Abstract
Background
Brassica juncea is an economically important vegetable crop in China, oil crop in India, condiment crop in Europe and selected for canola quality recently in Canada and Australia. B. juncea (2n = 36, AABB) is an allotetraploid derived from interspecific hybridization between B. rapa (2n = 20, AA) and B. nigra (2n = 16, BB), followed by spontaneous chromosome doubling.
Results
Comparative genome analysis by genome survey sequence (GSS) of allopolyploid B. juncea with B. rapa was carried out based on high-throughput sequencing approaches. Over 28.35 Gb of GSS data were used for comparative analysis of B. juncea and B. rapa, producing 45.93% reads mapping to the B. rapa genome with a high ratio of single-end reads. Mapping data suggested more structure variation (SV) in the B. juncea genome than in B. rapa. We detected 2,921,310 single nucleotide polymorphisms (SNPs) with high heterozygosity and 113,368 SVs, including 1-3 bp Indels, between B. juncea and B. rapa. Non-synonymous polymorphisms in glucosinolate biosynthesis genes may account for differences in glucosinolate biosynthesis and glucosinolate components between B. juncea and B. rapa. Furthermore, we identified distinctive vernalization-dependent and photoperiod-dependent flowering pathways coexisting in allopolyploid B. juncea, suggesting contribution of these pathways to adaptation for survival during polyploidization.
Conclusions
Taken together, we proposed that polyploidization has allowed for accelerated evolution of the glucosinolate biosynthesis and flowering pathways in B. juncea that likely permit the phenotypic variation observed in the crop.
Keywords: Brassica juncea, Comparative genome analysis, Flowering pathway, Genome survey sequencing, Glucosinolate biosynthesis
Background
The Brassicaceae family includes approximately 3,700 species in 350 genera with diverse characteristics, many of which are of agronomic importance as vegetables, condiments, fodder and oil crops [1]. The genus Brassica contains the majority of crop species of Brassicaceae family. Of particular importance are the cole crop and vegetable species B. rapa, B. oleracea, B. napus, and B. juncea as sources of oils and vegetables. Because of their agricultural importance, genome components of several Brassica species have been characterized in detail over the past few years [2-4]. The genomes of three diploid species, B. rapa (AA, 2n = 20), B. nigra (BB, 2n = 16), and B. oleracea (CC, 2n = 18), have been shown to contain triplicate homologous counterparts of corresponding segments in the Arabidopsis genome due to whole-genome triplication that occurred approximately 12–17 million years ago [1,5]. Additional natural allopolyploidization events in the last 10,000 years, have resulted in the creation of three allotetraploid hybrids, B. juncea (AABB, 2n = 36), B. napus (AACC, 2n = 38) and B. carinata (BBCC, 2n = 34) [6-10]. B. juncea is used as a vegetable in China and Southeastern Asia, and is a source of oil in India and Europe. The species possesses unique traits that include much wider morphological variation in leafy types, root type, stem type, seed stalk type and oil type [11]. B. juncea has been reported to contain higher glucosinolates than other Brassica species [12]. Glucosinolates are of higher value to human nutrition that may reduce the risk of cancer incidence. In addition, they are toxic to some soil-borne plant pathogens, hence, accounting for their selection [13,14].
The recent accomplishment of genome sequencing and annotation of B. rapa[5], combined with the available genome sequence data for model Arabidopsis in Brassicaceae[15], provide improved strategies for comparative genome analysis and breeding. Attempts to develop a unified comparative genomics system in the Brassicaceae have revealed 24 conserved genomic blocks [4], an extension to the 21 syntenic blocks identified in B. napus[16]. Comparative mapping studies between members of Brassica and Arabidopsis thaliana[16-22], and Arabidopsis thaliana and Capsella rubella[23], together with the identification of an ancestral karyotype (AK) [24], have stimulated interest in the evolutionary processes underlying diversification in the Brassicaceae. Since the allotetraploid species possess much larger genomes than their diploid counterparts in Brassica[2], we expect that novel gene/pathway interactions have emerged in the allotetraploid Brassica species through sub-functionalization and/or neo-functionalization of paralogs [25,26].
Low coverage genome survey sequences (GSS) can provide information about gene content, polymorphism, functional elements, repetitive elements and molecular markers [27-31]. In some studies, most of the coding sequence in a genome can be surveyed with less than 2 genome coverage [32]. It was possible to recover 38% of the coding fraction of the mouse-human alignment with only 0.66 × coverage of the pig genome [33]. With only 0.1 × coverage, it was possible to generate a considerable amount of biologically useful information and genomic resources for Megaselia scalaris, including identification of repetitive elements, the mitochondrial genome, microsatellites and identification of gene homologs [34]. These studies make a compelling case for low density sequencing in the genomic studies of non-model species.
Here, we employed high-throughput sequencing for comparative genome analysis of B. juncea and B. rapa to identify genome changes associated with polyploidization that might account for the phenotypic diversity of B. juncea. We showed clues of glucosinolate biosynthesis and flowering pathway evolution occurred in Brassica juncea, likely accounting for some of the phenotypic diversity that is observed. Furthermore, it provides a valuable resource for more focused investigations into the rate and distribution of genomic changes that accompany polyploidization in this species.
Results
Karyotype of B. juncea
According to the 'U-triangle’ theory of Brassicaceae[6], allotetraploid B. juncea originated from hybridization of B. rapa (AA, 2n = 20) and B. nigra (BB, 2n = 16). We identified genomic components of B. juncea by genomic in situ hybridization (GISH). The two predicted genomes (A and B) of the allotetraploid were distinguished using genomic DNA from B. rapa and B. nigra as probes representing the putative progenitor genomes. The 20 A and 16 B chromosomes detected suggest that the two genomes have remained somewhat distinct in B. juncea with no significant genome homogenization and no large-scale translocations between genomes (Figure 1).
Figure 1.

Genomic in situ hybridization analysis of genome component in B. juncea. Metaphase chromosome from root tip cell of B. juncea(A), detection of B genome chromosome in B. juncea chromosome (B), detection of A genome chromosome in B. juncea chromosome (C), A and B genomes with red and green fluorescence in B. juncea(D). Bar = 5 μm.
Comparative genome analysis of B. juncea and B. rapa
After quality evaluation of sequencing data (Additional file 1: Figure S1), a total of 28.35 Gb high quality data were collected for the B. juncea genome and used to compare with whole genome sequence of B. rapa. It was feasible to map 45.93% sequences of the B. juncea GSS data to the genome sequences of B. rapa. Of these, only 18.44% single-end reads were mapped to the genome sequences of B. rapa, which indicated more SV in the B. juncea genome compared to B. rapa. The identity of mapped sequences is 98.14%, which shows a close genetic relationship between B. juncea and B. rapa (Additional file 1: Table S1). The coverage depth and distribution on chromosomes suggest a high comparison ratio over the B. rapa genome (Additional file 1: Figure S2).
Polymorphism analysis identified 2,921,310 SNPs, including 58.53% transitions, 41.47% transversions and 58.19% heterozygosity. We showed the distributions of SNP-type in 10 chromosomes of B. rapa genome (Additional file 1: Table S2, Additional file 1: Figure S3). 44,053 SVs were detected as insertions and deletions, with approximately even distributions of SVs across the 10 chromosomes of B. rapa genome (Additional file 1: Table S3, Additional file 1: Figure S3). 69,315 Indel (1–3 bp) polymorphisms were also observed, of which 1 bp-sized Indels were most abundant in genome and 3 bp-sized Indels were most abundant in coding sequence (Additional file 1: Table S4, Additional file 1: Figure S3). Most SNPs and SVs (including 1–3 bp Indels) were located in exon, intron, transposon, intergenic, TEprotein, TandemRepeat region of genome, others were found in miRNA, tRNA and snRNA coding regions of genome (Table 1). These SNPs cause a relatively high ratio of non-synonymous mutations in genes; for example, 9680 genes were found with (≧10) non-synonymous SNPs. Moreover, 1448 genes coding regions were changed by frame-shift Indels, and we also found 5989 genes have SV within gene coding regions (Table 2). A number of gene functions were found to be altered by these mutations based on Non-Redundant Nucleotide Database (NT/NR), Cluster of Othologues Groups Proteins Database (COG) and Kyoto Encyclopedia of Genes and Genomes Database (KEGG) database searches (data not shown). Here, we have focused on glucosinolate biosynthesis and flowering pathways in particular.
Table 1.
Distribution of SNPs and SVs polymorphisms in genomic components in B . juncea
| Type | Exon | Intron | miRNA | tRNA | snRNA | TEprotein | Transposon | TandemRepeat | Intergenic |
|---|---|---|---|---|---|---|---|---|---|
|
SNP |
912059 |
276795 |
393 |
789 |
671 |
88942 |
629793 |
59754 |
952114 |
|
SV* |
3769 |
9038 |
14 |
33 |
51 |
2374 |
11901 |
2896 |
13977 |
| Indel* | 3294 | 17253 | 11 | 21 | 19 | 2314 | 15313 | 1459 | 29631 |
SV*: >3 bp.
Indel*: 1–3 bp.
Table 2.
Statistics of non-synonymous mutations by SNPs, genes with Frame-shift by Indels and genes with SVs in B . juncea
| Genes with Non-synonymous SNPs: total detected SNPs | Genes with( > =10) Non-synonymous SNPs: total detected SNPs | Genes with Frame-shift Indels: total detected Indel | Genes with SVs: total detected SVs |
|---|---|---|---|
| 35457: 278241 | 9608: 166533 | 1448: 1528 | 5989: 7059 |
Glucosinolate biosynthesis genes expression between B. juncea and B. rapa
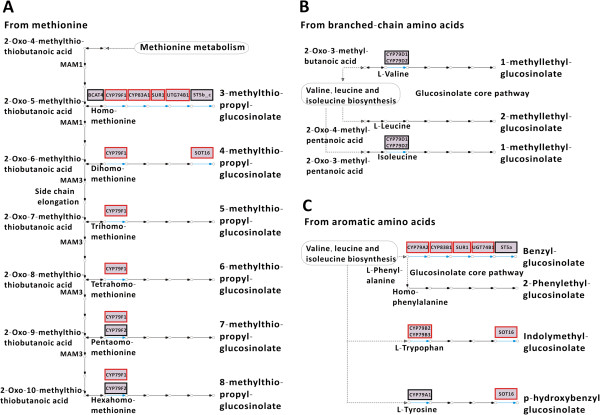
We constructed glucosinolate biosynthesis pathway in B. juncea by KEGG analysis. Three biosynthesis pathways were identified from different substrates including methionine, branched-chain amino acid and aromatic amino acid (Figure 2). Among glucosinolate biosynthesis-related genes, we found non-synonymous SNPs and deletion/insertion SV polymorphisms in CYP79F1 (CYP, cytochromes P450), CYP83A1, SUR1 (SUPERROOT1), UGT74B1 (UDP-glucose:thiohydroximate S-glucosyltransferase), SOT16 (sulfotransferase), CYP79A2, CYP83B1, CYP79B2 and CYP79B3 genes (Additional file 1: Table S5), which suggested different genes expressions and glucosinolate components and contents. Gene expression of 6 selected glucosinolate biosynthesis-related genes were investigated in leaves between B. juncea and B. rapa. CYP83A1, CYP79A2 and CYP79F1 expressions were up-regulated in B. juncea than B. rapa. CYP83B1 expression was down-regulated in B. juncea than B. rapa. There was no difference in CYP79B2 and SUR1 expressions between B. juncea and B. rapa (Figure 3). These mutations appear to cause differences in gene expression and glucosinolate content between B. juncea and B. rapa.
Figure 2.
The three glucosinolate biosynthesis pathway in B. juncea by KEGG analysis. Glucosinolate biosynthesis from methionine (A), glucosinolate biosynthesis from branched-chain amino acids (B) and glucosinolate biosynthesis from aromatic amino acid (C). The red frames show polymorphic genes from the non-synonymous polymorphism compared to B. rapa.
Figure 3.
Transcriptional patterns of glucosinolate biosynthesis related genes in B . juncea and B . rapa .
Glucosinolate component and content between B. juncea and B. rapa
We checked glucosinolate component and content between B. juncea and B. rapa by HPLC. Of glucosinolate component, sinigrin, gluconapin, glucobrassicanapin, glucobrassicin and 4-Methoxy glucobrassicin were detected in young leaves of B. juncea, of which sinigrin showed very high content with 19.58 μ mol/g DW in leaves. Only glucobrassicin, 4-Methoxy glucobrassicin and neoglucobrassicin were detected in young leaves of B. rapa (Figure 4).
Figure 4.
Glucosinolates components and contents in B . juncea and B . rapa
The flowering pathway in Brassica juncea
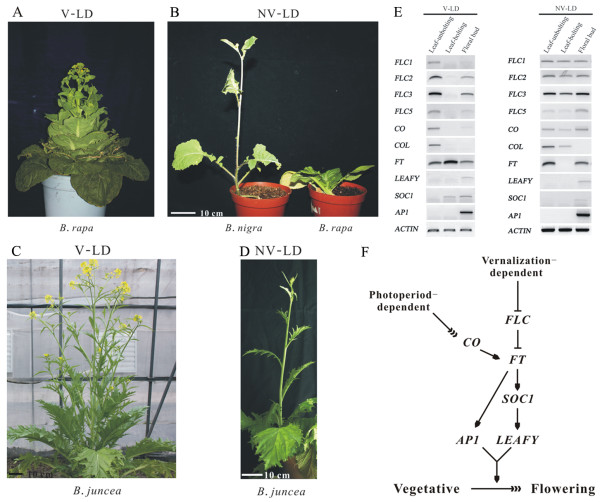
Flowering behavior is an essential feature affecting Brassicaecae crop production. For B. rapa (AA genome), seed vernalization and long-day photoperiod conditions are necessary for flowering (Figure 5-A, B), while only long-day photoperiod conditions promote B. nigra flowering, without any need for vernalization treatment (Figure 5-A, B). Interestingly, long-day photoperiod conditions lead to flowering in B. juncea regardless of vernalization conditions (Figure 5-C, D). We identified four FLOWERING LOCI C (FLC1, FLC2, FLC3 and FLC5) genes and other flowering pathway-related genes, including CONSTANS (CO), CONSTANS -like (COL), FLOWERING T (FT), LEAFY, SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO1) and AP1 (APETALA1), in B. juncea. Under vernalization and long-day photoperiod conditions, when FLCs gene expression is down-regulated, flowering occurs by an FLC-dependent pathway in B. juncea. Under non-vernalization and long-day photoperiod conditions, flowering occurs by a CONSTANS-dependent pathway, not FLC-dependent, since FLCs genes are still expressed during flowering (Figure 5-E). These results indicate that vernalization- and photoperiod-dependent flowering pathways coexist in the allotetraploid B. juncea (Figure 5-F).
Figure 5.
Vernalization- and photoperiod-dependent flowering pathways coexist in allotetraploid B. juncea. Vernalization and long-day photoperiod condition (A and C), Non-vernalization and long-day photoperiod condition (B and D), B. juncea flowering pathway related gene expression (E) and proposed flowering pathway in B. juncea(F)
Discussions
Allotetraploid B. juncea possesses unique traits that influence its utility as a vegetable crop in China and oil crop in India; these features emerged after natural hybridization between B. rapa and B. nigra and allopolyploidization. The A genome (B. rapa) [5] and C genome [35] sequences were recently completed, providing considerable momentum in molecular genetic studies of Brassica. Brassica A/B/C genome phylogeny and evolution is of considerable interest. Largely because of the vast phenotypic diversity available within the Brassicas[1,6].
The fate of duplicated genes can be defined as sub-functionalization, neo-functionalization or non-functionalized after polyploidization or whole genome duplication (WGD) in polyploid crops [36,37]. Biased gene expression between homologous gene is usually observed in allopolyploid plants, including Gossypium[38], Arabidopsis[39] and Tragopogon[40], resulting from genetic and epigenetic interactions between redundant genes, and these interactions can influence plant phenotypes and evolutionary fates of polyploid types [37]. Among the many models that attempt to explain how/why duplicated genes are retained after polyploidy [41], sub-functionalization is the most popular hypothesis even though it remains controversial [42]. Genome plasticity, redundancy and diversity are well described and discussed in polyploid Brassicaceae[43-45], and are thought to contribute to adaptive phenotypic variation [37,40,46]. For example, flowering time variation is affected by the replicated copies of the flowering time gene FLC in Brassicaceae[46]. Here, we preliminarily show that vernalization-dependent and photoperiod-dependent flowering pathways coexist in allopolyploid B. juncea, suggesting that the flowering pathways of B. rapa and B. nigra can express in independent vernalization environments in the allopolyploid B. juncea. Timing of flowering onset is an essential trait that affects crop production and plant life cycle. To meet the challenges of climate changes and adapt to a wider range of growing environments, plants adjust their flowering time or pathway during evolution. The coexistence of vernalization-dependent and photoperiod-dependent flowering pathways might indicate better adaptation for survival during evolution in B. juncea. On the other hand, with global warming, B. juncea may have more potential to be used as oil crops because of its flowering trait independent of vernalization status.
In this study, we employed high-throughput sequencing approach based on Illumina/Solexa platform to investigate 30 × genome survey sequences of B. juncea. After comparison to B. rapa genome, 45.93% genome survey sequences of B. juncea can be mapped to B. rapa genome, which indicate relative far phylogenetic relationship between A/B than A/C. This provides an opportunity that we can sequence this genome by diploid approaches. After comparative genome analysis between B. juncea and B. rapa, we find more SV in B. juncea genome, which may be resulted from polyploidy event. Moreover, based on the 30 × genome survey sequences of B. juncea, we observed huge polymorphisms between B. juncea and B. rapa including SNPs, SVs and Indels. The non-synonymous SNPs, frame-shift Indels and genes with SVs resulted from these polymorphisms caused a large number of pathways to be changed in B. juncea by KEGG analysis, for example, glucosinolate biosynthesis pathway. Higher expressions of CYP83A1 and CYP79F1 genes are associated with a higher content of aliphatic glucosinolate in B. juncea than B.rapa. Increased CYP83B1 gene expression is associated with a higher content of indole glucosinolate in B.rapa than B. juncea. However, we did not observe a higher content of aromatic glucosinolate resulting from observed higher expression of CYP79A2 in B. juncea than B. rapa. That may be reason that we did not observe higher expressions of SUR1 downstream of CYP79A2 and CYP79B2 in aromatic glucosinolate biosynthesis pathway in B. juncea. The advent of high-throughput sequencing (Next-generation sequencing, NGS) has revolutionized genomic and transcriptomic approaches to biology. These new sequencing tools are also valuable for discovering, sequencing and genotyping not only hundreds but thousands of markers across almost any genome of interest, even in species in which little or no genetic information is available [47].
Conclusions
In this study, we find the clues of glucosinolate biosynthesis and flowering pathways evolution in B. juncea based on comparative analysis between 30 × genome survey sequences of B. juncea and genome of B. rapa, which allow us to propose that polyploidization resulted in the evolution of glucosinolate biosynthesis and flowering pathways in B. juncea. The genome survey sequencings promote the whole genome sequencing processing in B. juncea. To conclude, next-generation sequencing, even low genome coverage is pushing forward the molecular genetics especially in non-model plant.
Methods
Plant materials
The inbred line of Brassica jucnea var tumida Tsen et Lee from our lab (Institute of Vegetable Science, Zhejiang University) was used to conduct genome survey sequencing in this study. Brassica rapa and Brassica nigra seeds were procured from the University of Warwich and Beijing Academy of Agriculture and Forestry Sciences, respectively.
Genome in situ hybridization of chromosome in B. juncea
Seeds of B. juncea, B. rapa and B. nigra were germinated at 28°C in dark. Root tips were harvested, in ice-bath for 24 hours and fixed in solution (Ethanol: Acetic acid = 3: 1) for 24 hours. The root tips were stained within 1% acetocarmine for 15 min and dropped on slide with 45% acetic acid then covered with a coverslip. The slides with samples were examined by microscope to find the metaphase stage of chromosome and then conserved. Total genomic DNA was isolated from young leaf tissue of B. rapa and B. nigra using a DNA extraction kit (QIAGEN, USA). The genomic DNA of B. rapa was labeled with biotin-16-dUTP by nick translation and the genomic DNA of B. nigra was labeled with digoxingenin-11-dUTP by nick translation (Roche, USA). For genomic in situ hybridization, slide pretreatment, chromosome denaturation with probe, hybridization and post-hybridization treatments were referred to the method [48]. The images were captured and analyzed using Zeiss Axioskop fluorescence microscope system (ZEISS, Germany).
Library construction, sequencing and re-sequencing
Genomic DNAs were isolated from young leaf tissue of B. juncea using a DNA extraction kit (Illunima, USA). Genomic Paired-end libraries with 170 bp and 500 bp insertion were constructed following a standard protocol provided by Illumina. The adapter ligation and DNA cluster preparation were performed and subjected to sequencing using Illumina Genome Analyzer (Illumina Hiseq2000, USA) according to the manufacturer’s standard protocol. Low-quality reads, reads with adaptor sequences and duplicated reads were filtered, and remaining high-quality data was used in the following assembly and analysis.
Comparative genome analysis
Genome sequence of B. rapa was used as reference to comparatively analyze the genome survey sequences (GSS) of B. juncea by using Burrows-Wheeler Aligner (BWA) program. Samtools, Pindel and Breakdancer software were used to analyze the molecular polymorphisms including SNP, SV and Indel polymorphisms by comparison of the survey genome of B. juncea and genome of B. rapa. BLAST software was used for gene annotation.
Glucosinolate biosynthesis gene expression
Total RNA was extracted from seedlings using an RNeasy Plant Mini Kit (QIAGEN, USA) following the manufacturer’s protocol. During extraction, total RNA was exhaustively treated with RNase-free Dnase (Qiagen, Germany). RNA concentration and quality were determined with a biophotometer (Eppendorf, Germany) and gel analysis. 1 μg total RNA was transcribed to synthesize cDNA first strand using a Reverse Transcriptase M-MLV Kit (Takara, Japan). The expression of 6 selected genes was assayed in B. juncea and B. rapa by quantitative real-time PCR (qPCR) on ABI Step One (Applied Biosystems, USA). qPCR reaction were performed using 2.5 μl cDNA template, 6.5 μl of Fast start universal SYBR Green Master (Roche Germany), and 2.0 μM primer, in a total 20 μl reaction system. The relative quantification of the target gene was determined using the ΔΔCT method. All PCR reactions were run in triplicate on each plate as technical replicates and three independent biological replicates were used. Gene fragment of CYP83A1, CYP79A2, CYP83B1, CYP79B2, CYP79F1, SUR1 and 25S were cloned from B. juncea and B. rapa and conserved sequences of these genes were used for primer design. 25S was used as an internal control gene to evaluate relative gene expression level. Primers used in this study are listed in Additional file 1: Table S6.
Glucosinolate content measurement
Duplicates of the freeze-dried powder (0.25 g) in 10 ml glass tubes were preheated for 5 min in 75°C water bath. And 4 ml of 70% boiling methanol (75°C) were added and extracted at 75°C in a water bath for 10 min. For internal standardization 100 μl of 5 mM sinigrin (Sigma-Aldrich Co., MO, USA) were added to one of the duplicates before extraction. Then 1 ml of 0.4 M barium acetate were rapidly added and the vials vortexed for several seconds. After centrifugation at 4,000 rpm for 10 min at room temperature, the supernatants were collected and the pellets were re-extracted twice with 3 ml of 70% boiling methanol (75°C). Three supernatants were combined and made up to a final volume of 10 ml with 70% methanol. 5 ml extracts were loaded onto a 1 ml mini-column (JT Baker, USA) containing 500 μl of activated DEAE Sephadex™ A-25 (Amersham Biosciences, Sweden), and allowed to desulphate overnight with aryl sulfatase (Sigma-Aldrich Co., MO, USA). The resultant desulpho (ds)-GS were eluted with 2.5 ml of ultra pure water produced by Milli-Q system (Millipore Co., USA) and stored at -20°C prior to separation by high performance liquid chromatography (HPLC).
Samples of 40 μl were analyzed in a Shimadzu HPLC system (LC-10AT pump, CTO-10A column oven, SCL-10A VP system controller, Shimadzu, Kyoto, Japan) consisting of a UV–VIS detector (SPD-10A) set at 229 nm and a prontosil ODS2 column (250 × 4 μm, 5 μm, Bischoff, Germany). The mobile phase consisted of ultrapure water (A) and acetonitrile (Tedia, USA) (B). The mobile phase was in the following gradient: H2O (2 min), a linear gradient of 0-20% acetonitrile (32 min), 20% acetonitrile (6 min), followed by 100% acetonitrile and 0% acetonitrile prior to the injection of the next sample.
Identification of flowering pathway in B. juncea
For vernalization and long-day treatment, B. juncea and B. rapa were grown in glass greenhouse during winter season starting 4th week of October. Under these conditions, B. rapa began flowering in March and B. juncea in April. For non-vernalization and long-day treatment, B. juncea, B. nigra and B. rapa were grown in a growth chamber under conditions of 25°C and photoperiod of 16 light: 8 dark. Semi-RT-PCR method was employed to study the flowering pathway-related gene expression, including FLC1/2/3/5, CO, COL, FT, LEAFY, SOC1, AP1. ACTIN gene from B. juncea was used as an internal control gene to evaluate relative gene expression level. Degenerate primers of flowering pathway-related genes were referred to publications [49-51], NCBI Accessions JQ314107, JN699544 and cloned gene fragment. Primers of ACTIN gene was designed by NCBI Accessions HM565958. Primers used in this study are listed in Supporting Information Additional file 1: Table S6.
Competing interests
The authors have declared that no competing interests exist.
Authors’ contributions
JY and MZ conceived and designed the experiments. JY, NS, XZ, XQ and ZH performed the experiments and data analysis. JY wrote the paper and MZ edited the paper. All authors read and approved the final manuscript.
Supplementary Material
Statistical comparison of sequencing reads of B. juncea with genome of B. rapa. Table S2: Statistic of SNPs between mapped sequences of B. juncea and B. rapa genome sequence. Table S3: Statistic of SVs between mapped sequences of B. juncea and B. rapa genome sequence. Table S4: Statistic of Indel (1–3 bp) between mapped sequences of B. juncea and B. rapa genome sequence. Table S5: Polymorphism information on glucosinolate biosynthesis related genes between B. juncea and B. rapa.Table S6: Primer sequences used for qRT-PCR and RT-PCR. Figure S1: Estimation of high-throughput sequencing quality including insert size, quality distribution, nucleotide content and cycle average quality distribution. Figure S2: Comparison of in-depth distribution of sequencing reads from B. juncea on chromosome of B. rapa. Figure S3: Distribution of SNP and SV polymorphisms on chromosome of B. rapa. Figure S4: Distribution of 1–3 bp Indels in genome and coding sequence (CDS) region of B. rapa.
Contributor Information
Jinghua Yang, Email: yangjinghua@zju.edu.cn.
Ning Song, Email: 916482841@qq.com.
Xuan Zhao, Email: xuanxuan8709@163.com.
Xiaohua Qi, Email: xhqi@yzu.edu.cn.
Zhongyuan Hu, Email: huzhongyuan@zju.edu.cn.
Mingfang Zhang, Email: mfzhang@zju.edu.cn.
Acknowledgements
We thank to Mr. H He from Biomarker Technologies Inc. for comparative genome analysis. We thank to Dr. YX Zang from Zhejiang A&F University for measuring the contents of glucosinolates. We express gratitude to Prof. Dr. Y Mukai and G Suzuki from Osaka Kyoiku University for GISH analysis. We also thank to Prof. Sally A. Mackenzie from University of Nebraska-Lincoln for her critical comments and editing of this paper.
Funding
This work was supported by grants from Qianjiang Talents Project of Science Technology Department of Zhejiang Province (2012R10024), the National Natural Science Foundation of China (30971994).
References
- Beilstein MA, Al-Shehbaz IA, Kellogg EA. Brassicaceae phylogeny and trichome evolution. Am J Bot. 2006;93(4):607–619. doi: 10.3732/ajb.93.4.607. [DOI] [PubMed] [Google Scholar]
- Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, Drabek J, Lopez R, Price HJ. Evolution of genome size in Brassicaceae. Ann Bot-London. 2005;95(1):229–235. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Beaulieu JM, Meister A, Leitch IJ. The Dynamic Ups and Downs of Genome Size Evolution in Brassicaceae. Mol Biol Evol. 2009;26(1):85–98. doi: 10.1093/molbev/msn223. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11(11):535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan Journal of Botany. 1935;7:389–452. [Google Scholar]
- O'Neill CM, Bancroft I. Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 2000;23(2):233–243. doi: 10.1046/j.1365-313x.2000.00781.x. [DOI] [PubMed] [Google Scholar]
- Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ. et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18(6):1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJ, Kim JS, Kwon SJ, Lim KB, Choi BS, Kim JA, Jin M, Park JY, Lim MH, Kim HI. et al. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell. 2006;18(6):1339–1347. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JH, Kwon SJ, Yang TJ, Seol YJ, Jin M, Kim JA, Lim MH, Kim JS, Baek S, Choi BS. et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009;10(10):R111. doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XH, Zhang MF, Yang JH. Molecular phylogeny of Chinese vegetable mustard (Brassica juncea) based on the internal transcribed spacers (ITS) of nuclear ribosomal DNA. Genet Resour Crop Ev. 2007;54(8):1709–1716. doi: 10.1007/s10722-006-9179-0. [DOI] [Google Scholar]
- Antonious GF, Bomford M, Vincelli P. Screening Brassica species for glucosinolate content. J Environ Sci Health B. 2009;44(3):311–316. doi: 10.1080/03601230902728476. [DOI] [PubMed] [Google Scholar]
- Verkerk R, Schreiner M, Krumbein A, Ciska E, Holst B, Rowland I, De Schrijver R, Hansen M, Gerhauser C, Mithen R. et al. Glucosinolates in Brassica vegetables: the influence of the food supply chain on intake, bioavailability and human health. Mol Nutr Food Res. 2009;53:S219–S265. doi: 10.1002/mnfr.200800065. [DOI] [PubMed] [Google Scholar]
- Bending GD, Lincoln SD. Characterization of volatile sulphur-containing compounds produced during decompositioin of Brassica juncea tissues in soil. Soil Biology & Biochemistry. 1999;31:695–703. doi: 10.1016/S0038-0717(98)00163-1. [DOI] [Google Scholar]
- Kaul S, Koo HL, Jenkins J, Rizzo M, Rooney T, Tallon LJ, Feldblyum T, Nierman W, Benito MI, Lin XY. et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171(2):765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U, Lydiate DJ. Comparative genome mapping in Brassica. Genetics. 1996;144(4):1903–1910. doi: 10.1093/genetics/144.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics. 1998;150(3):1217–1228. doi: 10.1093/genetics/150.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan TH, DelMonte TA, Reischmann KP, Hyman J, Kowalski SP, McFerson J, Kresovich S, Paterson AH. An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Research. 2000;10(6):776–788. doi: 10.1101/gr.10.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babula D, Kaczmarek M, Barakat A, Delseny M, Quiros CF, Sadowski J. Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: complexity of the comparative map. Mol Genet Genomics. 2003;268(5):656–665. doi: 10.1007/s00438-002-0782-2. [DOI] [PubMed] [Google Scholar]
- Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, Gupta V, Pradhan AK, Pental D. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics. 2008;9:113. doi: 10.1186/1471-2164-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens L, Zou F, Lydiate D, Parkin I, Osborn T. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics. 2003;164(1):359–372. doi: 10.1093/genetics/164.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin K, Acarkan A, Mbulu RS, Clarenz O, Schmidt R. The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 2004;135(2):735–744. doi: 10.1104/pp.104.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. P Natl Acad Sci USA. 2006;103(13):5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154(1):459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XL, Zhang JZ. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169(2):1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinkovitch MC, Helaers R, Depiereux E, Tzika AC, Gabaldon T. 2x genomes–depth does matter. Genome Biol. 2010;11(2):R16. doi: 10.1186/gb-2010-11-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Xu J, Xia S, Gu J, Yang Y, Fu J, Qian X, Zhang S, Wu J, Liu K. Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor Appl Genet. 2009;118(6):1121–1131. doi: 10.1007/s00122-009-0967-8. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, Delcher AL, Pop M, Wang W, Fraser CM. et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301(5641):1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC. et al. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5(4):e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Narechania A, Sabot F, Stein J, Vu GT, Graner A, Ware D, Stein N. Low-pass shotgun sequencing of the barley genome facilitates rapid identification of genes, conserved non-coding sequences and novel repeats. BMC Genomics. 2008;9:518. doi: 10.1186/1471-2164-9-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck J, Miller W, Gorrell JH, Muzny D, Gibbs RA. Analysis of the quality and utility of random shotgun sequencing at low redundancies. Genome Res. 1998;8(10):1074–1084. doi: 10.1101/gr.8.10.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Schierup MH, Jorgensen FG, Gorodkin J, Panitz F, Staerfeldt HH, Christensen OF, Mailund T, Hornshoj H, Klein A. Pigs in sequence space: a 0.66X coverage pig genome survey based on shotgun sequencing. BMC Genomics. 2005;6:70. doi: 10.1186/1471-2164-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen DA, Noor MAF. What can you do with 0.1 × genome coverage? A case study based on a genome survey of the scuttle fly Megaselia scalaris (Phoridae) BMC Genomics. 2009;10:382. doi: 10.1186/1471-2164-10-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Zhao MX, Wang XW, Tong CB, Huang SM, Tehrim S, Liu YM, Hua W, Liu SY. Bolbase: a comprehensive genomics database for Brassica oleracea. BMC Genomics. 2013;14:664. doi: 10.1186/1471-2164-14-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Chen ZJ. Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol. 2010;13(2):153–159. doi: 10.1016/j.pbi.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nature Reviews Genetics. 2005;6(11):836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Chaudhary B, Flagel L, Stupar RM, Udall JA, Verma N, Springer NM, Wendel JF. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (gossypium) Genetics. 2009;182(2):503–517. doi: 10.1534/genetics.109.102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16(7):1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggs RJA, Elliott NM, Zhang LJ, Koh J, Viccini LF, Soltis DE, Soltis PS. Tissue-specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol. 2010;186(1):175–183. doi: 10.1111/j.1469-8137.2010.03205.x. [DOI] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Freeling M. The evolutionary position of subfunctionalization, downgraded. Genome Dyn. 2008;4:25–40. doi: 10.1159/000126004. [DOI] [PubMed] [Google Scholar]
- Lukens LN, Quijada PA, Udall J, Pires JC, Schranz ME, Osborn TC. Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol J Linn Soc. 2004;82(4):665–674. doi: 10.1111/j.1095-8312.2004.00352.x. [DOI] [Google Scholar]
- Leitch AR, Leitch IJ. Genome plasticity and the diversity of polyploid plants. Science. 2008;320:481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- Marhold K, Lihová J. Polyploidy, hybrization and reticulated evolution: lessons from the Brassicaceae. Plant Systematics and Evolution. 2006;259:143–174. doi: 10.1007/s00606-006-0417-x. [DOI] [Google Scholar]
- Osborn TC. The contribution of polyploidy to variation in Brassica species. Physiologia Plantatum. 2004;121:531–536. doi: 10.1111/j.1399-3054.2004.00360.x. [DOI] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12(7):499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Howell EC, Kearsey MJ, Jones GH, King GJ, Armstrong SJ. A and C genome distinction and chromosome identification in brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics. 2008;180(4):1849–1857. doi: 10.1534/genetics.108.095893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynov VV, Khavkin EE. Polymorphism of the CONSTANS gene in Brassica plants. Russ J Plant Physl+ 2005;52(2):242–248. doi: 10.1007/s11183-005-0037-2. [DOI] [Google Scholar]
- Vorobiev VA, Martynov VV, Pankin AA, Khavkin EE. Polymorphism of the LEAFY gene in Brassica plants. Russ J Plant Physl+ 2005;52(6):814–820. doi: 10.1007/s11183-005-0120-8. [DOI] [Google Scholar]
- Schranz ME, Quijada P, Sung SB, Lukens L, Amasino R, Osborn TC. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics. 2002;162(3):1457–1468. doi: 10.1093/genetics/162.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical comparison of sequencing reads of B. juncea with genome of B. rapa. Table S2: Statistic of SNPs between mapped sequences of B. juncea and B. rapa genome sequence. Table S3: Statistic of SVs between mapped sequences of B. juncea and B. rapa genome sequence. Table S4: Statistic of Indel (1–3 bp) between mapped sequences of B. juncea and B. rapa genome sequence. Table S5: Polymorphism information on glucosinolate biosynthesis related genes between B. juncea and B. rapa.Table S6: Primer sequences used for qRT-PCR and RT-PCR. Figure S1: Estimation of high-throughput sequencing quality including insert size, quality distribution, nucleotide content and cycle average quality distribution. Figure S2: Comparison of in-depth distribution of sequencing reads from B. juncea on chromosome of B. rapa. Figure S3: Distribution of SNP and SV polymorphisms on chromosome of B. rapa. Figure S4: Distribution of 1–3 bp Indels in genome and coding sequence (CDS) region of B. rapa.