Abstract
Philadelphia chromosome-positive (Ph+) AML is a controversial diagnosis, as others propose it represents CML in blast phase (CML-BP). NPM1 mutations occur in 25-35% of AML but are absent in CML patients. Conversely, ABL1 mutations occur in 25% of Imatinib-naïve CML-BP but are not described in AML patients. We analyzed for NPM1 and ABL1 mutations in 9 Ph+ AML and 5 CML-BP patients initially presented in BP. In 6 Ph+ AML cases, we screened for a panel of gene mutations using Sequenome®-based methods including AKT1, AKT2, AKT3, BRAF, EGFR, GNAQ, GNAS, IDH1, IDH2, KRAS, MET, NRAS, PIK3CA, and RET. Two of 9 (22%) Ph+ AML patients had NPM1 mutations and were alive 36 and 71 months after diagnosis. All Ph+ AML were negative for ABL1 and other gene mutations. One (20%) CML-BP patients had ABL1 mutation; no patients had NPM1 mutations. These data suggest that Ph+ AML is distinct from CML-BP.
Keywords: acute myeloid leukemia, BCR-ABL1, blast phase, chronic myelogenous leukemia, NPM1, Philadelphia chromosome
INTRODUCTION
Patients with Ph+ acute myeloid leukemia (AML) account for 0.5-3.0% of all AML cases and, by definition, have no evidence of chronic myelogenous leukemia (CML)either before the onset of AML or after successful therapy for AML. 1-5 Nevertheless, the pathogenesis of Ph+ AML remains controversial as many investigators believe these neoplasms represent de no myeloid blast phase of CML. The most recent version of the World Health Organization (WHO) classification does not recognize AML associated with t(9;22)(q34;q11.2)/BCR-ABL1.6 In a review article discussing the rationale for the WHO classification, Vardiman and colleagues wrote, “Although BCR-ABL1–positive AML has been reported, criteria for its distinction from CML initially manifesting in a blast phase are not entirely convincing.”7 Therefore, other means of distinguishing Ph+ AML from CML-BP, if they truly are different entities, would be helpful.
Gene mutations are known to occur with various types of AML and are involved in pathogenesis. Nucleophosmin (NPM1), mapped to chromosome 5q35, is one of the most commonly mutated genes in AML patients..8-10NPM1 is mutated in 25-35% of all AML cases, with a higher frequency of 45-64% in AML patients with normal karyotype.10NPM1 mutations, if not counterbalanced by other gene mutations (e.g. FLT3), are associated with favorable prognosis.8,9 In contrast, others have shown that NPM1 consistently is wild type in patients with CML, including CML-BP.8-12
ABL1 mutations occur in a proportion of patients with CML as well as Ph+ acute lymphoblastic leukemia.13 Furthermore, ABL1 mutations are more common in CML-BP than in CML in chronic phase, with a frequency as high as 80% in CML-BP patients.14-16 Of note, ABL1 mutations are not restricted to patients with prior Imatinib exposure. In a study of unselected Imatinib-naïve CML-BP patients, 5 of 19 patients had ABL1 mutations.15ABL1 mutations have not been reported Ph+ AML patients .
In this study, we hypothesized that analysis of NPM1 and ABL1 genes, often mutated in AML and CML-BP patients, respectively, might yield insights into the relationship between Ph+ AML and CML-BP. We also screened 6 cases of Ph+ AML for a number of other gene mutations that our laboratory screens for in the workup of malignant neoplasms of all types. Our results suggest that Ph+ AML is a clinicopathologic entity distinct from CML-BP.
MATERIAL AND METHODS
Study Group
Following approval by the Institutional Review Board, the database of the Department of Hematopathology at The University of Texas MD Anderson Cancer Center was searched for Ph+ AML patients seen from January 1998 until the time of writing. Cases were excluded from this study if there was: a clinical history of an antecedent hematologic disorder suggestive of CML in chronic or accelerated phase; evidence of a CML-like picture following therapy for AML; a history of chemotherapy and/or radiation therapy; presence of splenomegaly or basophilia (defined as >2% of basophils in peripheral blood) suggestive of a myeloproliferative neoplasm; and evidence of biphenotypic or bilineage leukemia as defined by the 2008 WHO classification.6 Ph+ AML patients formed a study group.
For comparison, we searched for patients who were diagnosed with CML BC at initial presentation during the same period of time and had no clinical history of an antecedent hematologic disorder suggestive of CML in chronic or accelerated phase. The comparison group included patients with confirmed t(9;22)(q34;q11.2) and peripheral blood basophilia combined with either splenomegaly or additional cytogenetic abnormalities considered to be typical for CML (trisomy 8, isochromosome of the long arm of chromosome 17, trisomy 19, or an extra copy of Ph). Cases with lymphoid subtype of CML BP and cases fulfilling diagnostic criteria of the 2008 WHO classification for acute biphenotypic or bilineage leukemia6 were excluded.
In addition, we also identified a group of patients who was diagnosed with CML-BP during the same period of time at our institution and had a well documented antecedent chronic phase during the same study period. Cases with lymphoid subtype of CML BP and cases fulfilling diagnostic criteria of the 2008 WHO classification for acute biphenotypic or bilineage leukemia6 were excluded.
Morphologic, Cytochemical, and Immunophenotypic, and Cytogenetic Analysis
Bone marrow aspirate smears were stained with Wright-Giemsa and blast counts were performed manually. Bone marrow aspirate smears were also analyzed cytochemically for myeloperoxidase (MPO) and alpha-naphthyl butyrate esterase using previously reported methods.17
Four-color flow cytometric immunophenotypic immunophenotypic analysis was performed using a FACScalibur cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA) and analyzed using the CellQuest software package (Becton Dickinson Immunocytometry Systems) as previously described.17 Antibodies specific for following antigens were used: CD3, CD7, CD10, CD13, CD19, CD20, CD33, CD34, CD45, CD56, CD64, CD117, HLA-DR, MPO, and TdT. All antibodies were obtained from Becton-Dickinson Biosciences (San Jose, CA). Blasts were gated for analysis using CD45 expression and light side-scatter characteristics. Blasts were considered positive for antigens using an arbitrary but standard cutoff level of at least 20% blasts that expressed the antigen compared with an isotype control.
Conventional cytogenetic analysis was performed on bone marrow aspirate specimens at the time of initial presentation using standard GTG-banding as described previously.18
NPM1 and ABL1 Gene Mutation Analysis
Exon 12 of NPM1 was amplified using PCR and two primers 5'-GATGTTGAACTATGCAAAGAGACA-3' (forward) and 5'-AACCAAGCAAAGGGTGGAGTT-3' (reverse). PCR products were purified using the MinElute PCR purification Kit (QIAGEN, Valencia, CA) and directly sequenced using the reverse primer GGCATTTTGGACAACACA and fluorescent dye chain-terminator chemistry (Sanger sequencing) with an ABI PRISM 3100 or 3130 genetic analyzer (Applied Biosystems, Foster City, CA). The ABI GeneMapper software program (Applied Biosystems) was used to analyze the raw NPM1 gene mutation analysis data.
Mutational analysis of the ABL1 kinase domain was performed using nested PCR followed by Sanger sequencing as described previously. 19 In the first round, the BCR-ABL1 fusion transcripts b2a2, b3a2, and e1a2 were amplified. In the second round, the analysis consisted of two separate PCRs that covered codons 221-380 and codons 350-500 of the ABL1 kinase domain, respectively. PCR products were sequenced using standard dideoxy chain-termination DNA sequencing with an ABI PRISM 3700 genetic analyzer (Applied Biosystems). The results were analyzed using the SeqScape and sequencing analysis software programs (Applied Biosystems). All ABL1 mutations were confirmed by sequencing of forward and reverse strands, with a sensitivity rate of 20% mutation-bearing transcripts in the analyzed population established in periodic dilution studies.
Analysis of Other Genes in Ph+ AML
BCR-ABL1 fusion transcripts were assessed using real-time quantitative reverse transcriptase (RT)-PCR analysis of peripheral blood and/or bone marrow aspirate specimens as described previously.20
The FLT3 gene was assessed for internal tandem duplication (ITD) and the codon 835/836 point mutation using polymerase chain reaction (PCR)-based methods also as described previously.21 PCR-based DNA high-resolution melting curve analysis was used to screen for IDH gene mutations as described previously.22 Mutations in codons 87-138 of exon 4 of IDH1 and exon 4 of IDH2 were specifically assessed.
A PCR-based DNA primer extension analysis with a MassARRAY system (Sequenom, San Diego, CA, USA) was used to screen for a number of gene mutations as described previously.23 The genes that were assessed are part of a routine screening panel applied to the study of all newly diagnosed tumors. The panel is focused on known “hot spots” for gene mutation and includes: mutations in codons 17, 49, 173, 517, 179, and 536 of the AKT1 gene; codons 17, 49, 175, and 523 of the AKT2 gene; codons 17, 49, 171, and 511 of the AKT3 gene; codons 464, 466, 469, 594, 597, 600, and 601 of the BRAF gene; codon 858 of the EGFR gene; codon 209 of the GNAQ gene; codons 201 and 227 of the GNAS gene; codon 132 of the IDH1 gene; codon 172 of the IDH2 gene; codons 12, 13, 61, and 146 of the KRAS gene; codons 375, 848, 988, 1010, 1112, 1124, 1248, 1253, and 1268 of the MET gene; codons 12, 13, and 61 of the NRAS gene; codons 60, 88, 110, 111, 345, 405, 418, 420, 453, 539, 542, 545, 546, 909, 1021, 1025, 1043, 1046, 1047, and 1049 of the PIK3CA gene; and codon 918 of the RET gene were detected
RESULTS
Clinical data
The clinical data of the study group and the comparison group are listed in Table 1. Among 2241 AML patients at our institution during the study period, we identified 12 (0.54%) patients Ph+ AML patients (0.54%). DNA extracted from bone marrow samples at presentation was available for 9 patients, the study group. The Ph+ AML patients were five men and four women, ages 22 to 76 years (median, 57). During the study period, we also identified 18 patients with CML who presented in myeloid blast phase and had no documented chronic phase or accelerated phase (CML-BP group). A total of 5 patients with readily available DNA extracted from bone marrow samples obtained at the time of CML-BP diagnosis were included in this study. CML-BP group had four men and one woman, ages 26 to 81 years (median, 44). All patients received intensive chemotherapy regimens; 4 of 9 Ph+ AML patients and all CML-BP patients also received Imatinib. The median follow-up was 19 months (range, <1 month – 113 months). Six Ph+ AML patients and three CML-BP patients achieved complete remission (CR), including two Ph+ AML patients who did not receive Imatinib; one Ph+ AML patient and one CML-BP patient relapsed. Five Ph+ AML patients and 3 CML-BP patients died, including one Ph+ AML patient in CR.
Table 1.
| Pt # | Dx | Age | Sex | CR | relapse | FU (months) | Death | WBC | Hb | Plt | PB blasts | PB basos | Transcript | NPM1 | FLT3 | ABL1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dx | CR | ||||||||||||||||
| 1 | Ph AML | 22 | f | yes | yes | 8 | dead | 84.7 | 6.6 | 28 | 72% | 0% | ND | no | wild | wild | wild |
| 2 | Ph AML | 76 | m | no | no | 1 | dead | 16.1 | 9 | 41 | 64% | 0.5% | b3a2 | NA | wild | wild | wild |
| 3 | Ph AML | 71 | m | no | no | 1 | dead | 49.8 | 10.8 | 164 | 40% | 0% | e1a2 | NA | wild | wild | wild |
| 4 | Ph AML | 70 | m | no | no | 6 | dead | 47.7 | 13.6 | 99 | 47% | 0% | b2a2 | NA | wild | wild | wild |
| 5 | Ph AML | 50 | m | yes | no | 71 | alive | 120.6 | 12.3 | 43 | 68% | 1% | b3a2 | no | mutant | wild | wild |
| 6 | Ph AML | 48 | f | yes | no | 36 | alive | 108.9 | 9.4 | 306 | 40% | 1% | b3a2 b2a2 | no | mutant | wild | wild |
| 7 | Ph AML | 66 | f | yes | no | 32 | alive | 38.3 | 10.2 | 266 | 36% | 0% | e1a2 | no | wild | wild | wild |
| 8 | Ph AML | 49 | f | yes | no | 28 | alive | 7.4 | 12.1 | 319 | 20% | 1% | b3a2 b2a2 | no | wild | wild | wild |
| 9 | Ph AML | 57 | m | no | NA | <1 | dead | 99.7 | 8.4 | 18 | 92% | 0% | e1a2 | NA | wild | ITD | wild |
| 10 | CML BP | 46 | m | yes | yes | 14 | dead | 60.3 | 6.5 | 33 | 46% | 6% | b2a2 | yes | wild | wild | E459K |
| 11 | CML BP | 43 | f | yes | no | 113 | alive | 60.3 | 7.4 | 165 | 14% | 13% | b3a2 | no | wild | wild | wild |
| 12 | CML BP | 81 | m | no | NA | 9 | dead | 16.7 | 9.7 | 21 | 0% | 10% | b3a2 | NA | wild | wild | wild |
| 13 | CML BP | 44 | m | no | NA | 30 | dead | 41 | 10.6 | 270 | 9% | 11% | b3a2 | NA | wild | wild | wild |
| 14 | CML BP | 26 | m | yes | no | 23 | alive | 212 | 5.9 | 377 | 43% | 8% | b3a2 | yes | wild | wild | wild |
Laboratory data
The laboratory data at presentation are listed in Table 1. Compared to CML-BP patients, Ph+ AML patients had higher percentage of peripheral blasts (median, 47% vs. 14%, p=0.03) and by definition, lower percentage of basophils (median, 0% vs. 10%, p=0.001). There was no statistically significant difference in white blood count, hemoglobin, or platelet count.
RT-PCR analysis for BCR-ABL1 fusion transcript
The results of RT-PCR studies for BCR-ABL1 fusion transcript at presentation and in CR are listed in Table 1. At presentation, p210 protein product was detected in 5 of 8 Ph+ AML patients tested and all CML-BP patients; p190 BCR ABL1 protein product was detected in 3 Ph+ AML patients and was not detected in any CML-BP patients. At the time of CR, no BCR-ABL1 fusion transcript was detected in all 5 Ph+ AML who achieved CR, while BCRABL1 fusion transcript persisted in 2 of 3 CML-BP patients who achieved CR. Third CMLBP patient (patient#11) received allogeneic stem cell transplantation in CR and was tested for BCR-ABL1 fusion transcript after a successful engraftment, when the bone marrow chimerism studies showed 100% donor cells; no BCR-ABL1 fusion transcripts studies were performed in CR
Other gene mutation testing
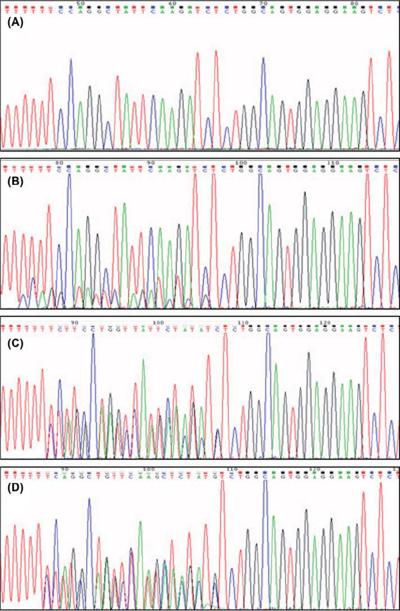
Two of 9 (22%) Ph+ AML patients had NPM1 mutations (Figure 1). Both patients had a four-base-pair (TCTG) insertion consistent with a type A mutation. Of note, both patients were alive and in molecular remission 36 and 71 months after diagnosis, respectively. No NPM1 mutations were detected in all CML-BP patients
Figure 1.
Polymerase chain reaction-capillary electrophoresis for NPM1 mutational analysis. A (case 1) shows wild type NPM1 sequence. B (case 5) and C (case 6) show mutant NPM1 sequence. A positive control is shown in D.
One Ph+ AML patient had FLT3 ITD, whereas the other 8 Ph+ AML patients and all CMLBP patients had wild-type FLT3.
All 9 Ph+ AML patients and all but one CML-BP had no evidence of ABL1 gene mutations. One CML-BP patient has E459K ABL1 gene mutation at presentation.
In 6 Ph+ AML patients tested analyzed by Sequenome-based methods no mutations were identified in AKT1, AKT2, AKT3, BRAF, EGFR, GNAQ, GNAS, IDH1, IDH2, KRAS, MET, NRAS, PIK3CA, or RET.
The bone marrow findings
The bone marrow findings are listed in Table 2. The only features discriminating CML-BP patients from Ph+ AML patients was presence of dwarf megakaryocytes, which were observed in 4 of 5 CML-BP patients, but were not detected in all but one Ph+ AML patient (p= .0319). There was no statistically significant difference in bone marrow cellularity, number of megakaryocytes, or blast percentage.
Tabe 2.
| Pt # | Dx | Cellularity % | Megs | Druf Megs | BM blasts | MPO | Esterase | FAB | Cytogenetics at diagnosis |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Ph AML | 70-80 | decreased | no | 97 | 6% | neg | M1 | 46,XX,t(9;22)(q34;q11.2)[20] |
| 2 | Ph AML | 50-60 | decreased | no | 65 | neg | neg | M0 | 46,XY,t(9;22)(q34;q11.2)[19] |
| 3 | Ph AML | 40-50 | usual | no | 63 | 68% | neg | M4 | 46,XY,der(7)inv(7)(q11.2q36)del(7)(q22q31), t(9;22)(q34;q11.2)[19]/46,XY[1] |
| 4 | Ph AML | 90-100 | decreased | no | 82 | 36% | 2% | M2 | 46,XY,t(9;22)(q34;q11.2)[1]/45,XY, -7,t(9;22)(q34;q11.2)[18]/46,XY[1] |
| 5 | Ph AML | 95-100 | decreased | no | 87 | 70% | 5% | M1 | 46,XY,t(9;22)(q34;q11.2)[20] |
| 6 | Ph AML | 90-100 | usual | no | 60 | 40% | neg | M2 | 46,XX,t(9;22)(q34;q11.2)[20] |
| 7 | Ph AML | 70-80 | usual | no | 69 | 70 | 40% | M4 | 46,XX,t(9;22)(q34;q11.2)[18]/46,XX[1] |
| 8 | Ph AML | 50-60 | increased | yes | 33 | 1 | 2% | M7 | 46,XX,t(9;22)(q34;q11.2)[20] |
| 9 | Ph AML | 80-90 | absent | NA | 88 | 1 | neg | M0 | 46,XY,der(9)inv(9)(p12q13)t(9;22)(q34;q11.2)[3] /46,XY, inv(9)[7] |
| 10 | CML BP | 95-100 | decreased | yes | 76 | 6 | neg | M2 | 46,XY,t(9;22)(q34;q11.2)[17]/46,XY[3] |
| 11 | CML BP | 70-80 | decreased | yes | 36 | 40 | neg | M2 | 46,XX,t(8;11)(p11.2;p15),t(9;22)(q34;q11.2)[20] |
| 12 | CML BP | 95-100 | decreased | no | 64 | 1 | neg | M0 | 55,XY,+8,t(9;22)(q34;q11.2),+13,+14,+14,+16, +18,+19,+20,+der(22)t(9;22)[6]/46,XY[14] |
| 13 | CML BP | 30-40 | increased | yes | 65 | neg | neg | M2 | 46,X,-Y,+8,t(9;22)(q34;q11.2)[6]/46,X, - Y,+8,t(9;22)(q34;q11.2),inv(16)(p13q22)[7]/ 47,XY,+Y[2]/46,XY[5] |
| 14 | CML BP | 95-100 | increased | yes | 38 | 20 | neg | M7 | 46,XY,t(3;21)(q26.2;q22),del(7)(q22), t(9;22)(q34;q11.2) [2]/46,XY,t(3;21)(q26.2;q22), t(4;15)(p16;q22),del(7)(q22),t(9;22)(q34;q11.2)[5]/ 46,XY,t(3;14)(q27;q24),t(3;21)(q26.2;q22),del(7) (q22),t(9;22)(q34;q11.2)[5]/46,XX[11] |
Cytogenetic data
The results of conventional cytogenetic studies at presentation are listed in Table 2. The translocation t(9;22)(q34;q11.2) was detected in all patients and was the sole cytogenetic abnormality in 6 Ph+ AML patients and in one CML-BP patient. Cytogenetically normal metaphases were observed, in addition to the abnormal ones, in 3 Ph+ AML patients and in 4 CML-BP patients. Trisomy 8 was detected as an additional abnormality in two CML-BP patients.
CML-BP with preceding CP group
During the study period we also identified 138 patients with CML who subsequently developed myeloid blast phase (CML-BP). A total of 37 patients with readily available DNA extracted from bone marrow samples obtained at the time of CML-BP diagnosis were available for this study. These patients included 23 men and 14 women, ages 23 to 81 years (median, 50). There was no evidence of NPM1 mutation in all 37 CML-BP with preceding CP patients. Eighteen of 37 (49%) patients had ABL1 mutations. The most common mutation was T315I (detected in five patients) followed by Y253H (detected in four patients). Other mutations identified included: E255K, E255V, E279K, F317L, G250E, N331S, Q252H, H396R, and V299L. Three patients had two ABL1 mutations simultaneously: Q252H and E255K, T315I and E255K, and T315I and H396R.
DISCUSSION
The frequency of adult Ph+ AML in our study (0.54%) is at the low end of the range of 0.5-3.0% in reported the literature.1,3-5 We believe the stringent exclusion criteria used in this study may explain the lower frequency.
Comparing bone marrow findings in Ph+ AML and CML-BP, we found that the only discriminating feature was the presence of dwarf megakaryocytes, which were found in all but one CML-BP patients and only one Ph+ AML. In contrast to the previous study by Soupir and colleagues,3 we did not find a statistically significant difference in bone marrow cellularity.
Six of nine (67%) Ph+ AML patients in our study had t(9;22)(q34;q11.2) as a sole cytogenetic abnormality, which is similar to patients with Ph+ AML in previous studies1,3 but markedly different from patients with CML-BP who demonstrate additional cytogenetic abnormalities in up to 60-65% of patients.11,24,25 Of three Ph+ AML patients with cytogenetic abnormalities other than Ph in our study, two had abnormalities involving chromosome 7. This finding is in accord with a report by Paietta et al who described a high frequency of abnormal chromosome 7 in Ph+ AML patients.26 No cytogenetic abnormalities known to occur frequently in CML-BP patients, such as an isochromosome of the long arm of chromosome 17, gain of an extra copy of Ph, trisomy 19, and trisomy 8, were present in our Ph+ AML patients.3,4,11,20,27 Of interest, normal metaphases were detected in both groups and therefore could not be regarded as a reliable way to discriminate Ph+ AML from CML-BP..
The frequency of NPM1 mutations in the Ph+ AML patients (2/9; 22%) was similar to that reported in the general AML population,25-35%.8,9 No NPM1 mutations were identified in the CML-BP group. Conversely, frequency of ABL1 mutations in the CML-BP patients (1/5; 20%) was similar to that reported in for CML-BP patients in the literature.15 No Ph+ AML patients had ABL1 mutations. We suggest that these data support the interpretation that Ph+ AML is a clinicopathologic entity distinct from CML-BP. Admittedly, the number of Ph+ AML patients is small, but this is a truly rare entity and 9 cases is an appreciable number of patients in this context.
We suggest that the findings we present add to other evidence suggesting that Ph+ AML is a distinct entity. Others have shown that Ph+ AML patients are less likely to have splenomegaly or peripheral basophilia, and show lower bone marrow cellularity, and have a lower myeloid to erythroid ratio than CML-BP patients.3 At the cytogenetic level, the most frequent abnormalities observed in CML-BP patients, including gain of an extra Ph, trisomy 8, trisomy 19, and an isochromosome 17q, are consistently absent in cases of Ph+ AML reported.3,11,20,27
Although the “two-hit theory” of leukemogenesis suggests leukemia cells require class I and II mutations,28 this is theoretically a minimum requirement. The co-existence of BCR-ABL1 and NPM1 mutations in 2 Ph+ AML patients in this study raises the possibility of synergistic effect between two class I mutations. Bacher and colleagues recently identified an AML patient with leukemic subclones carrying BCR-ABL1 and NPM1 mutations, supporting this concept of synergistic class I mutation in leukemia.2
Isocitrate dehydrogenase 1 (IDH1) and IDH2 genes encode cytoplasmic/peroxisomal IDH1 and mitochondrial IDH2 enzymes, respectively. Investigators have identified somatic heterozygous mutations of IDH1 and IDH2 in a subset of AML cases.22,29,30IDH mutations are most common in AML patients with normal karyotype and NPM1 mutations.30 Data regarding IDH1 and IDH2 mutations in CML-BP patients are scarce with minor discordance. One study did not detect any IDH1 or IDH2 mutations in 91 CML patients, including 20 patients with CML-BP.31 Another study detected IDH2 mutations in 3 of 75 patients with CML-BP32 whereas all 75 patients had wild-type IDH1.32 Overall, the frequency of IDH mutations is clearly low in CML and CML-BP and our data support these findings as all 37 patients lacked evidence of either IDH1 or IDH2 mutation. No data regarding IDH1 and IDH2 mutations in Ph+ AML patients was available in the literature until this study; all 9 Ph+ AML cases were negative.
In summary, we have presented molecular evidence to support the concept that Ph+ AML is distinct from CML-BP. Ph+ AML cases carried NPM1 at a similar to that in AML patients in general, and lacked ABL1 mutations. The important potential clinical implication of discriminating of Ph+ AML from CML-BC is that the current practice for CML patients requires indefinite therapy with imatinib,33 but there is no data to support the need of indefinite therapy with imatinib for patients with Ph+ AML. However, a biger study of Ph+ AML patients with a long BCR-ABL1 fusion transcript surveliance is required to establish a standard of care for Ph+ AML patients.
ACKNOWLEDGEMENTS
We thank Sherry Pierce for her critical help in our database search; Yujia Huo, Michael Fernandez, and Hongbo Lu for their help with sequence analysis; Geneva Williams for manuscript preparation, and Don Norwood for editing the manuscript.
This work was supported in part by National Institutes of Health grants CA016672 and 2P50 CA100632-06.
Footnotes
DECLARATION OF INTERESTS
The authors declare no conflict of interest.
REFERENCES
- 1.Keung YK, Beaty M, Powell BL, et al. Philadephia chromosome positive myelodysplastic syhndrome and acute myeloid leukemia – retrospective study and review of the literature. Leuk Res. 2004;28:579–586. doi: 10.1016/j.leukres.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 2.Bacher U, Haferlach T, Alpermann T, et al. Subclones with the t(9;22)/BCL-AML1 rearrangement occur in AML and seem to cooperate with distinct genetic alterations. Br J Haematol. 2011;152:713–720. doi: 10.1111/j.1365-2141.2010.08472.x. [DOI] [PubMed] [Google Scholar]
- 3.Soupir CP, Vergilio J, Dal Cin P, et al. Philadelphia chromosome-positive acute myeloid leukemia. A rare aggressive leukemia with clinicopatholgical features distinct from chronic myeloid leukemia in myeloid blast crisis. Am J Clin Pathol. 2007;127:642–650. doi: 10.1309/B4NVER1AJJ84CTUU. [DOI] [PubMed] [Google Scholar]
- 4.Bloomfield CD, Lindquist LL, Brunning RD, et al. The Philadelphia chromosome in acute leukemia. Virchows Arch B Cell Pathol. 1978;29:81–91. doi: 10.1007/BF02899340. [DOI] [PubMed] [Google Scholar]
- 5.Cuneo A, Ferrant A, Michaux JL, et al. Philadelphia chromosome–positive acute myeloid leukemia: cytoimmunologic and cytogenetic features. Haematologica. 1996;81:423–427. [PubMed] [Google Scholar]
- 6.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC; Lyon, France: 2008. [Google Scholar]
- 7.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 8.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 9.Boissel N, Renneville A, Biggio V, et al. Prevalence, clinical profile, and prognosis of NPM mutations in AML with normal karyotype. Blood. 2005;106:3618–3620. doi: 10.1182/blood-2005-05-2174. [DOI] [PubMed] [Google Scholar]
- 10.Falini B, Mecucci C, Tiacci E, et al. GIMEMA Acute Leukemia Working Party. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 11.Grossmann V, Kohlmann A, Zenger M, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25:557–560. doi: 10.1038/leu.2010.298. [DOI] [PubMed] [Google Scholar]
- 12.Piccaluga PP, Sabattini E, Bacci F, et al. Cytoplasmic mutated nucleophosmin (NPM1) in blast crisis of chronic myeloid leukemia. Leukemia. 2009;23:1370–1371. doi: 10.1038/leu.2009.95. [DOI] [PubMed] [Google Scholar]
- 13.Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100:1014–1018. doi: 10.1182/blood.v100.3.1014. [DOI] [PubMed] [Google Scholar]
- 14.Soverini S, Hochhaus A, Nicolini FE, et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: recommendations from an expert panel on behalf of European LeukemiaNet. Blood. 2011;1185:1208–1215. doi: 10.1182/blood-2010-12-326405. [DOI] [PubMed] [Google Scholar]
- 15.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCRABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor Imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 16.Willis SG, Lange T, Demehri S, et al. High-sensitivity detection of BCR-ABL kinase domain mutations in Imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood. 2005;106:2128–2137. doi: 10.1182/blood-2005-03-1036. [DOI] [PubMed] [Google Scholar]
- 17.Oyarzo MP, Lin P, Glassman A, et al. Acute myeloid leukemia with t(6;9)(p23;q34) is associated with dysplasia and a high frequency of flt3 gene mutations. Am J Clin Pathol. 2004;122:348–358. doi: 10.1309/5DGB-59KQ-A527-PD47. [DOI] [PubMed] [Google Scholar]
- 18.Onciu M, Schlette E, Medeiros LJ, et al. Cytogenetic findings in mantle cell lymphoma cases with a high level of peripheral blood involvement have a distinct pattern of abnormalities. Am J Clin Pathol. 2001;116:886–892. doi: 10.1309/JQMR-323G-71Y9-M7MB. [DOI] [PubMed] [Google Scholar]
- 19.Yin CC, Cortes J, Galbincea J, et al. Rapid clonal shifts in response to kinase inhibitor therapy in chronic myelogenous leukemia are identified by quantitation mutation assays. Cancer Sci. 2010;101:2005–2010. doi: 10.1111/j.1349-7006.2010.01627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortes J, Talpaz M, O'Brien S, et al. Molecular responses in patients with chronic myelogenous leukemia in chronic phase treated with Imatinib mesylate. Clin Cancer Res. 2005;11:3425–3432. doi: 10.1158/1078-0432.CCR-04-2139. [DOI] [PubMed] [Google Scholar]
- 21.Chen W, Jones D, Medeiros LJ, Luthra R, et al. Acute myeloid leukaemia with FLT3 gene mutations of both internal tandem duplication and point mutation type. Br J Haematol. 2005;130:726–728. doi: 10.1111/j.1365-2141.2005.05666.x. [DOI] [PubMed] [Google Scholar]
- 22.Patel KP, Barkoh BA, Chen Z, et al. Diagnostic testing for IDH1 and IDH2 variants in acute myeloid leukemia an algorithmic approach using high-resolution melting curve analysis. J Mol Diagn. 2011;13:678–86. doi: 10.1016/j.jmoldx.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009 doi: 10.1002/0471142905.hg0212s60. chapter 2:unit 2.12. [DOI] [PubMed] [Google Scholar]
- 24.Kantarjian HM, Keating MJ, Talpaz M, et al. Chronic myelogenous leukemia in blast crisis. Analysis of 242 patients. Am J Med. 1987;83:445–454. doi: 10.1016/0002-9343(87)90754-6. [DOI] [PubMed] [Google Scholar]
- 25.Griesshammer M, Heinze B, Hellmann A, et al. Chronic myelogenous leukemia in blast crisis: retrospective analysis of prognostic factors in 90 patients. Ann Hematol. 1996;73:225–230. doi: 10.1007/s002770050233. [DOI] [PubMed] [Google Scholar]
- 26.Paietta E, Racevskis J, Bennett JM, et al. Biologic heterogeneity in Philadelphia chromosome-positive acute leukemia with myeloid morphology: the Eastern Cooperative Oncology Group experience. Leukemia. 1998;12:1881–1885. doi: 10.1038/sj.leu.2401229. [DOI] [PubMed] [Google Scholar]
- 27.Berger R. Differences between blastic chronic myeloid leukemia and Ph-positive acute leukemia. Leuk Lymphoma. 1993;11(suppl 1):235–237. doi: 10.3109/10428199309047892. [DOI] [PubMed] [Google Scholar]
- 28.Gilliland DG. Hematologic malignancies. Current Opinion in Hematology. 2001;8:189–191. doi: 10.1097/00062752-200107000-00001. [DOI] [PubMed] [Google Scholar]
- 29.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 30.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roche-Lestienne C, Marceau A, Labis E, et al. Mutation analysis of TET2, IDH1, IDH2 and ASXL1 in chronic myeloid leukemia. Leukemia. 2011 doi: 10.1038/leu.2011.139. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.Soverini S, Score J, Iacobucci I, et al. IDH2 somatic mutations in chronic myeloid leukemia patients in blast crisis. Leukemia. 2011;25:178–181. doi: 10.1038/leu.2010.236. [DOI] [PubMed] [Google Scholar]
- 33.Barton MK. Discontinuation of imatinib may be possible in chronic myelogenous leukemia. CA Cancer J Clin. Mar-Apr. 2011;61(2):65–6. doi: 10.3322/caac.20110. [DOI] [PubMed] [Google Scholar]