Abstract
We have previously examined characteristics of maternal chromosomes 21 that exhibited a single recombination on 21q and proposed that certain recombination configurations are risk factors for either meiosis I (MI) or meiosis II (MII) nondisjunction. The primary goal of this analysis was to examine characteristics of maternal chromosomes 21 that exhibited multiple recombinant events on 21q to determine whether additional risk factors or mechanisms are suggested. In order to identify the origin (maternal or paternal) and stage (MI or MII) of the meiotic errors, as well as placement of recombination, we genotyped over 1,500 SNPs on 21q. Our analyses included 785 maternal MI errors, 87 of which exhibited two recombinations on 21q, and 283 maternal MII errors, 81 of which exhibited two recombinations on 21q. Among MI cases, the average location of the distal recombination was proximal to that of normally segregating chromosomes 21 (35.28 vs. 38.86 Mb), a different pattern than that seen for single events and one that suggests an association with genomic features. For MII errors, the most proximal recombination was closer to the centromere than that on normally segregating chromosomes 21 and this proximity was associated with increasing maternal age. This pattern is same as that seen among MII errors that exhibit only one recombination. These findings are important as they help us better understand mechanisms that may underlie both age-related and nonage-related meiotic chromosome mal-segregation.
Introduction
The failure of chromosomes to properly segregate during meiosis results in the production of aneuploid gametes (e.g., those that have an abnormal number of chromosomes). Aneuploidy is the leading cause of pregnancy loss and intellectual and developmental disability; as a result, it is important that we identify factors that increase the risk for meiotic chromosome mal-segregation (Hassold et al. 1996). Trisomy 21, caused mainly by mal-segregation of chromosome 21 primarily within oocytes (Antonarakis 1991; Freeman et al. 2007), has long been used as a model to identify risk factors associated with meiotic chromosome mal-segregation as it is the most frequent autosomal trisomy observed among live-born individuals (Hassold et al. 1996; Hassold et al. 1995; Hassold and Jacobs 1984). To date, both the absence of exchange and the presence of a single exchange within the telomeric region of 21q have been found to be associated with an increased risk for maternal meiosis I (MI) errors. Additionally, the presence of a single exchange within the pericentromeric region of 21q has been found to be associated with maternal meiosis II (MII) errors (Lamb et al. 1997). In attempt to understand how these factors contribute to meiotic chromosome mal-segregation, we recently examined the association between maternal age (i.e., the age of the mother at time of conception) and the location of recombination along 21q (Oliver et al. 2008). Results suggested that the risk for a maternal MI error among women that exhibited either no exchange on 21q or a single telomeric exchange along 21q were independent of maternal age. In contrast, the association with maternal MII errors increased with increasing maternal age among chromosomes 21 that exhibited a single pericentromeric exchange. These Wndings begin to provide insight into candidate meiotic processes and proteins associated with recombination that increase the vulnerability of bivalents to nondisjunction.
Since the completion of our most recent study (Oliver et al. 2008), we have more accurately defined the number and location of recombinant events by expanding our genotyping panel from approximately 25 short tandem repeat (STR) markers to over 1,500 SNPs located on chromosome 21. Our first goal was to confirm or refute previous findings on the association between maternal age and the location of single recombination events (Ghosh et al. 2009; Oliver et al. 2008) using dense SNP genotyping data so that we could precisely define the location of recombination as well as the number of recombinant events on 21q.
Our second goal was to examine the characteristics of chromosomes 21 that exhibit more than one recombinant event to determine whether the placement of the recombinant events differed from those on normally segregating chromosomes 21 and whether placement played a role in promoting nondisjunction. Results from our analyses confirmed our previous findings associated with single recombinants as outlined above. With respect to patterns associated with observed double recombinants, we found a reduction in the distance between recombinant events, due to altered placement of the distal recombinant event only (i.e., the recombinant event closest to the telomere) among maternal MI errors. Among MII errors, the location of the most proximal recombination was closer to the centromere in older women. This pattern is essentially same as we have observed for single events among MII errors.
Results
Comparison of datasets
For this study, we genotyped over 1,500 SNPs along chromosome 21 in the proband with trisomy 21 due to a maternal nondisjunction error and their parents (trios) to obtain a comprehensive recombination profile along the nondisjoined chromosome (see “Materials and methods”). A subset of families (MI, N = 369 and MII, N = 139) from our previous study (Oliver et al. 2008) in which STR genotypes were used to identify the location of recombination were also genotyped for SNPs along 21q and included in the present analysis. We found that both the average number of recombinant events and mean maternal ages for samples taken from our previous STR dataset were similar to our current SNP dataset for both maternal MI and MII errors (Table 1). As a result, we used the combined SNP and STR data for our analysis of the location of recombination.
Table 1.
Comparison of the subset of cases that were previously characterized by Oliver et al. (2008) using chromosome 21 STR markers and newly ascertained cases characterized using only chromosome 21 SNPs (“new cases”) The combined dataset includes all STR and SNP marker information on the total number of families (“all”)
| Sample set | Number of trios | Average number of recombinant events |
Standard deviation for number of recombinant events |
Mean maternal age |
Standard deviation for mean maternal age |
|---|---|---|---|---|---|
| 2008 MI cases | 369 | 0.43 | 0.63 | 32.20 | 6.51 |
| New MI cases | 416 | 0.52 | 0.69 | 32.81 | 5.98 |
| MI All | 785 | 0.47 | 0.66 | 32.52 | 6.24 |
| 2008 MII cases | 139 | 1.23 | 0.47 | 33.40 | 6.83 |
| New MII cases | 141 | 1.28 | 0.45 | 33.28 | 6.64 |
| MII all | 280 | 1.26 | 0.46 | 33.34 | 6.72 |
Maternal MI errors
To confirm our previous finding that a single telomeric recombinant event is associated with MI errors, we analyzed all families using the combined genetic marker set. We found that the average location of recombinant events among maternal MI errors with only one recombination was 37.56 Mb. This location was significantly more distal than that for normally disjoined chromosomes (27.53 Mb, p < 0.0001, Table 2). This pattern was the same when all events were included (i.e., both single and double recombinant events), although this difference was primarily driven by the single events (Table 2).
Table 2.
Average location of recombinant events and 95% upper and lower confidence limits (UCL and LCL) for all meiotic outcome groups stratified by number of observed recombinations along chromosome 21
| Meiotic outcome group |
Average location of recombination (Mb) among single events |
Average location of most proximal recombination (Mb) among all events |
||||||
|---|---|---|---|---|---|---|---|---|
| N | Mean | LCL | UCL | N | Mean | LCL | UCL | |
| MI errors | 222 | 37.56 | 36.66 | 38.46 | 309 | 33.00 | 31.84 | 34.17 |
| MII errors | 202 | 22.60 | 21.52 | 23.69 | 283 | 21.53 | 20.66 | 22.40 |
| FHS | 606 | 27.42 | 26.67 | 28.17 | 764 | 25.75 | 25.09 | 26.41 |
| AGRE | 579 | 27.65 | 26.92 | 28.37 | 743 | 25.97 | 25.33 | 26.61 |
| GENEVA | 87 | 27.60 | 25.60 | 29.60 | 107 | 26.04 | 24.25 | 27.83 |
| All | 1,272 | 27.53 | 27.03 | 28.04 | 1,614 | 25.87 | 25.43 | 26.32 |
The FHS, AGRE and GENEVA meiotic outcome groups represent normally disjoined chromosomes 21
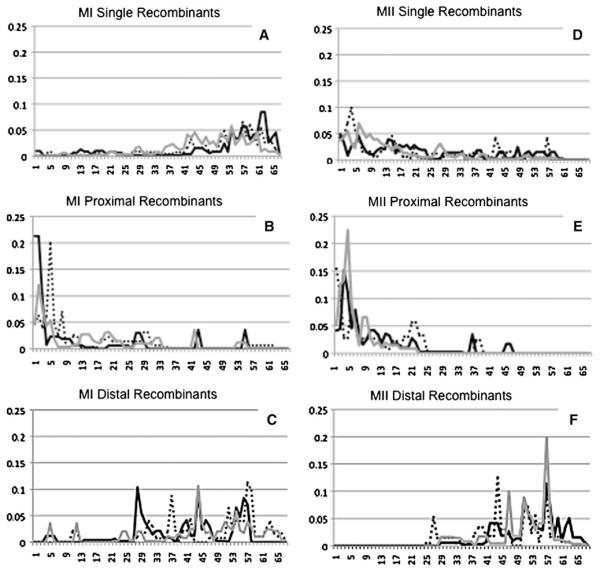
We also confirmed the negative correlation between maternal age and the location of the single recombinant event using linear regression. We found that as maternal age increased, the mean location of the single recombinant event was more medially located (r2 = 0.02, p = 0.02; Fig. 1a). We then extended our analyses to specifically examine patterns among nondisjoined chromosomes with two observed recombinant events on chromosome 21. There was no difference between the average location of the proximal recombinant event among MI errors compared with normal transmissions (p = 0.97); however, the average location of the distal recombinant event was more centromeric among abnormally segregating chromosomes (p < 0.0001, Table 3). Thus the average distance between the two observed recombinant events was reduced among the MI errors compared with normally segregating chromosomes (15.39 vs. 19.16 Mb, p = 0.0002, Table 3). Importantly, this reduction in the distance between recombinant events was due to the more proximal location of distal recombinant event among the MI doubles. Further examination of the frequency distribution of locations of the distal recombinant events showed that this observation was not due to outliers (data not shown).
Fig. 1.
Distribution of Recombinations by age and stage of mal-segregation 21q was divided into 500 kb segments and the proportion of recombinations within each 500 kb segment was calculated. On the X-axis is the segment in which the recombination occurred, on the Y-axis is the proportion of recombinations per segment. Data from the young group (<29 years in maternal age) is plotted in as a solid black line, data from the middle group (29-34 years in maternal age) is plotted as a black dotted line and data from the oldest group (≥34 years in maternal age) is plotted as a gray line
Table 3.
Average location of recombinant events and 95% upper and lower confidence limits (UCL and LCL) for all meiotic outcome groups among those chromosomes 21 with two observed recombinations
| Meiotic outcome group |
N | Average location of proximal recombination (Mb) |
Average location of distal recombination (Mb) |
Average distance between recombinant events (Mb) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | LCL | UCL | Mean | LCL | UCL | Mean | LCL | UCL | ||
| MI errors | 75 | 19.89 | 18.26 | 21.53 | 35.28 | 33.59 | 36.97 | 15.39 | 13.53 | 17.24 |
| MII errors | 75 | 18.17 | 17.11 | 19.24 | 38.22 | 37.17 | 39.27 | 20.05 | 18.64 | 21.45 |
| FHS | 158 | 19.37 | 18.63 | 20.10 | 38.62 | 37.89 | 39.34 | 19.25 | 18.31 | 20.20 |
| AGRE | 164 | 20.07 | 19.22 | 20.92 | 39.23 | 38.55 | 39.91 | 19.16 | 18.18 | 20.14 |
| GENEVA | 20 | 19.24 | 16.77 | 21.72 | 37.73 | 34.69 | 40.76 | 18.48 | 14.98 | 21.98 |
| All | 342 | 19.70 | 19.15 | 20.24 | 38.86 | 38.37 | 39.36 | 19.16 | 18.50 | 19.83 |
The FHS, AGRE and GENEVA meiotic outcome groups represent normally disjoined chromosomes 21
With respect to maternal age, we found evidence for a marginally significant positive correlation between maternal age and the location of the proximal recombinant event (r2 = 0.05, p = 0.05, Fig. 1b) among the MI errors. However we did not find a significant correlation between maternal age and the location of the distal recombinant event (r2 = 0.01, p = 0.37, Fig. 1c). We also did not find an association between the distance between recombinant events and maternal age (p = 0.39).
Maternal MII errors
We found that the average location of recombinant events among maternal MII errors with only one recombination was 22.60 Mb. This was significantly more proximal to that of normally segregating chromosomes 21 that exhibit only one recombination (27.53 Mb, p < 0.0001, Table 2). This pattern was the same when all events were included (i.e., both single and double recombinants). We also confirmed that there was a significant negative correlation between maternal age and the location of recombination using linear regression. Thus as maternal age increased, the mean location of the single recombinant event was more proximally located (r2 = 0.05, p = 0.001; Fig. 1d).
Upon extending our analyses to specifically examine MII errors with two observed recombinant events, we found that the average location of the proximal recombinant event was closer to the centromere compared with that on normally disjoining chromosomes 21 (Table 3, p = 0.005). The average location of distal recombinant events was not different from normal distal recombinant events (p = 0.29, Table 3).
With respect to maternal age, we found a statistically significant correlation between maternal age and the location of the proximal (r2 = 0.10, p = 0.007, Fig. 1e) but not the distal recombination (r2 = 0.004, p = 0.60, Fig. 1f) among MII errors. We found no association between the distance between events and maternal age (p = 0.11).
Discussion
Technological advances in detecting genetic variation have increased our ability to detect and precisely locate recombination breakpoints. To better define regions of recombination in trisomy 21 samples, we have expanded our genotyping panel from approximately 25 STR markers to over 1,500 chromosome 21 specific SNPs. In addition, due to the increased number of families with trisomy 21 enrolled, we now have, for the first time, an adequate sample size of cases of trisomy 21 with two observed recombinant events on 21q to examine their potential role in nondisjunction. These steps have enabled us to confirm our previous findings on the association between maternal age and the location of recombination among cases of trisomy 21 that exhibit only one observed recombinant. Considering that the risk for nondisjunction increases significantly with increased maternal age, our findings remain consistent with the interpretation that a single telomeric exchange increases the risk of a maternal MI error for women of all ages and that a single pericentromeric exchange increases the risk of MII errors primarily among older ooctyes.
In this study, we examined the location of observed double recombinant events among MI errors, MII errors and normally segregating chromosomes to ask whether altered placement of multiple events, intuitively very stable chiasma configurations, is associated with nondisjunction errors. Among MI errors, we found that the average location of the observed distal recombinant event was more proximal than that of normal meiotic outcomes, although the more centromeric event did not differ in location. Thus, the average distance between observed recombination events is reduced in MI errors, but only driven by the distal recombinant event. The location of this distal event was not associated with maternal age. Among cases that have only one recombinant, we and others have proposed that the minimal amount of the sister chromatid cohesion complex remaining distal to the exchange event is expected to increase the risk for MI errors (Orr-Weaver 1996). Such events are predicted to be too far from the kinetochore to aid in amphitelic attachment of chromosomes to the meiotic spindle during MI (Hawley et al. 1994; Koehler et al. 1996; Nicklas 1974; Oliver et al. 2008; Ross et al. 1996). This pattern places risk on the bivalent irrespective of the age of the oocyte (Oliver et al. 2008). This same mechanism cannot be applied to MI errors with multiple exchanges that span the chromosome. The reduced distance between observed multiple recombinant events may be indicative of altered properties that normally help stabilize the bivalent. However, the observation that the reduced distance is only due to the more distal recombinant event points more to localized features that guide exchanges. Perhaps the altered placement of the distal recombinant nullifies the potential “good effect” the proximal recombinant event leading to MI errors. Alternatively, reduced interference may lead to bivalents with more than two recombinant events. Perhaps the most distal event was unobserved in these MI errors. Bivalents with too many recombinant events may be difficult to segregate (Dernburg 2001). However, we would have expected to observe several bivalents with three recombinant events and we did not.Although we cannot identify a mechanism to explain our observation at this time, it may be important to consider negative interference, a phenomenon that is rare in meiosis compared with positive interference (Broman and Weber 2000; Munz 1994; Zhao et al. 1995), as a feature associated with an increased risk for the nondisjunction of chromosomes 21 within oocytes.
For MII errors with two recombinant events, the location of the most proximal recombinant event was closer to centromere with increasing maternal age (Fig. 1e). This pattern is essentially same as we have observed for single events among MII errors (Fig. 1d). Interestingly, among MI errors with multiple exchanges, there was a marginally significant positive correlation between maternal age and the location of the proximal recombinant event. It will be important to determine whether this is due to a similar process as that in MII errors. We think that the observation that a pericentromeric exchange is associated with both single and multiple exchange bivalents that undergo MII errors helps to distinguish whether this event is a risk factor or a protective factor for nondisjunction. That is, we suggested previously that a pericentromeric exchange may create a suboptimal conformation that is made worse by maternal age-related risk factors. Alternatively, a pericentromeric exchange may protect the bivalent from maternal age-related risk factors allowing the proper segregation of homologs, but not sister chromatids. An example of the former would be that a pericentromeric exchange compromises the centromeric cohesion complex, exacerbating the normal degradation of this important complex with age. The effect of degradation of centromere or sister chromatid cohesin complexes or of spindle proteins with age of the oocyte (Pan et al. 2008; Steuerwald et al. 2001) may lead to premature sister chromatid separation as first suggested by Angell (1991). If a pericentromeric exchange is protective and helps to stabilize the maternal age-compromised bivalent through MI, but not MII, there would be an enrichment of MII errors among the older oocytes. Under this model, we would not necessarily expect to see an increased frequency of pericentromeric recombinants among bivalents with other stabilizing configurations, i.e., those with two exchanges instead of just one (Robinson et al. 1998; Thomas et al. 2001). The fact that we did leads us to think that a pericentromeric events is a risk factor for nondisjunction, not a protective factor, during the long protracted state of MI.
Collectively these findings are important as they help us better understand mechanisms that may underlie both age-related and nonage-related meiotic chromosome malsegregation. Our next step will be to determine whether the distribution of genomic features differ between the refined breakpoints of recombination in maternal errors when compared to those of the euploid population. Potentially the presence or absence of certain genomic features or the differential use of those factors by proteins involved in initiating recombination (e.g., PRDM9, Baudat et al. 2010) may help explain why chromosomes that successfully undergo recombination fail to properly segregate.
Materials and methods
Ethics statement
All recruitment sites obtained the necessary Institutional Review Board approvals from their institutions.
Trisomic population
Study sample
Families with an infant with full trisomy 21 were recruited through a multisite study of risk factors associated with chromosome mal-segregation (Freeman et al. 2007; Lamb et al. 1996, 1997). Parents and the infant donated a biological sample (either blood or buccal) from which DNA was extracted. Only families in which DNA was available from both biological parents and the child with trisomy 21 were included in the present analysis which includes 785 maternal MI and 283 maternal MII cases of trisomy 21; 416 of the maternal MI and 141 of the maternal MII cases were included in a previous study reported by Oliver et al. (2008) (Table 1).
Genotyping and quality control
Samples were genotyped at 1,536 SNP loci on 21q by the Center for Inherited Disease Research using the Illumina Golden Gate Assay. The most centromeric SNP was rs2259403 and the most telomeric was rs46909248. In order to assess the quality of our genotyping data, Mendelian inconsistencies and sample mix-ups were identified using RelCheck among the trios. In addition, parental genotyping data were used to identify poorly performing SNPs. SNPs that met the following criteria were excluded from our analyses: minor allele frequency (MAF) < 0.01, deviation from Hardy–Weinberg Equilibrium (HWE) (p < 0.01), heterozygosity >0.60, or >10% missingness. We also excluded SNPs on a family-by-family basis if >50% of the genotype data for a proband had low intensity levels.
Determining stage and origin of meiotic chromosome mal-segregation
The parental origin of the meiotic error was determined by establishing the contribution of parental alleles to the proband with trisomy 21. Only cases of maternal origin were included in our analyses. Once the maternal origin of the meiotic error was established, markers located in the pericentromeric region (13,615,252 bp–16,784,299 bp) of 21q were used to infer the stage of the meiotic error, MI or MII. If parental heterozygosity was retained in the trisomic offspring, we concluded a MI error. If parental heterozygosity was reduced to homozygosity, we concluded a MII error. In this assay, we cannot distinguish between the different types of underlying errors that might lead to an MII error. For example, sister chromatids that fail to separate during anaphase of MII or an error that is initiated in MI and not resolved properly in MII both lead to the contribution of sister chromatids to the gamete. Also, if sister chromatids prematurely separate in MI, some configurations will lead to both sister chromatids segregating to the same pole in MII. Similarly, if homologues pairs fail to separate in MI and then go through a reductional division at MII, sister chromatids will be present in the resulting oocyte. Lastly, when all informative markers in the parent of origin were reduced to homozygosity, the origin of non-disjunction was inferred to be a postzygotic, mitotic error and excluded from the study.
Identifying the location of recombination
After genotyping quality control measures were implemented and SNP data were combined with STR data from our previous studies (Oliver et al. 2008), we defined the location of recombinant events. Recombination breakpoints were deWned by switches from nonreduction (N) or reduction (R) or vice versa of maternal heterozygosity to proband homozygosity for each marker along the nondisjoined chromosome 21 (e.g., NNNNNNNNNNRRRRRRRRRR). In this example, the location of neighboring markers indicating the first change from N to R (highlighted in bold) would indicate the location of our recombination breakpoints. In order to ensure that the switch from nonreduction to reduction or vice versa was not due to a genotyping error a minimum of either one informative STR or eight consecutive informative SNPs flanking the recombination breakpoint were required (the example for informative SNPs is shown above). An exception to this rule occurred when the most proximal or distal informative markers on 21q indicated the presence of recombinant event. In these instances, a minimum of either one informative STR or four consecutive informative SNPs were required to define the breakpoints of recombination (e.g., NNNNNNNNRRRR—telomere). The presence of a double recombinant event was defined by a minimum of either one informative STR or eight consecutive informative SNPs flanking the recombination breakpoint on each side for both events (e.g., NNNNNNNNRRRRRRRRNNNNNNNN).
Euploid population
Study sample
SNP genotyping data for normally segregating chromosomes 21 were taken from families recruited for (1) the Autism Genetic Research Exchange (AGRE) (N = 743) (Weiss et al. 2008), (2) the Framingham Heart Study (FHS) (N = 764) (Dawber et al. 1951) and (3) the GENEVA dental caries study (N = 107) (Polk et al. 2008). All families were two-generation families with a minimum of three children. This was necessary to define specific recombination profiles for each parent child transmission.
Genotyping and quality control
The AGRE samples were genotyped for SNPs genome-wide using the Infinium (R) HumanHap550-Duo Bead-Chip. The AGRE data included genotypes at 5,20,017 markers genome-wide, however 11,473 markers were excluded from the analysis due to deviation HWE (p < 10−7) or Mendelian errors. After quality control measures were completed, there was genotype information for 7,810 SNPs on 21q for the AGRE dataset. The FHS samples were genotyped for SNPs genome-wide using the Genome-Wide Human SNP Array 5.0. The FHS data included genotypes at 500,568 markers. However, 22,000 markers were excluded from the analysis due to deviation from HWE (p < 10−7) or Mendelian errors. After quality control measures were completed, there was genotype information for 6,705 SNPs on 21q for the FHS dataset. The GENEVA samples were genotyped using the Illumina 610-Quad Array. The GENEVA dataset included genotypes at 620,901 SNPs. 58,610 markers were excluded from the analysis due to deviation from HWE (p < 10−5), a MAF <0.02 or Mendelian errors. After quality control measures were completed, there was genotype information for 8,189 SNPs on 21q from the GENEVA population. All SNP locations were based on human NCBI Build 36 (hg18).
Identifying the location of recombination
For the AGRE, FHS and GENEVA datasets, genotype data from members of two-generation families with three or more children were used to infer the location of recombination along the maternal chromosome 21. Our approach and software are described in Chowdhury et al. (2009). Briefly, parental genotypes were used to identify informative markers. Then, using these markers, genotypes of the children were compared to identify alleles inherited identical-by-descent from the mothers and fathers. Between two sibs, a switch from sharing the same maternal allele to the different maternal allele was scored as a maternal recombination event.
Statistical analyses
We used linear regression to determine whether the number of recombinations, maternal age and the amount of interference between recombinant events were significant predictors of the location of recombination among our MI and MII errors. Unfortunately, we did not have maternal ages for the normal meiotic outcome group. However, studies to date indicate that the maternal age effect is only minimally associated with rate and location of recombination (Broman et al. 1998; Kong et al. 2004; Oliver et al.2008), nothing on the same scale as that for chromosome nondisjunction events. For the present analysis, all predictors and the outcome variables were continuous variables. The location of recombination in megabases was defined as the midpoint of the interval within which the recombination was localized. A stepwise regression using backward elimination was implemented in order to identify predictors that contributed to the overall significance of the model. Results from the best model (i.e., the model with the highest r2 and smallest number of predictors) are presented. t tests were performed to compare the mean location of recombinant breakpoints between normally and abnormally segregating chromosomes and 95% confidence intervals are provided. All analyses were conducted using Statistical Analysis Software version 9.1.
Acknowledgments
We would like to thank our lab personnel, recruiters and the families that participated in this study. This work was supported by the National Institutes of Health (1T32MH087977, R01 HD057029, R01 HD38979, R01 HL083300, R01-DE 014899 and U01-DE018903); the Center for Inherited Disease Research (HHSN268200782096C); and the Children’s Healthcare of Atlanta Cardiac Research Committee.
Contributor Information
Tiffany Renee Oliver, Department of Human Genetics, Emory University School of Medicine, 615 Michael St, Suite 301, Whitehead Bldg, Atlanta, GA 30322, USA; Department of Biology, Spelman College, Atlanta, GA, USA.
Stuart W. Tinker, Department of Human Genetics, Emory University School of Medicine, 615 Michael St, Suite 301, Whitehead Bldg, Atlanta, GA 30322, USA
Emily Graves Allen, Department of Human Genetics, Emory University School of Medicine, 615 Michael St, Suite 301, Whitehead Bldg, Atlanta, GA 30322, USA.
Natasha Hollis, Department of Human Genetics, Emory University School of Medicine, 615 Michael St, Suite 301, Whitehead Bldg, Atlanta, GA 30322, USA.
Adam E. Locke, Department of Human Genetics, Emory University School of Medicine, 615 Michael St, Suite 301, Whitehead Bldg, Atlanta, GA 30322, USA
Lora J. H. Bean, Department of Human Genetics, Emory University School of Medicine, 615 Michael St, Suite 301, Whitehead Bldg, Atlanta, GA 30322, USA
Reshmi Chowdhury, Department of Pediatrics, University of Pennsylvania, Philadelphia, PA, USA.
Ferdouse Begum, Department of Human Genetics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA; Department of Biostatistics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA.
Mary Marazita, Department of Human Genetics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA; Division of Oral Biology, Center for Craniofacial and Dental Genetics, School of Dental Medicine, University of Pittsburgh, Pittsburgh, PA, USA.
Vivian Cheung, Departments of Pediatrics and Genetics, University of Pennsylvania, Philadelphia, PA, USA; Howard Hughes Medical Institute, University of Pennsylvania, Philadelphia, PA, USA; Department of Genetics, University of Pennsylvania, Philadelphia, PA, USA.
Eleanor Feingold, Department of Human Genetics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA; Department of Biostatistics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA.
Stephanie L. Sherman, Department of Human Genetics, Emory University School of Medicine, 615 Michael St, Suite 301, Whitehead Bldg, Atlanta, GA 30322, USA
References
- Angell RR. Predivision in human oocytes at meiosis I: a mechanism for trisomy formation in man. Hum Genet. 1991;86:383–387. doi: 10.1007/BF00201839. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Down Syndrome Collaborative Group Parental origin of the extra chromosome in trisomy 21 as indicated by analysis of DNA polymorphisms. N Engl J Med. 1991;324:872–876. doi: 10.1056/NEJM199103283241302. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Weber JL. Characterization of human crossover interference. Am J Hum Genet. 2000;66:1911–1926. doi: 10.1086/302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009;5:e1000648. doi: 10.1371/journal.pgen.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF. Here, there, and everywhere: kinetochore function on holocentric chromosomes. J Cell Biol. 2001;153:F33–F38. doi: 10.1083/jcb.153.6.f33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SB, Allen EG, Oxford-Wright CL, Tinker SW, Druschel C, Hobbs CA, O’Leary LA, Romitti PA, Royle MH, Torfs CP, Sherman SL. The National Down Syndrome Project: design and implementation. Public Health Rep. 2007;122:62–72. doi: 10.1177/003335490712200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Feingold E, Dey SK. Etiology of Down syndrome: evidence for consistent association among altered meiotic recombination, nondisjunction, and maternal age across populations. Am J Med Genet A. 2009;149A:1415–1420. doi: 10.1002/ajmg.a.32932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold TJ, Jacobs PA. Trisomy in man. Annu Rev Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- Hassold T, Sherman S, Hunt PA. The origin of trisomy in humans. Prog Clin Biol Res. 1995;393:1–12. [PubMed] [Google Scholar]
- Hassold T, Abruzzo M, Adkins K, Griffin D, Merrill M, Millie E, Saker D, Shen J, Zaragoza M. Human aneuploidy: incidence, origin, and etiology. Environ Mol Mutagen. 1996;28:167–175. doi: 10.1002/(SICI)1098-2280(1996)28:3<167::AID-EM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Hawley RS, Frazier JA, Rasooly R. Separation anxiety: the etiology of nondisjunction in files and people. Hum Mol Genet. 1994;3:1521–1528. doi: 10.1093/hmg/3.9.1521. [DOI] [PubMed] [Google Scholar]
- Koehler KE, Hawley RS, Sherman S, Hassold T. Recombination and nondisjunction in humans and files. Hum Mol Genet. 1996;5(Spec No):1495–504. doi: 10.1093/hmg/5.supplement_1.1495. [DOI] [PubMed] [Google Scholar]
- Kong A, Barnard J, Gudbjartsson DF, Thorleifsson G, Jonsdottir G, Sigurdardottir S, Richardsson B, Jonsdottir J, Thorgeirsson T, Frigge ML, Lamb NE, Sherman S, Gulcher JR, Stefansson K. Recombination rate and reproductive success in humans. Nat Genet. 2004;36:1203–1206. doi: 10.1038/ng1445. [DOI] [PubMed] [Google Scholar]
- Lamb NE, Freeman SB, Savage-Austin A, Pettay D, Taft L, Hersey J, Gu Y, Shen J, Saker D, May KM, Avramopoulos D, Petersen MB, Hallberg A, Mikkelsen M, Hassold TJ, Sherman SL. Susceptible chiasmate configurations of chromosome 21 predispose to nondisjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14:400–405. doi: 10.1038/ng1296-400. [DOI] [PubMed] [Google Scholar]
- Lamb NE, Feingold E, Savage A, Avramopoulos D, Freeman S, Gu Y, Hallberg A, Hersey J, Karadima G, Pettay D, Saker D, Shen J, Taft L, Mikkelsen M, Petersen MB, Hassold T, Sherman SL. Characterization of susceptible chiasma configurations that increase the risk for maternal nondisjunction of chromosome 21. Hum Mol Genet. 1997;6:1391–1399. doi: 10.1093/hmg/6.9.1391. [DOI] [PubMed] [Google Scholar]
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. Chromosome segregation mechanisms. Genetics. 1974;78:205–213. doi: 10.1093/genetics/78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TR, Feingold E, Yu K, Cheung V, Tinker S, Yadav-Shah M, Masse N, Sherman SL. New insights into human nondisjunction of chromosome 21 in oocytes. PLoS Genet. 2008;4:e1000033. doi: 10.1371/journal.pgen.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. Meiotic nondisjunction does the two-step. Nat Genet. 1996;14:374–376. doi: 10.1038/ng1296-374. [DOI] [PubMed] [Google Scholar]
- Pan H, Ma P, Zhu W, Schultz RM. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev Biol. 2008;316:397–407. doi: 10.1016/j.ydbio.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polk DE, Weyant RJ, Crout RJ, McNeil DW, Tarter RE, Thomas JG, Marazita ML. Study protocol of the Center for Oral Health Research in Appalachia (COHRA) etiology study. BMC Oral Health. 2008;8:18. doi: 10.1186/1472-6831-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson W, Kuckinka BD, Bernascoi F, Brondum-Neilsen K, Christian S, Horsthemke B, Langlois S, Ledbetter D, Michaelis R, Petersen M, Schinzel A, Schuffenhauer S, Schulze A, Hassold T. Maternal meiosis I nondisjunction of chromosome 15: dependence of the maternal age effect on the level of recombination. Hum Mol Genet. 1998;7:1011–1109. doi: 10.1093/hmg/7.6.1011. [DOI] [PubMed] [Google Scholar]
- Ross LO, Maxfield R, Dawson D. Exchanges are not equally able to enhance meiotic chromosome segregation in yeast. Proc Natl Acad Sci USA. 1996;93:4979–4983. doi: 10.1073/pnas.93.10.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuerwald N, Cohen J, Herrera RJ, Sandalinas M, Brenner CA. Association between spindle assembly checkpoint expression and maternal age in human oocytes. Mol Hum Reprod. 2001;7:49–55. doi: 10.1093/molehr/7.1.49. [DOI] [PubMed] [Google Scholar]
- Thomas NS, Ennis S, Sharp AJ, Durkie M, Hassold TJ, Collins AR, Jacobs PA. Maternal sex chromosome non-disjunction: evidence for X chromosome-specific risk factors. Hum Mol Genet. 2001;10:243–250. doi: 10.1093/hmg/10.3.243. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Zhao H, Speed TP, McPeek MS. Statistical analysis of crossover interference using the Chi-square model. Genetics. 1995;139:1045–1056. doi: 10.1093/genetics/139.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]