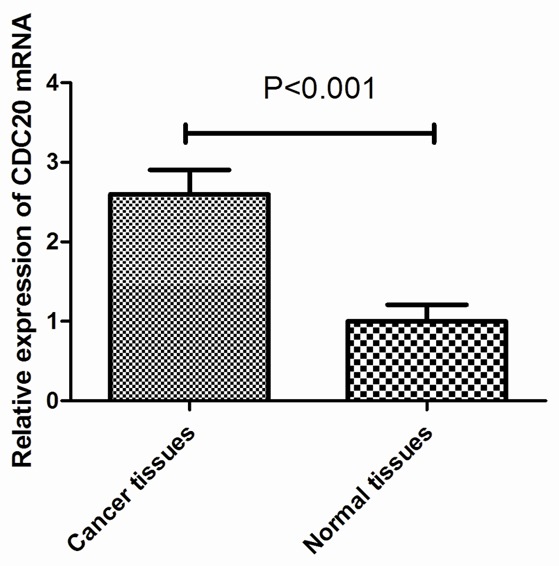
Upregulation of CDC20 mRNA expression in GC tissues. qRT-PCR showed that the mean expression value of CDC20 mRNA in cancer tissues was significantly higher than the value in the corresponding noncancerous tissues (2.63 ± 0.84 vs 0.98 ± 0.28, P<0.001).
Correlation of CDC20 expression with clinicopathologic factors
Immunohistochemistry was performed in 131 archived paraffin-embedded GC samples and 60 adjacent noncancerous samples. Overall, 51.9% (68/131) of the GC tissues showed high CDC20 expression, and only 18.3% (11/60) of the adjacent noncancerous tissues showed high CDC20 expression. The difference in CDC20 staining between the GC and adjacent normal tissues was statistically significant (P<0.05). Figure 2 shows representative cases with different expression levels of CDC20 protein. Moreover, as shown in Table 1, CDC20 protein expression was positively correlated with tumor size (P=0.02), histological grade (P=0.037), lymph node involvement (P=0.009), and TNM stage (P=0.015); however, no significant associations were detected for CDC20 expression with patient sex and age (all P>0.05).
Figure 2.
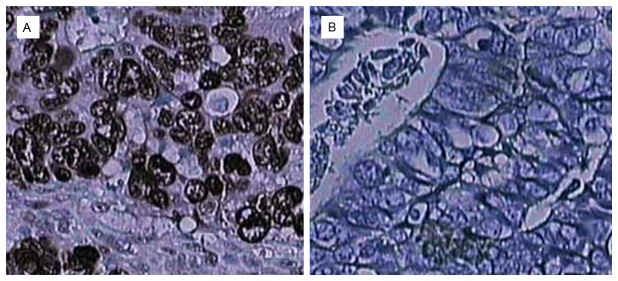
Immunohistochemical staining of CDC20 in GC specimens. A: High CDC20 expression. B: Low CDC20 expression.
Prognostic significance of CDC20 expression in GC
To investigate the prognostic value of CDC20 for GC, we assessed the association between CDC20 expression and survival duration using a Kaplan-Meier analysis with a log-rank test. Kaplan-Meier analysis indicated that CDC20 expression was significantly associated to the survival of GC patients. The overall 5-year survival rate of patients with high CDC20 expression was significantly shorter than those with low CDC20 expression (P<0.001; Figure 3). Thus, the expression of CDC20 protein could affect the prognosis of GC patients. To evaluate the possibility of CDC20 used as an independent risk factor for poor prognosis, conventional clinicopathological factors and CDC20 protein levels were assessed by Cox’s univariate and multivariate hazard regression model (Table 2). Univariate analysis indicated that tumor size, histological grade, lymph node involvement, TNM stage and CDC20 protein expression were significantly associated with overall survival of GC patients. By multivariate analysis, we showed that CDC20 protein expression, together with tumor size, histological grade, lymph node involvement, and TNM stage was an independent prognostic factor for overall survival of GC patients.
Figure 3.
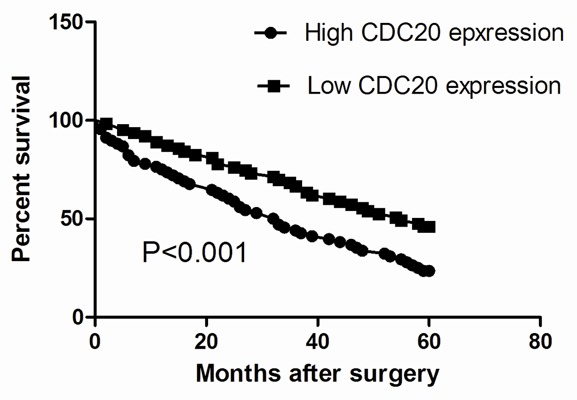
Kaplan-Meier survival curves of overall survival in GC patients based on CDC20 expression. Patients with high CDC20 expression had poorer overall survival rate as compared to those with low CDC20 expression.
Table 2.
Univariate analysis and multivariate analysis identifies factors influencing the overall survival rate of GC patients
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| CDC20 expression | 1.473 | 0.785-2.213 | <0.001 | 1.508 | 0.673-2.302 | <0.001 |
| Age | 1.395 | 0.698-2.163 | 0.591 | 1.406 | 0.711-2.223 | 0.634 |
| Gender | 1.506 | 0.803-2.376 | 0.615 | 1.386 | 0.527-2.061 | 0.586 |
| Tumor size | 1.442 | 0.705-2.114 | 0.032 | 1.335 | 0.503-2.116 | 0.028 |
| Histologic grade | 1.394 | 0.604-2.252 | 0.011 | 1.422 | 0.703-2.012 | 0.017 |
| Lymph node involvement | 1.364 | 0.756-2.253 | 0.007 | 1.198 | 0.687-2.068 | 0.008 |
| Tumor stage | 1.477 | 0.824-2.382 | 0.012 | 1.386 | 0.705-2.213 | 0.009 |
Discussion
Surgical resection is the most effective treatment for GC patients with curative potential, yet the clinical outcomes of these patients are generally poor, largely due to a high ratio of postoperative metastasis or recurrence [14]. In China, the overall 5-year survival rate of GC patients is only about 40% [15]. Multi-modal and individualized treatments are often based on the TNM staging status. However, the treatment res-ponse and prognosis vary in GC patients of the same stage using the same therapeutic strategy. Therefore, finding a suitable marker to predicate the prognosis of GC is necessary. Aberrant expression of CDC20 has been detected in several types of human malignant tumors and closely associated with oncogenic transformation and tumor progression [9-13]. However, the expression of CDC20 in the development and progression of GC are not clear.
Our findings confirmed the remarkable up-regulation of CDC20 in GC tissues and indicated significant associations for CDC20 expression with prognosis-related features, including that tumor size, histological grade, lymph node involvement, and TNM stage. Thus, it is likely that CDC20 plays important roles in GC progression. Consistent with our results, previous study showed that overexpression of CDC20 is strongly associated with progression in diverse human cancers, such as colorectal cancer [9], pancreatic cancer [10] and non-small cell lung cancer [11]. Predicting the clinical outcomes of patients with malignant tumors may provide valuable information for better treatment stratification and personalized therapeutic regimens. However, no clinical data has indicated the potential benefit of CDC20 as a biomarker for predicting prognosis of GC patients. In the current study, multivariate survival analysis based on the Cox proportional hazard model revealed that, among all the factors analyzed, CDC20 expression was a significant independent prognostic factor for overall survival of GC patients following curative resection. These findings clearly demonstrate that aberrant expression of CDC20 may be closely associated with GC progression in our patient cohort, and suggest its potential as a prognostic biomarker of GC outcome.
In conclusion, our results indicate that high CDC20 expression in GC may be important for tumor progression and thus serves as an independent biomarker for poor survival. Therefore, high CDC20 expression identifies high-risk patients and is a potential novel therapeutic target for GC.
Acknowledgements
This work was supported by the Program of Health Department of Jiangsu Provincea (No. Z201311).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Zhang YZ, Zhang LH, Gao Y, Li CH, Jia SQ, Liu N, Cheng F, Niu DY, Cho WC, Ji JF, Zeng CQ. Discovery and validation of prognostic markers in gastric cancer by genome-wide expression profiling. World J Gastroenterol. 2011;17:1710–7. doi: 10.3748/wjg.v17.i13.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia YF, Xiao DJ, Ma XL, Song YY, Hu R, Kong Y, Zheng Y, Han SY, Hong RL, Wang YS. Differentiated embryonic chondrocyte-expressed gene 1 is associated with hypoxia-inducible factor 1α and Ki67 in human gastric cancer. Diagn Pathol. 2013;8:37. doi: 10.1186/1746-1596-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin J, Jin T, Quan M, Piao Y, Lin Z. Ezrin overexpression predicts the poor prognosis of gastric adenocarcinoma. Diagn Pathol. 2012;7:135. doi: 10.1186/1746-1596-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sotoudeh K, Hashemi F, Madjd Z, Sadeghipour A, Molanaei S, Kalantary E. The clinicopathologic association of c-MET overexpression in Iranian gastric carcinomas; an immunohistochemical study of tissue microarrays. Diagn Pathol. 2012;7:57. doi: 10.1186/1746-1596-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinstein J, Jacobsen FW, Hsu-Chen J, Wu T, Baum LG. A novel mammalian protein, p55CDC, present in dividing cells is associated with protein kinase activity and has homology to the Saccharomyces cerevisiae cell division cycle proteins Cdc20 and Cdc4. Mol Cell Biol. 1994;14:3350–63. doi: 10.1128/mcb.14.5.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–41. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 9.Wu WJ, Hu KS, Wang DS, Zeng ZL, Zhang DS, Chen DL, Bai L, Xu RH. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J Transl Med. 2013;11:142. doi: 10.1186/1479-5876-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu WJ, Hu KS, Wang DS, Zeng ZL, Zhang DS, Chen DL, Bai L, Xu RH. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J Transl Med. 2013;11:142. doi: 10.1186/1479-5876-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kaji M. Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J Surg Oncol. 2012;106:423–30. doi: 10.1002/jso.23109. [DOI] [PubMed] [Google Scholar]
- 12.Bie L, Zhao G, Cheng P, Rondeau G, Porwollik S, Ju Y, Xia XQ, McClelland M. The accuracy of survival time prediction for patients with glioma is improved by measuring mitotic spindle checkpoint gene expression. PLoS One. 2011;6:e25631. doi: 10.1371/journal.pone.0025631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mondal G, Sengupta S, Panda CK, Gollin SM, Saunders WS, Roychoudhury S. Overexpression of Cdc20 leads to impairment of the spindle assembly checkpoint and aneuploidization in oral cancer. Carcinogenesis. 2007;28:81–92. doi: 10.1093/carcin/bgl100. [DOI] [PubMed] [Google Scholar]
- 14.Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, Van Cutsem E. Gastric cancer. Crit Rev Oncol Hematol. 2009;71:127–64. doi: 10.1016/j.critrevonc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Zhao ZS, Wang YY, Chu YQ, Ye ZY, Tao HQ. SPARC is associated with gastric cancer progression and poor survival of patients. Clin Cancer Res. 2010;16:260–8. doi: 10.1158/1078-0432.CCR-09-1247. [DOI] [PubMed] [Google Scholar]


