Abstract
Purpose
The design of electronic health records (EHR) to translate genomic medicine into clinical care is crucial to successful introduction of new genomic services, yet there are few published guides to implementation.
Methods
The design, implemented features, and evolution of a locally developed EHR that supports a large pharmacogenomics program at a tertiary care academic medical center was tracked over a 4-year development period.
Results
Developers and program staff created EHR mechanisms for ordering a pharmacogenomics panel in advance of clinical need (preemptive genotyping) and in response to a specific drug indication. Genetic data from panel-based genotyping were sequestered from the EHR until drug-gene interactions (DGIs) met evidentiary standards and deemed clinically actionable. A service to translate genotype to predicted drug response phenotype populated a summary of DGIs, triggered inpatient and outpatient clinical decision support, updated laboratory records, and created gene results within online personal health records.
Conclusion
The design of a locally developed EHR supporting pharmacogenomics has generalizable utility. The challenge of representing genomic data in a comprehensible and clinically actionable format is discussed along with reflection on the scalability of the model to larger sets of genomic data.
Introduction
The use of diagnostic gene tests within clinical care has risen rapidly in the United States as the cost of genotyping drops precipitously1 and new research supports the value of testing.2 Pharmacogenomics is poised to experience similar growth as many routinely prescribed drugs now have increasingly well validated relationships to adverse events or reduced efficacy when gene variants are present.3–5 Additionally, genotyping technologies have advanced to the point that panel assays involving hundreds of genes are economical, raising the prospect of testing patients once and using stored genomic data repeatedly over a lifetime. With 119 Food and Drug Administration (FDA)-approved drugs currently including germline or tumor pharmacogenomic information in their labels, the potential for a patient to be exposed to a drug with published pharmacogenomic associations is significant. We have previously demonstrated that the opportunities to use variants from a pharmacogenomic panel test are high, with 65% of ambulatory care patients followed longitudinally at our institution exposed to at least one medication with an established pharmacogenomic association within a five year timeframe.6
The promise of translating pharmacogenomics to clinical practice is highly dependent on the ability to communicate the value of genomic data to practicing clinicians and to manage genomic data across a fractured care delivery system.7 The use of health information technology (HIT), including electronic health records (EHRs) and clinical decision support (CDS) is considered indispensable. However, there is little published experience on how to best apply these technologies to clinical pharmacogenomics.8,9 Several NIH-funded consortia are filling the gaps. The Clinical Pharmacogenetics Implementation Consortium (CPIC) has defined and published best practices for knowledge management and clinical decision support. CPIC recommendations are extensively annotated, supported with graded evidence, and freely available.10–14 In addition, the multi-institute consortia Electronic Medical Records and Genomics (eMERGE) Network and the Pharmacogenomics Research Network’s (PGRN) Translational Pharmacogenomics Project (TPP) are actively piloting efforts to integrate genomic information with EHRs, both to facilitate translation of pharmacogenomics to the clinical setting as well as capitalize on the wealth of clinical data contained in the EHR for research.
EHR Design Principles for a Pharmacogenomics Implementation
PREDICT was established as a quality improvement program in 2010 to apply clinically significant gene variants designated by the FDA as pertinent to decisions involving drug selection and dosing.15 EHR features were developed with the expectation that panel-based pharmacogenomic testing will become pervasive, and genomic considerations will routinely influence prescribing. Accordingly, the design of supportive EHR functions have followed ten objectives (Table 1), which seek to give universal, comprehensible and timely access to clinically significant genetic variants. Displays of pharmacogenomics results were created to be highly visible, in an effort to prevent priority results from being “buried” among other laboratory data. Preemptive identification of patients who were expected (based on statistical prediction models) to benefit from panel-based gene variant data to tailor future therapies was incorporated into outpatient workflow. While all gene variant data were stored long term, selective clinically actionable drug-gene combinations that met the burden of evidence for a significant drug genome interaction (DGI), attained institutional approval for release, and for which we had developed CDS logic to guide the physician, were promoted to the EHR. The design for disseminating results features a single source for both genetic variant data and genotype to drug phenotype interpretation, reinforcing the consistency and reliability of genotype reporting. Knowledge and data sources were constructed using service-based software architecture such that both genetic variant data and the DGI knowledge base could be easily updated and the updates would propagate to all linked systems. Finally, the EHR mechanisms for reporting the results and delivering CDS were initially designed to serve a small set of targeted DGIs but easily scale to support a large quantity of pharmacogenomic variants.
Table 1. Design Objectives for a Pharmacogenomics-Enabled Electronic Health Record (EHR).
The following objectives were prospectively addressed in the design and implementation of pharmacogenomics CDS within VUMC’s EHR.
|
A Locally-Developed EHR Perspective
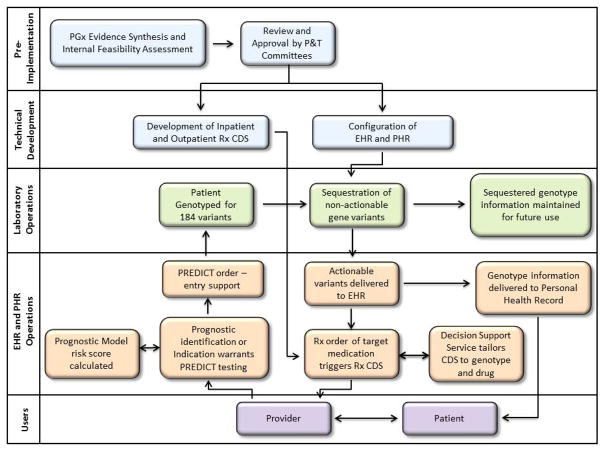
Early in the course of designing the translation of genomic medicine to clinical practice, the biomedical informatics and genomic professions have projected the need to store, manage, interpret, present, and share genetic results.17 Institutions with locally developed clinical information systems are well-suited for pharmacogenomics implementation, as they wield greater control over the underlying architecture and interoperability of their HIT compared to institutions with vendor installed systems. Historically, “homegrown” EHRs have been recognized for providing a test-bed for new HIT ideas, for evaluation of clinical effectiveness, and for providing proof of concept implementations for the wider informatics community.18–21 Additionally, locally developed EHRs have the advantage of access to an engaged user base with whom on-site developers can work directly to obtain feedback and produce iterative improvements that can refine usability and features. However, as clinical information needs have expanded, the financial and human capital required to create, maintain, and certify locally developed EHRs can become daunting, even for large academic medical centers and integrated health systems; few centers have maintained this capability, even with decades of previous investment in technical infrastructure and programming. A second potential disadvantage is that development work is not easily shared or exported, reflecting the current monolithic model of EHRs, in which large ecosystems of HIT from a single vendor or institution are interoperable internally but have limited facility to interact externally. For example, three of the eMERGE pharmacogenomics implementation sites have partial or full development efforts on site; yet these are proceeding relatively independently because of the difficulty in standardizing EHR implementations. The eMERGE and TPP sites do share and disseminate best practices in design and knowledge management, giving other sites or vendors a path to follow. In the remainder of the manuscript, we describe the EHR and related functions designed and implemented to support pharmacogenomics from ordering to clinical use (Figure 2).
Figure 2. EHR Development and Operational Processes.
Pharmacogenomics implementation requires pre-implementation research and assessment, technical development of informatics infrastructure, and integration with laboratory and clinical operations. Accessibility to users, both patient and provider, is integral. (PGx, pharmacogenomics; P&T, Pharmacy and Therapeutics; Rx, prescription; CDS, Clinical Decision Support; EHR, electronic health record; PHR, personal health record)
Implementation of Preemptive and Indication-based Pharmacogenomic Test Ordering
Pharmacogenomic variant data are ideally incorporated into the initial drug selection and dosing; after the patient has achieved a stable dose or drug selection through experience or sequential drug trials, the genomic information contributes diminishing returns to clinical outcomes for the majority of prescribing scenarios currently covered by PREDICT, including the drugs warfarin, simvastatin, clopidogrel, tacrolimus and thiopurines.3–5,22–25 As an example, warfarin dosing is stochastically adjusted in response to serial international normalized ratio (INR) measurements, and thus the clinical impact of genetic data, such as VKORC1 and CYP2C9 variant status, is thought to wane considerably after a stable INR is achieved, an event which generally occurs within the first two weeks of therapy. Similarly, the risk of in-stent thrombosis in CYP2C19 variant patients who are prescribed clopidogrel is highest in the first thirty days following placement of a stent. To this end, the program has prioritized testing in advance of or concurrent with drug initiation to maximize the impact of the genotype data on clinical care.
Consequently, two pharmacogenomics ordering strategies were created - preemptive and indication-triggered testing. For preemptive genotyping, the EHR was modified to display an alert when a statistical risk score for all patients scheduled for clinical encounters in primary care or cardiology. The risk score predicts the probability of receipt of simvastatin, warfarin or clopidogrel over a 3-year time horizon and the trigger score was set to 40% - a threshold that saturated the capacity of the molecular diagnostics laboratory. When a patient’s chart is flagged, the system creates a draft order for the PREDICT test within the outpatient order entry system, which requires confirmation by the treating clinician. For indication based testing, the PREDICT panel test was incorporated into order sets or pre-procedure planning prior to cardiac catheterization (to capture catherization patients who receive intracoronary stents and antiplatelet therapy such as clopidogrel) and certain orthopedic procedures (e.g., joint replacements) for which warfarin based anticoagulation is standard. Notably, pre-emptive genotyping eliminates delays in obtaining the genotype, which has a minimum 2 day and median 5 day turnaround time.
We suggest significant cost savings using a pre-emptive panel-based genotyping strategy compared to serial single gene tests, given the decreasing cost of genotyping, possible exposure to multiple different medications with pharmacogenomic indications, and very high cost of severe adverse events.6,26,27 Use of multiplexed gene tests over a patient’s lifetime is likely to be less expensive relative to the potential benefit, particularly in patients with a common set of cardiovascular risk factors likely to need associated therapies. However, no health economic studies have determined the value of panel-based genetic tests outside of oncology, and there is a paucity of evidence relating panel-based genetic tests to health care spending. VUMC has supported the PREDICT program costs with institutional funds, including assay costs, reagents, labor, instrumentation for processing, empiric research among patients and providers, development of patient informational materials, decision-support tools which provide point-of-care interventions and drug/dosing guidance based on test results, and education and training given the associated dearth of knowledge and familiarity among prescribers.15 A key goal of this investment is to catalyze further pharmacoeconomic analyses of this approach.
EHR Storage Model for Sequestration and Repository
National data standards for genetics are in early stages; a model to exchange genetic testing results is proposed by Health Level 7 (HL7)28 with contributions by PGRN-affiliated academic groups29,30 and EHR vendors.31,32 In the absence of established standards in 2010, and to meet the immediate needs of the program, PREDICT developers created a coded storage model to meet local requirements for clinical decision support and distribution to multiple clinical information systems. Future adaptation to emerging standards such as HL7 is planned to support communication with external systems. Genetic variant data produced by the Illumina VeraCode® Absorption, Distribution, Metabolism, and Excretion (ADME) Core Panel for PREDICT are provided either as a Portable Document Format (PDF) or as plain text. As the former does not provide computable results, automatic parsing of the text format is required to extract the gene name, variant result in star nomenclature, and a call rate, which indicates the ability of the panel to yield a result at a specific variant. In the event of a call rate less than 98.7%, the test result is manually reviewed and generally retested by Molecular Diagnostics Laboratory staff; otherwise it is released to an Oracle® database, which initially sequesters all results from the main EHR storage.
The Oracle® database is exposed to downstream systems through a filtered view limited to actionable approved variants. An automated script queries the filtered database view hourly to extract new or updated entries and, if discovered, creates a new or updated entry in the genotype section of the Patient Summary Service (PSS), a central web service that is available to all components of the EHR and CDS (Figure S1). Examples of four components of the EHR that use PSS are shown in Figure 3. PSS serves as a single source of patient specific knowledge for medications, diagnoses, allergies, and other significant family and social history, and this infrastructure was expanded to manage genomic variants and their interpretations.
Figure 3. Task-specific views of genomic results present in the EHR.
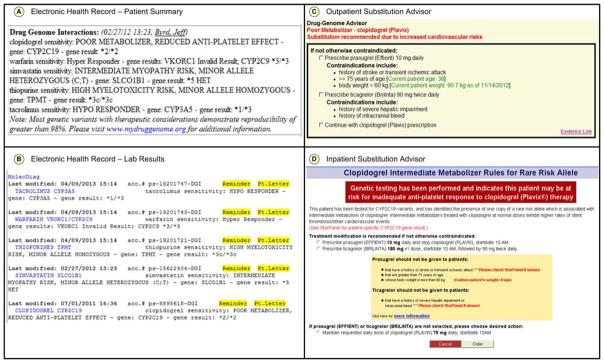
The Patient Summary, which serves as the front page of each patient’s record, includes a Drug Genome Interaction section detailing the patient’s genotype in star allele nomenclature as well as phenotype and implcations for prescribing (Panel A). Genomic results and phenotypes are also available in the Lab Results section of the EHR (Panel B). When a drug is ordered for a patient with an actionable genotype, Clinical Decision Support (CDS), such as the representative Outpatient Substitution Advisor, is presented to the ordering clinician (Panel C). Similarly, parallel mechanisms offer CDS in the inpatient setting (Panel D).
Genotype to Phenotype Translation
Although the advantages of multiplexed genetic testing are becoming increasingly apparent, there are clear challenges associated with managing panel-based genetic data. Raw genotype output is not typically delivered in a standardized format and does not include phenotypic interpretations, which may be drug and patient specific. In order for the genetic results to be useful for clinical implementation through PREDICT, results were individually categorized to create a translation layer, which assigns a coded phenotype category and generates the DGI text string used for display in the EHR and CDS, when triggered (Figure S1). The assigned phenotypes are drawn from a translation table which relates the raw genotype text string to drug and metabolism effect categories (Table 2). Translations are made based on actionable variants, defined as variants that have been reviewed and approved for clinical implementation by the VUMC’s Pharmacy and Therapeutics (P&T) committee; however, a large proportion of variants on the PREDICT platform are not actionable due to insufficient evidence. For CYP2C9, for example, only 2 of the 13 variants tested on the platform have been approved for implementation. Genetic variants that are not deemed actionable are sequestered within a separate database, outside of the EHR, and are not accessible by patients or providers. The genotype data will only be released into the EHR as new genotypes are deemed actionable and new DGIs are incorporated into clinical care.15
Table 2. Example genotype to phenotype translations.
Translation entries exist for all encountered genotype combinations and phenotype categories as below, which ultimately drive decision support. There are a total of 971 unique, observed diplotype genotype entries currently, mapping to 19 phenotypes.
| Gene name(s) | Raw Genotype Result | Simplified Genotype | Phenotype | |
|---|---|---|---|---|
| Phenotype Category | Phenotype Detail | |||
| CYP2C19 | (*17 VAR) | *17/*17 | Clopidogrel Sensitivity | rapid metabolizer |
| CYP2C19 | (*4 VAR) | *4/*4# | Clopidogrel Sensitivity | poor metabolizer |
| SLCO1B1 | *1A/*1A | *1/*1 | Simvastatin Sensitivity | normal risk |
| SLCO1B1 | *1B HET;(*2 HET);*5 HET | *1/*5 | Simvastatin Sensitivity | intermediate risk |
| VKORC/CYP2C9 | VKORC1 -1639G>A No Call, CYP2C9 *1A/*1A | VKORC1 indeterminate; CYP2C9 *1/*1 | Warfarin Sensitivity | normal responder |
| VKORC/CYP2C9 | VKORC1 -1639G>A No Call, CYP2C9 *2 HET;(*11 HET);*15 No Call | VKORC1 indeterminate; CYP2C9 *1/*2 | Warfarin Sensitivity | hyper responder |
| VKORC/CYP2C9 | VKORC1 c.-1639 VAR, CYP2C9 *2 HET | VKORC1 -1639 AA; CYP2C9 *1/*2 | Warfarin Sensitivity | hyper responder |
| VKORC/CYP2C9 | VKORC1 NMD, CYP2C9 *2 No Call | VKORC1 -1639 GG; CYP2C9 indeterminate | Warfarin Sensitivity | indeterminate |
denotes a Rare Variant
The model for the current genotype to phenotype translation table is to assign a value to every result produced by the ADME platform, even if rare. For variants without sufficient evidence to be deemed actionable, a category labeled “indeterminate” was created (see Table 2). For purposes of CDS implementation, no change to usual care is recommended for indeterminate genotypes. Other pharmacogenomics implementation sites have used similar approaches,33 and several consortia have been established to develop and maintain consistent guidelines for translation of genotype test results, including CPIC and the TPP.34 The translated interpretations are viewable by providers via the EHR and incorporated into the EHR advisors; however they are not tailored to background level of provider pharmacogenomic knowledge. Thus, developing phenotype interpretations that are meaningful and clinically useful for providers presents its own set of challenges.
EHR Representations of Genotype and Phenotype
The centralized service architecture of the genotype to phenotype translation layer allows simultaneous population of multiple clinical information systems, supporting the clinician through EHR views and patients through their access to a PHR hosted on a patient portal (Figure S2). For each client system, the service responds to requests for new or updated genomic results. Whenever a phenotype assignment is changed (such as when CYP2C19*3 heterozygotes were added to an actionable “poor metabolizer” status for clopidogrel), the translation table within the service is updated manually, which triggers automatic revision of the results displayed in the EHR and PHR. Following the principle of high visibility and universal access, four task-specific views of genomic results are supported in the EHR (Figure 3; Panels A–D). First, the program team created a space for genomic variants to be visible within the patient summary that serves as the “front page” of the electronic chart and adjacent to the medication list. Much like an “allergy” section, this space is intended to communicate significant genomic variant information while a target medication was contemplated and prior to initiating a prescription. During review of the design, clinicians and the Pharmacy and Therapeutics committee required the display of any pharmacogenomic result whether indicating a variant or not, such that there was a quick method of determining if a patient had already been tested. This current presentation format does not scale to many implemented DGIs, thus a redesign is in progress.
Secondly, the phenotype delivered by PSS triggers clinical decision support within the outpatient e-prescribing environment as well as the inpatient computerized physician order entry (CPOE) environment when a prescription or medication order conflicts with the phenotype status (Figure S1). For example, providers prescribing clopidogrel in the presence of an intermediate or poor metabolizer phenotype will receive therapeutic guidance to switch to an alternate antiplatelet therapy (Table S1). Finally, new pharmacogenomic information is released from the laboratory. This mechanism (along with the patient summary) supported reconsideration of patient therapy whenever new DGIs are released. Among the challenges encountered, EHR designers must decide how to represent risk; the potential impact of phenotype labeling and the utility of adding quantitative risk measures to these brief interpretations are currently unknown.
Display of Genomic Results in Personal Health Records
PREDICT genetic results are released into the patient’s EHR to guide therapy and clinical decision making. In addition, given the burgeoning body of literature suggesting the importance of empowering patients with health information and increased efforts surrounding the HITECH act,35 PREDICT genetic results have also been made available to patients through VUMC’s patient portal, My Health At Vanderbilt (MHAV), a resource that allows patients to view EHR data, message their healthcare providers, and read general health information tailored to their medical history. Through PREDICT, we have added content in MHAV related to a patient’s genetic test results (Figure S2). The first release of genomic results contained a simplified copy of what was displayed to providers in the EHR: the genetic test result with a brief interpretation, e.g., “CYP2C19, one copy of the variant, poor metabolizer of clopidogrel.” Feedback from focus groups overwhelmingly indicated that patients preferred detailed, descriptive background information related to drug side effects and how genetics may affect a patient’s risk for adverse events. Based on this feedback, more comprehensive narratives with graphics are being developed and provided at a seventh grade reading level.
Evolution of PREDICT Since Launch
PREDICT was launched in September 2010 with genotype-tailored dosing guidance for clopidogrel.15 The decision to focus on clopidogrel was made following an FDA “black box” warning alerting physicians and patients to the role of CYP2C19 variants in medication response.3 The FDA did not indicate how to incorporate CYP2C19 variants into clinical decision making; however an efficacious alternative, the antiplatelet drug prasugrel, was not affected by CYP2C19 genotype.36–39 Thus, the initial clopidogrel advisor was designed to activate when patients were homozygous for CYP2C19*2 or *3 allele and displayed recommendations to increase clopidogrel maintenance dose to 150mg daily or switch to prasugrel barring any contraindications.
Since launching the program, over 75 manuscripts have been published with the potential to influence genotype to phenotype mappings or the content of the clopidogrel CDS. Following publication of a large meta-analysis3 and our internal analysis,40 both showing significant reduction in clopidogrel efficacy in individuals heterozygous for CYP2C19 variants, we added such individuals to the program. Moreover, new, rare CYP2C19 variants were determined to impair clopidogrel metabolism,14 and new, effective alternatives to clopidogrel were released on the market. These advances warranted modifications to both the genotype-phenotype translations as well as the clopidogrel CDS recommendations. Updating the knowledgebase and changing the user interface for the clinical decision support to add additional choices required comparatively less effort than the initial development partially because of the separation of these components into Enterprise Services (Figure S1). However, modifications to the phenotype map often changed the risk status of patients who were already genotyped, requiring providers to reconsider the initial drug selection or dosing. For each of these scenarios, we organized a communication plan, identifying affected patients and manually notifying providers using secure electronic messaging within the EHR.41
The program continues to expand and incorporate CDS for additional DGIs into the EHR, including recommendations for warfarin, simvastatin, thiopurines and tacrolimus. Two of the released DGIs are relevant to pediatric populations and required the development of guidelines applicable to both adult and pediatric populations, as well as DGI specific suppression of genetic results and EHR advisors for those DGIs that were not applicable to a pediatric population (e.g., warfarin advisors). Infrastructure available at the time of these deployments allowed for a simple, alternative set of text for adult and pediatric patients. This required changes in both the database model as well as the presentation layer to determine, based on the age of the patient, which text was appropriate for display.
Discussion
The design and implementation of EHR features to support a large multi-DGI pharmacogenomics program required iterative refinements, in part because there is little published guidance on how to leverage HIT to translate genomic medicine to clinical practice. We described our initial design choices and subsequent changes in an effort to inform other institutions that are contemplating or have initiated a similar effort. One of the major successes in the last five years is the formation of cooperative efforts from pioneering institutions associated with the PGRN to organize and curate the pharmacogenomics knowledgebase relating genomic variation to therapeutic decision making in the form of clear, accessible guidelines.10–14 Similar efforts to share implementation practices among members of the TPP and the eMERGE network have made substantial progress.34 Overall, the gap between the conceptual model of personalized medicine and actual clinical implementation is closing but remains wide for most health systems.42 The PREDICT implementation approach is distinct because of the scope of drug-genome interactions that are targeted for adult and pediatric populations, the duration of the program, and the emphasis on preemptive testing. Additionally, the ability to leverage on-site developers familiar with the locally developed EHR allowed efficient implementation. Although the specific form of this implementation is institution specific, the abstracted challenges described in this manuscript are generalizable.33
We found the major challenges for incorporating PREDICT relate to the complexity of raw genotype data and the lack of existing standards to store and transmit genomic data. Genotyping platforms do not output results in a coded reference standard and are not accompanied by interpretations. Integrating with downstream EHR tasks required parsing of the gene result report and a translation layer able to contend with undefined variants. Manufacturers of genotyping instruments can improve the ease of implementation by adhering to coded standards (as they are developed) and providing more detailed documentation of potential genomic output. Secondly, we sought to preemptively map all variants but discovered rare variants that were undefined; an automated process within the EHR infrastructure to track and examine new, undefined variants would be valuable to ensure the timely updating of a translation table and could eventually serve as a tool for discovery of potential variant function. Thirdly, EHR integration of genomic data requires a process to manage the release of new or materially updated drug-genome data as thousands of patient records are affected. Such releases also require significant communication and education efforts to inform providers of emerging or changing evidence. Finally, the scalability of EHR integration is challenged by several technical factors, including limited screen ‘real-estate’ to display significant variants and inflexible models of displaying results that may not yet be pertinent to patient care.
Limitations of the PREDICT EHR Model
The application of pharmacogenomic testing to clinical care is complex and requires established and comprehensive infrastructures to support implementation. With quickly advancing genotyping (and genome sequencing) technologies, emerging evidence, and changes in therapies, these infrastructures must be prepared to accommodate rapid modifications and an explosion in genetic variants. While PREDICT represents one viable model for implementation of pharmacogenomic information into the EHR, there are limitations and challenges that offer opportunity for improvement and fine-tuning of the program. Despite attention to the succinct and understandable interpretation of genomic results, the EHR displays may not be sufficient for providers without specific pharmacogenomic training. The brief interpretations provided presume a baseline knowledge of pharmacogenomics and are not intended to be educational. Furthermore, PREDICT affects providers in multiple specialties, creating even greater provider education challenges. The provider EHR displays are not currently customizable by specialty, health care role or baseline knowledge, but such flexibility may be needed as the number of implemented DGIs increase. Moreover, results may be returned outside of the context of a clinical encounter, for example, when a DGI is released into the EHR many years after the patient’s initial genetic testing. Similarly, while significant effort has been made to develop understandable and meaningful PHR displays, further research is warranted to elucidate more effective methods of communicating complex genomic information to patients. Additionally, there is currently no infrastructure in place to automatically and reliably deliver genetic results to providers outside of Vanderbilt’s EHR system; thus, some patients may be tested through PREDICT but not benefit from future decision support after they return to their primary providers outside of the Vanderbilt network. While PREDICT recommendations are based on the most up-to-date evidence and expert opinions, incorporating genomic information with clinically relevant non-genomic factors in CDS recommendations is currently outside of the scope of the program.
Pharmacogenomic Adoption: the way forward
The challenges and lessons learned from PREDICT implementation highlight the need for improved EHR integration and interoperability. For patients not receiving care exclusively at VUMC, improved communication and transfer of genetic results to external providers is the first step towards this integration and is necessary to advance genotype-tailored decision-making. Clinical notification of high-priority genetic results (e.g., those associated with life-threatening adverse events or with prolonged clinical utility) could be achieved by leveraging national electronic messaging infrastructures and will pave the way for full EHR integration. Pharmacogenomic adoption is limited by provider knowledge and usability of EHR-displayed genomic information. Maintaining awareness of evolving pharmacogenomic evidence and emerging therapies and incorporating this information into clinical practice requires procedures for systematic evidence review and an informatics infrastructure that enables prompt modifications of genomic advisors within the EHR system.15 Improved advisors and information displays that can be modified easily and incorporated within the EHR with very little informatics support will be vital as existing DGIs are updated and additional DGIs continue to be implemented. Moreover, portability of internally developed CDS across EHR systems will be critical for dissemination of clinical pharmacogenomics. We believe that use of internet-based web services to encapsulate genetic results and securely communicate relevant guideline-based recommendations and knowledge across institutional boundaries will compel efficient and widespread clinical adoption of pharmacogenomic evidence in real-world medical practice.
Figure 1. PREDICT EHR Development Timeline.
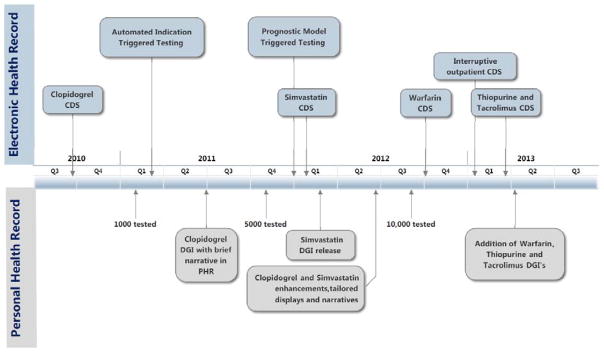
PREDICT has undergone a 4-year process of design, implementation, and iterative refinement. Several milestones, including new drug genome interaction implementation as well as high-impact EHR design features, are highlighted. (DGI, drug genome interaction; CDS, Clinical Decision Support; EHR, electronic health record; PHR, personal health record)
Acknowledgments
Sources of funding supporting this work include funding from Vanderbilt University, the Centers for Disease Control and Prevention (U47CI000824), the National Heart Lung and Blood Institute and the National Institute for General Medical Sciences (U19HL065962), the National Human Genome Research Institute (U01HG006378) and the National Center for Advancing Translational Sciences (UL1 RR024975). Its contents are solely the responsibility of the authors and do not necessarily represent official views of the Centers for Disease Control and Prevention or the National Institutes of Health.
Footnotes
Conflict of Interest Notification
The authors declare no conflict of interest.
References
- 1.Gullapalli RR, Desai KV, Santana-Santos L, Kant JA, Becich MJ. Next generation sequencing in clinical medicine: Challenges and lessons for pathology and biomedical informatics. J Pathol Informatics. 2012;3 doi: 10.4103/2153-3539.103013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson JA, Burkley BM, Langaee TY, et al. Implementing Personalized Medicine: Development of a Cost-Effective Customized Pharmacogenetics Genotyping Array. Clin Pharmacol Ther. 2012;92:437–439. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mega JL, Simon T, Collet JP, et al. Reduced-Function CYP2C19 Genotype and Risk of Adverse Clinical Outcomes Among Patients Treated With Clopidogrel Predominantly for PCI: A Meta-Analysis. Jama J Am Med Assoc. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The International Warfarin Pharmacogenetics Consortium. Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SEARCH Collaborative Group. SLCO1B1 Variants and Statin-Induced Myopathy A Genomewide Study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 6.Schildcrout JS, Denny JC, Bowton E, et al. Optimizing Drug Outcomes Through Pharmacogenetics: A Case for Preemptive Genotyping. Clin Pharmacol Ther. 2012;92:235–242. doi: 10.1038/clpt.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilke R, Xu H, Denny J, et al. The Emerging Role of Electronic Medical Records in Pharmacogenomics. Clin Pharmacol Ther. 2011;89:379–386. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Donnell PH, Bush A, Spitz J, et al. The 1200 Patients Project: Creating a New Medical Model System for Clinical Implementation of Pharmacogenomics. Clin Pharmacol Ther. 2012;92:446–449. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson JA, Cavallari LH, Beitelshees AL, et al. Pharmacogenomics: Application to the Management of Cardiovascular Disease. Clin Pharmacol Ther. 2011;90:519–531. doi: 10.1038/clpt.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Relling M, Klein T. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Thiopurine Methyltransferase Genotype and Thiopurine Dosing. Clin Pharmacol Ther. 2011;89:387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilke RA, Ramsey LB, Johnson SG, et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012;92:112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson J, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 Genotypes and Warfarin Dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy: 2013 Update. Clin Pharmacol Ther. 2013 doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulley JM, Denny JC, Peterson JF, et al. Operational Implementation of Prospective Genotyping for Personalized Medicine: The Design of the Vanderbilt PREDICT Project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny JC, Giuse DA, Jirjis JN. The Vanderbilt Experience with Electronic Health Records. Semin Colon Rectal Surg. 2005;16:59–68. [Google Scholar]
- 17.Altman RB, Klein TE. Challenges for biomedical informatics and pharmacogenomics. Annu Rev Pharmacol Toxicol. 2002;42:113–133. doi: 10.1146/annurev.pharmtox.42.082401.140850. [DOI] [PubMed] [Google Scholar]
- 18.Chaudhry B, Wang J, Wu S, et al. Systematic Review: Impact of Health Information Technology on Quality, Efficiency, and Costs of Medical Care. Ann Intern Med. 2006;144:742–752. doi: 10.7326/0003-4819-144-10-200605160-00125. [DOI] [PubMed] [Google Scholar]
- 19.Silow-Carroll S, Edwards JN, Rodin D. Using electronic health records to improve quality and efficiency: the experiences of leading hospitals. Issue Brief Commonw Fund. 2012;17:1–40. [PubMed] [Google Scholar]
- 20.Shortliffe EH, Davis R, Axline SG, et al. Computer-based consultations in clinical therapeutics: explanation and rule acquisition capabilities of the MYCIN system. Comput Biomed Res Int J. 1975;8:303–320. doi: 10.1016/0010-4809(75)90009-9. [DOI] [PubMed] [Google Scholar]
- 21.Zeng Q, Cimino JJ, Zou KH. Providing Concept-oriented Views for Clinical Data Using a Knowledge-based System. J Am Med Informatics Assoc Jamia. 2002;9:294–305. doi: 10.1197/jamia.M1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferder NS, Eby CS, Deych E, et al. Ability of VKORC1 and CYP2C9 to predict therapeutic warfarin dose during the initial weeks of therapy. J Thromb Haemost. 2010;8:95–100. doi: 10.1111/j.1538-7836.2009.03677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez AH, Shi Y, Schildcrout JS, et al. Predicting warfarin dosage in European Americans and African Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012;13:407–418. doi: 10.2217/pgs.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgs JE, Payne K, Roberts C, Newman WG. Are patients with intermediate TPMT activity at increased risk of myelosuppression when taking thiopurine medications? Pharmacogenomics. 2010;11:177–188. doi: 10.2217/pgs.09.155. [DOI] [PubMed] [Google Scholar]
- 25.Hesselink DA, van Schaik RHN, van der Heiden IP, et al. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003;74:245–254. doi: 10.1016/S0009-9236(03)00168-1. [DOI] [PubMed] [Google Scholar]
- 26.NHGRI. DNA Sequencing Costs. genomegov. at http://www.genome.gov/sequencingcosts.
- 27.Ghate SR, Biskupiak J, Ye X, Kwong WJ, Brixner DI. All-cause and bleeding-related health care costs in warfarin-treated patients with atrial fibrillation. J Manag Care Pharm Jmcp. 2011;17:672–684. doi: 10.18553/jmcp.2011.17.9.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shabo Shvo A. Meaningful use of pharmacogenomics in health records: semantics should be made explicit. Pharmacogenomics. 2010;11:81–87. doi: 10.2217/pgs.09.161. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Q, Freimuth RR, Lian Z, et al. Harmonization and semantic annotation of data dictionaries from the Pharmacogenomics Research Network: A case study. J Biomed Inform. 2013;46:286–293. doi: 10.1016/j.jbi.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samwald M, Coulet A, Huerga I, et al. Semantically enabling pharmacogenomic data for the realization of personalized medicine. Pharmacogenomics. 2012;13:201–212. doi: 10.2217/pgs.11.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman M, Arnoldi C, Chuang I. The clinical bioinformatics ontology: a curated semantic network utilizing RefSeq information. Pac Symp Biocomput Pac Symp Biocomput. 2005:139–150. doi: 10.1142/9789812702456_0014. [DOI] [PubMed] [Google Scholar]
- 32.Noy NF, Rubin DL, Musen MA. Making Biomedical Ontologies and Ontology Repositories Work. Ieee Intell Syst. 2004;19:78–81. [Google Scholar]
- 33.Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther. 2012;92:563–566. doi: 10.1038/clpt.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shuldiner AR, Relling MV, Peterson JF, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Overcoming Challenges of Real-World Implementation. Clin Pharmacol Ther. 2013 doi: 10.1038/clpt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blumenthal D. Launching HITECH. N Engl J Med. 2010;362:382–385. doi: 10.1056/NEJMp0912825. [DOI] [PubMed] [Google Scholar]
- 36.Marcucci R, Gori AM, Paniccia R, et al. Cardiovascular Death and Nonfatal Myocardial Infarction in Acute Coronary Syndrome Patients Receiving Coronary Stenting Are Predicted by Residual Platelet Reactivity to ADP Detected by a Point-of-Care Assay A 12-Month Follow-Up. Circulation. 2009;119:237–242. doi: 10.1161/CIRCULATIONAHA.108.812636. [DOI] [PubMed] [Google Scholar]
- 37.Hulot JS, Collet JP, Silvain J, et al. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration: a systematic meta-analysis. J Am Coll Cardiol. 2010;56:134–143. doi: 10.1016/j.jacc.2009.12.071. [DOI] [PubMed] [Google Scholar]
- 38.Trenk D, Hochholzer W, Fromm MF, et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol. 2008;51:1925–1934. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 39.Wiviott SD, Braunwald E, McCabe CH, et al. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2007;357:2001–2015. doi: 10.1056/NEJMoa0706482. [DOI] [PubMed] [Google Scholar]
- 40.Delaney JT, Ramirez AH, Bowton E, et al. Predicting Clopidogrel Response Using DNA Samples Linked to an Electronic Health Record. Clin Pharmacol Ther. 2012;91:257–263. doi: 10.1038/clpt.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jirjis J, Weiss JB, Giuse D, Rosenbloom ST. A Framework for Clinical Communication Supporting Healthcare Delivery. AMIA Annu Symp Proc. 2005;2005:375–379. [PMC free article] [PubMed] [Google Scholar]
- 42.Starren JWM. Crossing the omic chasm: A time for omic ancillary systems. JAMA. 2013;309:1237–1238. doi: 10.1001/jama.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]