Abstract
Epigenetics is emerging as an important field in cancer epidemiology that promises to provide insights into gene regulation and facilitate cancer control throughout the cancer care continuum. Increasingly, investigators are incorporating epigenetic analysis into the studies of etiology and outcomes. To understand current progress and trends in the inclusion of epigenetics in cancer epidemiology, we evaluated the published literature and the National Cancer Institute (NCI) supported research grant awards in this field to identify trends in epigenetics research. We present a summary of the epidemiological studies in NCI’s grant portfolio (from January 2005 through December 2012) and in the scientific literature published during the same period, irrespective of support from NCI. Blood cells and tumor tissue were the most commonly used biospecimens in these studies, although buccal cells, cervical cells, sputum, and stool samples also were used. DNA methylation profiling was the focus of the majority of studies, but several studies also measured microRNA profiles. We illustrate here the current status of epidemiologic studies that are evaluating epigenetic changes in large populations. The incorporation of epigenomic assessments in cancer epidemiology studies has and is likely to continue to provide important insights into the field of cancer research.
Keywords: Acetylation, biomarker, chromatin, epidemiology, epigenetics, genetics, histone, imprinting, methylation, microRNA, risk prediction, survival, treatment
Introduction
Cancer is both a genetic and epigenetic disease (1). Epigenetics is defined as heritable changes in gene expression that are not the result of changes in DNA sequence. Four major epigenetic mechanisms–DNA methylation, histone modification, nucleosome remodeling, and microRNAs (miRNAs)–control gene activity and have been shown to play a role in a number of complex diseases, including cancer (2, 3). DNA methylation in particular has been extensively assessed in breast, colon, esophageal, lung, pancreas, ovary, prostate, and other cancers (4–12). Through their effects on genomic stability and gene expression, epigenetic changes influence carcinogenesis from initiation through progression, throughout a person’s lifespan, and, in some cases, across generations (13). Epigenetic events that are relevant to cancer risk are believed to occur early in cancer development, thus may serve as potential “first hits” for tumorigenesis. Epigenetic marks reflect both an individual’s genetic background and exposure to different environmental factors and thus may be useful for understanding the impact of environmental exposures in carcinogenesis (14). Since epigenetic changes occur before or during early tumor development, they can be modulated by diet, drugs, and other external factors such as infectious agents, epigenenetic profiling may provide clues to mitigate an individual’s risk of cancer (15–17). Mill and Hijmans recently proposed that improved understanding of the mechanism of cancer progression can be understood by studying epigenetics in populations as a part of an integrated functional genomic study (18). Epigenetic changes in comparison with genetic ones are reversible and are acquired in a gradual manner and this feature provides a huge potential for cancer prevention strategies. Additionally, therapies targeting epigenetic mechanisms have been shown to modify or inhibit gene expression and some have shown modest effects in clinical research settings.
In order to understand the current state of the field of epigenetics in cancer epidemiology, we evaluated the research project grant (RPG) awards funded by the NCI and the published literature in PubMed for trends in epigenetic research in cancer epidemiology across the cancer control continuum in studies conducted in human populations. This report presents summary of our findings, particularly in the context of studying risk, and cancer-relevant exposures, including nutrition and infectious agents, as well as practical matters such as the type of cancers being studied, and the methods and techniques that are both emerging and commonly used. Overall, we sought to present an overview of the progress in the inclusion of epigenetics in cancer epidemiology studies, and to identify scientific questions related to epigenetics that cancer epidemiology can address.
Methods
Criteria and terms used for identifying cancer epigenetics and epidemiology grants and publications (search strategy and analysis)
NCI supported RPGs related to epigenetic epidemiology funded from January 01, 2005 to December 31, 2012 were included in the portfolio analysis and the scientific terms used in analyzing grants in different categories are shown in Table 1. The portfolio was analyzed using NCI’s Portfolio Management Application software version 13.4. Search and selection criteria used for the grant proposal to be classified as “epigenetic epidemiology” study were as follows: “One OR more terms from column1 from Table 1” AND “one OR more terms from column 2 from the Table 1 AND “Human.” Additionally, the criteria for inclusion of a project in the analysis were as follows: a) the focus of the project is cancer, b) study involves human subjects, c) focus of at least one of the specific aims in the project is cancer epigenetics, and d) had at least 100 cases. We excluded studies that focused solely on polymorphisms in genes encoding DNA methyltransferases or miRNAs. After applying these criteria and exclusions, 79 RPGs were identified for further analysis. The authors of this report coded the grant abstracts for and analyzed the data by study design, organ site, biospecimen type used, exposure evaluated (if applicable), and method/technology used for epigenetic analysis.
Table 1.
Grant portfolio search and coding categories
| “HUMAN” AND “NEOPLASM” AND one or more epigenetic terms from column first and one or more epidemiology terms from the second column | |
|---|---|
| EPIGENETICS TERMS | EPIDEMIOLOGY TERMS |
| Epigenetics | Epidemiology |
| Methylation | Case control |
| Epigenome | Cohort |
| Histone | Epidemiology methods |
| Acetylation | Observational studies |
| Imprinting | Risk assessment |
| MicroRNA | Causal association |
| Chromatin compaction | Association studies |
| CpG island methylator phenotype (CIMP) | Nested case control studies |
| Clinical trial | |
| Etiology | |
| Risk factors | |
| Susceptibility | |
| Prognosis | |
| Prevention | |
| Population-based | |
Additionally, we searched published literature on epigenetic epidemiology in PubMed from January 1, 2005 to December 31, 2012 using the following search criteria: (epigenesis, genetic[mh] OR epigenomics[mh] OR “DNA methylation” OR methylation[ti] OR “histone modification” OR CIMP[tiab] OR microRNAs[mh] OR “CpG Islands/genetics”[mh] OR methylation[ti]) AND neoplasms[mh] AND (epidemiologic studies[mh] OR risk[mh] OR “population-based”[tiab] OR “odds ratio”[tiab] OR hazard[tiab] OR cohort*[tiab]) AND Humans[Mesh]. The following elimination criteria were applied: a) studies with less than 100 cancer cases; b) studies published before 2005; c) experimental studies in animals; d) review articles and articles reporting meta-analyses; e) letters, commentaries, editorials and news articles; and f) studies that are not clearly epidemiological. This search yielded 486 publications that are relevant for analysis. As with the grant analysis outlined above, authors listed above coded the publications on and analyzed the data by study design, organ site, biospecimen type used, exposure evaluated (if applicable), and method/technology used for epigenetic analysis. To check all manual coding, each publication and grant was coded by two different authors and any discrepancies resolved.
Results
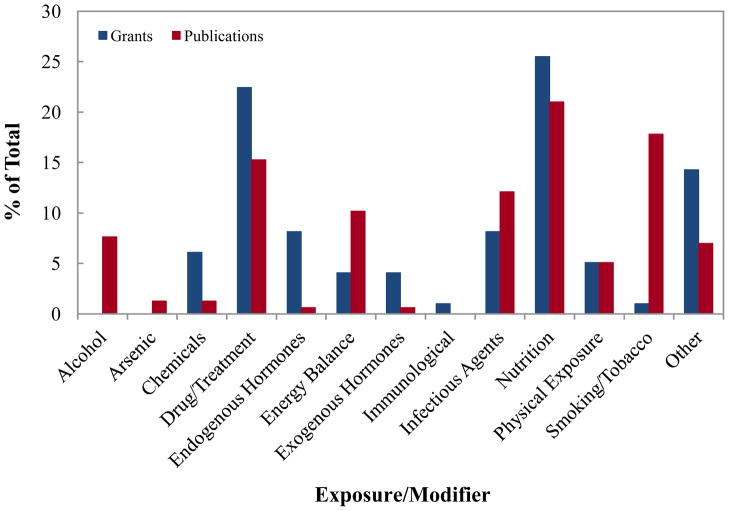
The type of exposures/modifiers proposed to be examined in NCI-supported grants and those examined in publications are shown in Figure 1. The grant proposals supported by NCI most often studied the influence of nutrition, drugs/treatment, or infectious agents. Other grants focused on energy balance, exogenous hormones, chemical exposures (e.g., pesticides), and physical exposures (e.g., radiation). In the literature, the majority of publications explored the effect of nutrition, smoking, drugs and treatments, and infectious agents on epigenetic processes.
Figure 1.
Number of NCI grants and PubMed indexed publications in cancer epigenetic epidemiology by exposure evaluated in the study. Different types of exposures are shown in the figure and discussed in the text.
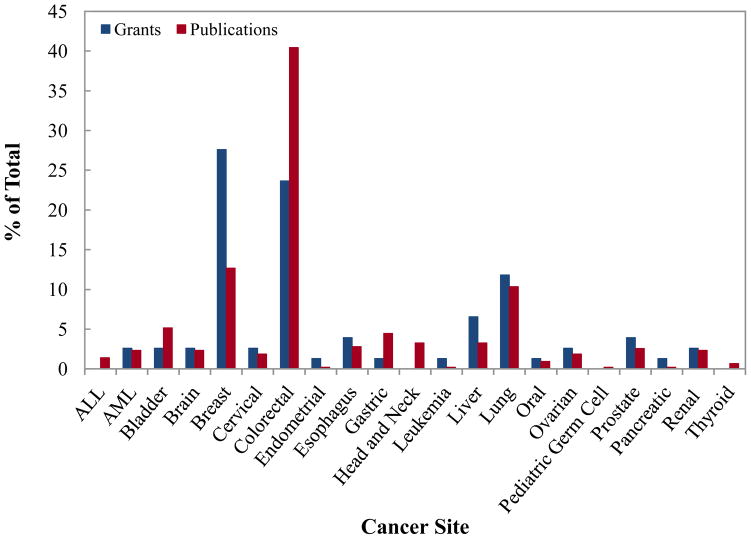
The results from the literature analysis revealed that the number of publications addressing epigenetic changes in cancers of colon were the largest followed by breast and lung (Figure 2). The NCI supported grant portfolio analysis indicated that in the field of epigenetics, breast cancer was the most frequently studied cancer type (Figure 2). The next two most frequently investigated cancers were colorectal cancer and lung cancer, in that order. Other organ sites examined for epigenetic changes included pancreas, ovary, liver, gastric, and head and neck cancers.
Figure 2.
Number of NCI grants and PubMed indexed publications in cancer epigenetic epidemiology by cancer site. Breast cancer site is the most studied cancer site in our grant portfolio whereas colorectal cancer site in publication analysis.
The portfolio analysis found that 67 of 79 grants planned to analyze methylation levels, mostly at selected individual loci (data not shown). Some of these grants also proposed to assess CIMP status, promoter methylation and microsatellite instability. Investigators proposed to assess histone modifications along with methylation level in cancers of the bladder, cervix, and prostate, and also in myelodysplastic syndrome. Non-coding RNAs, particularly miRNAs, were the focus of seven studies, with five studies planned to assess methylation at specific loci along with miRNAs.
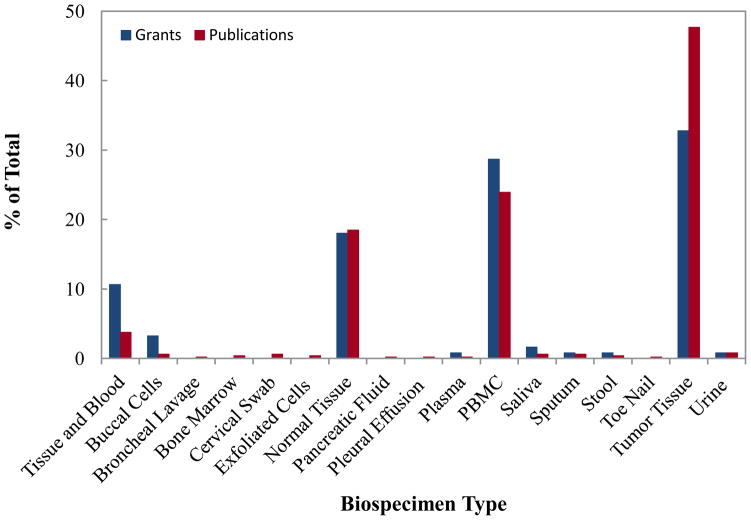
Regarding biospecimen types (Figure 3) used for epigenetic analysis, tumor tissues (40 grants) and blood (35 grants) were the most predominant specimen types collected, although a few grants proposed using buccal cells, paraffin-embedded tissues, plasma, sputum, mouthwash, urine, DNA from blood spot, stool, toenail and saliva for epigenetic analysis. Thirteen grant proposals used both blood and tumor tissue specimens for comparison analysis, while twenty two grant proposals used normal tissue specimens for comparison with tumor specific epigenetic markers. The portfolio analysis also found that 53 of 79 studies used tumor tissue for epigenetic analysis, while 13 studies planned to analyze epigenetic changes simultaneously in both leukocyte DNA and tumor DNA. Table 2 shows the types of biospecimens used for epigenetic analysis in published cancer epidemiology studies. Similar to the trend in grant proposals, both tissue samples and blood were the frequently utilized biospecimens for epigenetic analysis in published studies (19). The published literature also suggested that investigators used buccal cells, sputum, urine, cervical swab, and pancreatic fluid for epigenetic analysis in epidemiologic studies (Figure 3), albeit less frequently. Several investigators sought to determine whether blood cells could be used as a surrogate tissue for examining epigenetic profiles in tumor tissue (19, 20).
Figure 3.
Number of NCI grants and PubMed indexed publications in cancer epigenetic epidemiology by biospecimen utilized in the study. For epidemiologic studies, the most frequently collected sample is tumor tissue followed by blood.
Table 2.
Examples of the types of biospecimens examined in published cancer epidemiology studies of epigenetics, 2005–2012
| Biospecimens | Examples of types of cancer studied (all involved methylation marks unless noted in parentheses) |
|---|---|
| Blood | Bladder cancer (43), breast cancer (44, 45), cervical cancer (46), colon cancer (47), esophageal cancer (miRNA profile) (48), gastric cancer (49), head and neck cancer (50), leukemia (51), liver cancer (52), liver cancer (miRNA profile) (53), lung cancer (54), prostate cancer (55), renal cancer (56) |
| Duodenal secretion | Pancreatic cancer (57) |
| Exfoliated cells from oral rinse | Head and neck cancer (58) |
| Formalin fixed, paraffin embedded tissues | Lung cancer (histone modifications) (59), prostate cancer (59, 60), rectal cancer (61), renal cancer (histone modifications)(59) |
| Pancreatic secretion | Pancreatic cancer (62) |
| Salivary rinse | Lung cancer (63) |
| Sputum | Lung cancer (64, 65) |
| Tumor Tissue | Bladder cancer (66), brain cancer (miRNA profiling) (67), breast cancer (68), colon cancer (69, 70), esophageal cancer (71), gastric cancer (72), glioblastoma (73), head and neck cancer (74), laryngeal and hypolaryngeal cancer (75), liver cancer (76), lung cancer (59, 77, 78), neuroblastoma (79), oral cancer (80), ovarian cancer (81, 82), pancreatic cancer (57, 83), prostate cancer (59, 84), rectal cancer (85), renal cancer (59, 86) |
| Urine | Bladder cancer (87), prostate cancer (88, 89) |
| Uterine/cervix swab | Cervical cancer (46) |
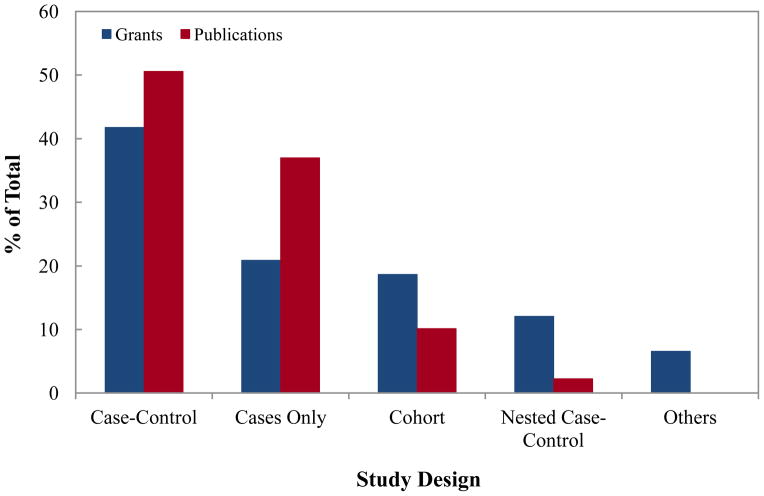
Case-control study design was the most predominantly used study design type both in RPGs and in publications (Figure 4). Investigators also used case only, cohort and nested case-control studies for epigenetic studies, but to a lesser extent. Eight grant proposals used either a mixed-methods approach or different approaches to address cancer etiology questions.
Figure 4.
Number of NCI grants and PubMed indexed publications in cancer epigenetic epidemiology by study design. The most common study design was case-control study in publications and grants.
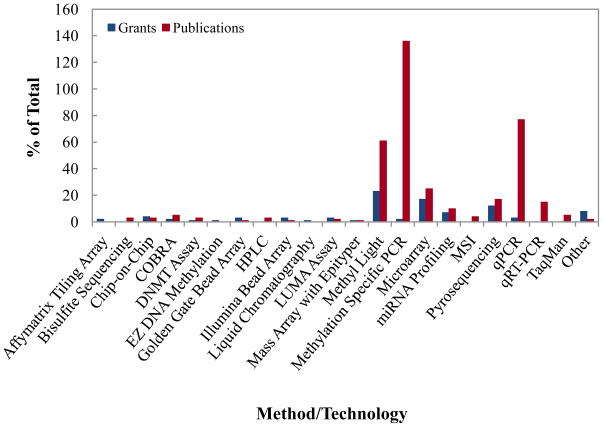
Several methodologies are currently used to generate a global view of DNA methylation, miRNA expression, and histone modifications in various cell types. As noted earlier, most of the projects examined both global and tumor specific DNA methylation changes (11, 20, 21). Investigators used several different type of technologies for methylation analyses (Figures 5), and the most commonly used were methylation-specific PCR, Methyl Light, and pyrosequencing based technologies (22). Ulrich et al. described a method which screens a broad scale of factors in relation to DNA methylation (23). In grants supported by NCI, Methyl Light and pyrosequencing were most commonly used technologies for methylation profiling; while methylation specific PCR, Methyl Light, was the routinely used technology in the published studies (Figure 5). Both grant awards and publications utilized mostly PCR-based miRNA analysis technologies. However, a recent trend is to use microarray or sequencing based technologies because of their high-throughput and broad dynamic range. Pyrosequencing of methylation regions was the preferred method in publications where global methylation was studied, see for example (21). Chromatin immonoprecipitation methods utilizing antibodies for specific histone modifications, such as ChIP-PCR (Chromatin Immunoprecipitation polymerase chain reaction), ChIP-on-ChIP (chromatin immunoprecipiation (Chip) on microarray Chip) and ChIP-SAGE (Chromatin immunoprecipitation combined with serial analysis of gene expression), were routinely used for understanding histone modifications and associated DNA sequences in both grant awards and published articles, while more recent publications utilized ChIP-Seq.
Figure 5.
Number of NCI grants and PubMed indexed publications in cancer epigenetic epidemiology by method or technology utilized in the study. Abbreviations: AML, acute myelogenous leukemia; ALL, acute lymphoblastic leukemia; Chip-on-Chip, chromatin immunoprecipiation (Chip) on microarray Chip; CIMP, CpG island methylator phenotype; COBRA, combined bisulfate restriction analysis; DNMT assay, DNA methyl transferase assay; HPLC, high pressure liquid chromatography; LUMA assay, LUminometric methylation assay; MSI, microsatellite instability; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
Overall, the major hypotheses being addressed in both the grant portfolio and the publications include i) whether diet or environmental exposure is associated with specific epigenetic marks, patterns and/or miRNA expression and whether epigenetic factors were related to cancer risk or survival from cancer; ii) whether an epigenetic profile in blood or other accessible biospecimens is related to the epigenetic profile observed in the tumor; and iii) whether an epigenetic pattern detected in tumor tissue differs from that of the normal tissue surrounding it.
In the publications studies, there are numerous examples of epidemiologic studies that reported association of methylation and risk of different cancers. Several examples will illustrate this. In a case-control study nested in the prospective Shanghai Women’s Health Study, 192 gastric cancer cases and 384 matched controls were used to analyze methylation of Alu and long interspersed nucleotide elements (LINE) and results indicated that hypomethylation of these repeat sequences was associated with increased risk of cancer (24). However, breast cancer was not associated with Alu and LINE hypomethylation, as reported in another group of participants from a cohort in New York (25). A case-control study design demonstrated an association of HPV16 DNA hypermethylation with high grade intraepithelial cervical cancer (26). Characterization of methylated promoter of other oncogenic papilloma viruses, HPV18, HPV31, and HPV41, indicated that methylation of these viruses was a general phenomenon and diagnostic assays, based on these results, could be developed (27). A case-control study showed a correlation between promoter methylation and testicular tumors (28). Constitutional BRCA1 promoter methylation determined in peripheral blood was shown to strongly predispose toward the development of tumors with features which resemble BRCA1 mutated tumors (29). Thus screening of peripheral blood for BRCA1 methylation may predict risk for breast cancer. In another study, analysis of tumor tissues indicated that IGFII promoter methylation was associated with ovarian cancer progression (30). In one nested case-control study, hypermethylation of runt related transcription factor 3 (RUNX3) was associated with advanced gastric lesions which were susceptible to H. pylori infection, a risk factor for gastric cancer (31). Inactivation of PTEN due to hypermethylation was associated with cervical cancer (32). Studies in samples from Colon Cancer Family Registry indicated that relatives of colorectal cancer with hypermethylation of MLH1 gene might be at increased risk of colorectal and gastric cancer and possibly ovarian and liver cancer (33).
Less commonly observed in the publications analysis are the effect of epigenetic factors on cancer survival, with very few examples illustrating this. Formalin-fixed tumor tissues from non-small cell lung cancer patients were analyzed for hypermethylation of selected genes (p16, MGM2, DAPK, RASSF1, CDH1, LET7-3-a, and PTEN) and results indicated that p16 hypermethylation was associated with a worse outcome in patients with age 60 years or younger but not in older than 60 years (12). Serum DNA shed from tumors was used to evaluate methylation of specific genes associated with breast cancer in a case-control study (34). Delgado-Cruzata et al. demonstrated that global DNA methylation levels measured in white blood cells may be a potential biomarker of breast cancer risk even within families at higher risk of cancer (21).
Discussion
Through our analyses of the NCI grant portfolio and the published literature in the use of epigenetic markers in epidemiologic research we identified a number of challenges and opportunities for the field (Table 3). There are tremendous potentials to integrate the knowledge of epigenetics research with genetic characteristics, environmental predisposition, and lifestyle factors in cancer epidemiologic studies. The field of cancer epidemiology research should leverage the use of epigenetic information when used appropriately to advance translational research and to enhance practice in cancer prevention, detection, diagnosis and treatment.
Table 3.
Trends, opportunities, and challenges in the cancer epigenetics and epidemiology field
| Trends | Opportunities | Challenges |
|---|---|---|
| Most of the studies have been conducted using methylation markers. The majority of the exposures evaluated for their impact on the epigenome were nutrition, smoking, drugs and treatments, and infectious agents. Most of the studies investigated epigenetic changes at specific individual loci, and very few studies explored changes at multiple loci or interactions among multiple loci. Only few investigators explored histone modifications along with methylation, nucleosome remodeling, or miRNA expression changes in cancer epidemiology. Most epigenetic studies have been conducted in blood which may not be an appropriate biospecimen. |
Integrate epigenetic research with genetics, environmental predisposition and lifestyle factors. Incorporate epigenetics into epidemiologic studies of cancer and the environment which could contribute greatly to our understanding of cancer risk and development. Determine the stability of epigenetic marks in repeated biospecimen samples from the same people over time. Explore the use of epigenomic information to better define cancer subcategories Develop improved strategies for epigenetic data analysis and interpretation Conduct studies that examine the relationship between epigenetic marks in germline DNA and tumor DNA. Characterize all the components of epigenome which might help in understanding the underlying mechanism of cancer risk and identifying new biomarkers of cancer initiation and development. Develop technologies which require small amount of samples compared to the amount currently used which might help in analyzing multiple biomarkers in small samples. Utilize exposomes with information of well defined factors (tobacco, diet, occupational exposures, environmental pollutants) and omics profiling (genomics, transcriptomics, epigenomics, and metabolomics) for evaluating environmental exposure and cancer risk. Understand the role of epigenetics in interaction of cancer-associated infectious agents with host factors. Utilize resources such as family registries in identifying cancers that tend to cluster in families. |
Follow individual’s epigenomic status which changes spatiotemporally and compartmentally in tissues and contribute to variations. Improve strategies for epigenetic data analysis and interpretation. Conduct large scale epidemiological studies to determine whether epigenetic changes detected using blood samples accurately reflect both inherent and acquired epigenetic changes that contribute to cancer risk and impact outcomes. Identify new chromatin abnormalities and their association with cancer. Develop high-throughput technologies for histone modifications and nucleosome remodeling. Distinguish between association and causality of epigenetic mark with disease. Evaluate relationships between epigenetic marks in germline vs. tumor DNA. Distinguish age-related epigenomic marks with cancer-associated marks. Synthesize momoclonal antibodies for histone modifications (currently available antibodies have low dynamic range). Develop technologies that use smaller amounts of sample for epigenomic profiling. Increase in funds to conduct studies on epigenome-wide association studies. Determine about how long longitudinal measurements should be taken in individuals at high risk before the disease develops. Two unresolved issues are: (i) whether epigenetic marks are transmitted intact from parent to offspring; (ii) can we develop an epigenetic transmission test comparable to the transmission disequilibrium test used in genetic epidemiology. Consider about confounding factors between the epigenome and increasing age and tissue heterogeneity. |
Breast, lung, colon, and prostate cancers were the most studied organ sites in grants and publications, while a few studies investigated other cancer sites. This result was anticipated because these are among the most common cancers and epidemiologist would likely have access to higher numbers of samples from these cancer types. The cancer site where NCI supported research is yielding many promising epidemiologic insights is colon cancer, particularly the identification of CpG island methylator phenotype (CIMP), a molecular subtype of colon cancer. Traditional histopathological examination cannot distinguish CIMP+ with CIMP− individuals (35, 36). In a prospective study of duration of smoking cessation and colorectal cancer risk was based on CIMP analysis and results indicated a protective effect of smoking cessation on a DNA methylation-related carcinogenesis pathway leading to CIMP+ phenotype (37). In the Netherland Cohort study, body size and physical activity were associated with risk of colorectal cancer in CIMP+ and CIMP− individuals (38). Central adiposity increased and high physical activity decreased the risk of colorectal cancer in this population. In a large prospective cohort of women (the Nurses’ Health Study) differential effects of B vitamins, alcohol, and methionine intake was observed in CIMP+ and CIMP− individuals (39). In another independent prospective study of older women, no link of alcohol intake was observed irrespective of the CIMP and BRAF mutation status (40). Examples discussed above also indicate that different risk factors have different effect on colorectal cancer in CIMP+ and CIMP− individuals. A recently completed prospective study involving 134,204 individuals indicating a protective effect of smoking cessation on a DNA-methylation related pathway leading to high CIMP colorectal cancer (37). Similar series of studies should be considered for other cancer types where CIMP phenotype has been reported such as bladder, ovarian and prostate cancer.
These epidemiologic findings complement other lines of evidence that epigenetic events can be driver events in the pathogenesis of colorectal cancer. Also, epigenetic events cooperate with genetic events in the progression of colonic mucosa to colorectal cancer. The number of genes affected by epigenetics is more than the number of genes mutated in colorectal cancer (41).
Our analysis indicated that most of the grants and publications that we examined evaluated either global or specific DNA methylation status and/or microRNA profiles. This is likely because the methodologies used to assess methylation and detect miRNAs are more amenable for the high-throughput, large scale studies performed in epidemiologic settings. In contrast, few investigators explored histone modifications along with methylation, nucleosome remodeling, or miRNA expression changes in cancer epidemiology. Histone modifications have been reported to play critical roles in cancer development, and aberrant patterns of histone modifications are linked to DNA methylation changes. Additionally, the studies that we examined did not utilize genome-wide agnostic approaches or integrated approaches to assess epigenetic changes that influence cancers although we anticipate an increase in these types of studies. Currently, ChIP based methods are being used for assessing chromatin structures on a genome-scale, but the limited availability of specific antibodies for histone modifications that efficiently precipitate chromatin, low dynamic range, and the labor-intensive, time-consuming, and costly nature of these studies preclude their use in epidemiology studies. In addition, technologies that use smaller amounts of biospecimens are needed for it to be feasible for epidemiologists to incorporate such experiments into their studies.
Our analysis showed that tumor tissues and blood were the most predominant specimen type used for epigenetic research in epidemiology. An individual’s epigenomic status changes spatiotemporally and compartmentally in tissues (42). Blood samples contain different types of cells with half-lives varying from a few hours to several years. Additionally, diverse cell types may have different inherent epigenetic characteristics and may harbor or sustain different levels of epigenetic changes (19). Large scale epidemiological studies are needed to determine whether epigenetic changes detected using blood samples accurately reflect both inherent and acquired epigenetic changes that contribute to cancer risk and impact outcomes (19, 42). Additionally, such studies could examine whether these inherent or acquired epigenetic changes reflect the pattern of epigenetic changes in the tumor tissues.
Future research in this area includes developing improved strategies for epigenetic data analysis and interpretation; determining the stability of epigenetic marks in repeated biospecimen samples from the same people over time; and studies that examine the relationship between epigenetic marks in germline DNA and tumor DNA. Resources that are particularly valuable include studies that have prospectively collected and stored biospecimens, and thus allow for the analysis of epigenetic profiles well before cancer starts developing. The serial sampling of biospecimens at multiple time points across the life course may provide additional value allowing insights into the temporal variation in epigenetic marks over time. Some important research resources that could be explored for such studies include some of the large cancer prevention trials (such as NCI’s Prostate, Lung, Colorectal and Ovarian cancer Screening Trial that obtained repeated biosamples during the trials. Some confounding factors need to be considered in studies of epigenomics and cancer risk, particularly age. Moreover since multiple types of exposures (e.g., alcohol, tobacco smoking) may modify one’s epigenomic profile, these exposures need to be controlled for when examining any one exposure. Studies that correlate in epigenetic characteristics in different tissues and body fluids in the same persons would be very useful to validate the use of easily accessible biospecimens such as white blood cells in studies of cancer risk.
In infectious-agent associated cancers (cervical cancer, liver cancer, gastric cancer, nasopharyngeal carcinoma, Kaposi’s sarcoma), epigenetic epidemiology has emerged as another promising area for future research. However, these studies face temporal causality problem. Additionally, the reversible and context dependent nature of epigenetic changes poses challenges to epidemiological studies. To overcome these challenges, pathogen-associated epigenetic studies should be accompanied by comprehensive longitudinal (multi-stage and multi-individual) and transgenerational data.
Investment of resources is needed in this area of cancer epigenetics and epidemiology. Epigenetics hold substantial potential for furthering our understanding of the molecular mechanisms of health related risks due to environmental exposure and individual susceptibility.
Footnotes
Conflict statement: This work was done by government staff. There was no grant for the work. There is no conflict.
References
- 1.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones P. Out of Africa and into epigenetics: discovering reprogramming drugs. Nature cell biology. 2011;13:2. doi: 10.1038/ncb0111-2. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP. Genome-scale approaches to the epigenetics of common human disease. Virchows Archiv : an international journal of pathology. 2010;456:13–21. doi: 10.1007/s00428-009-0847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khare S, Verma M. Epigenetics of colon cancer. Methods Mol Biol. 2012;863:177–85. doi: 10.1007/978-1-61779-612-8_10. [DOI] [PubMed] [Google Scholar]
- 5.Kumar D, Verma M. Methods in cancer epigenetics and epidemiology. Methods in molecular biology (Clifton, NJ) 2009;471:273–88. doi: 10.1007/978-1-59745-416-2_14. [DOI] [PubMed] [Google Scholar]
- 6.Verma M, Maruvada P, Srivastava S. Epigenetics and cancer. Critical reviews in clinical laboratory sciences. 2004;41(5–6):585–607. doi: 10.1080/10408360490516922. [DOI] [PubMed] [Google Scholar]
- 7.Verma M, Dunn BK, Ross S, Jain P, Wang W, Hayes R, et al. Early detection and risk assessment: proceedings and recommendations from the Workshop on Epigenetics in Cancer Prevention. Annals of the New York Academy of Sciences. 2003;983:298–319. doi: 10.1111/j.1749-6632.2003.tb05984.x. [DOI] [PubMed] [Google Scholar]
- 8.Dunn BK, Verma M, Umar A. Epigenetics in cancer prevention: early detection and risk assessment: introduction. Annals of the New York Academy of Sciences. 2003;983:1–4. doi: 10.1111/j.1749-6632.2003.tb05957.x. [DOI] [PubMed] [Google Scholar]
- 9.Verma M, Srivastava S. Epigenetics in cancer: implications for early detection and prevention. The lancet oncology. 2002;3:755–63. doi: 10.1016/s1470-2045(02)00932-4. [DOI] [PubMed] [Google Scholar]
- 10.Feinberg AP. Epigenetics at the epicenter of modern medicine. JAMA : the journal of the American Medical Association. 2008;299:1345–50. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- 11.Iwagami S, Baba Y, Watanabe M, Shigaki H, Miyake K, Ishimoto T, et al. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Annals of surgery. 2013;257:449–55. doi: 10.1097/SLA.0b013e31826d8602. [DOI] [PubMed] [Google Scholar]
- 12.Bradly DP, Gattuso P, Pool M, Basu S, Liptay M, Bonomi P, et al. CDKN2A (p16) promoter hypermethylation influences the outcome in young lung cancer patients. Diagnostic molecular pathology : the American journal of surgical pathology, part B. 2012;21:207–13. doi: 10.1097/PDM.0b013e31825554b2. [DOI] [PubMed] [Google Scholar]
- 13.Brasset E, Chambeyron S. Epigenetics and transgenerational inheritance. Genome biology. 2013;14:306. doi: 10.1186/gb-2013-14-5-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wild CP, Scalbert A, Herceg Z. Measuring the exposome: A powerful basis for evaluating environmental exposures and cancer risk. Environmental and molecular mutagenesis. 2013;54:480–99. doi: 10.1002/em.21777. [DOI] [PubMed] [Google Scholar]
- 15.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. International journal of epidemiology. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa FF. Epigenomics in cancer management. Cancer management and research. 2010;2:255–65. doi: 10.2147/CMR.S7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verma M. Cancer control and prevention: nutrition and epigenetics. Current opinion in clinical nutrition and metabolic care. 2013;16:376–84. doi: 10.1097/MCO.0b013e328361dc70. [DOI] [PubMed] [Google Scholar]
- 18.Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nature reviews Genetics. 2013;14:585–94. doi: 10.1038/nrg3405. [DOI] [PubMed] [Google Scholar]
- 19.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics : official journal of the DNA Methylation Society. 2011;6:828–37. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang WY, Su LJ, Hayes RB, Moore LE, Katki HA, Berndt SI, et al. Prospective study of genomic hypomethylation of leukocyte DNA and colorectal cancer risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:2014–21. doi: 10.1158/1055-9965.EPI-12-0700-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgado-Cruzata L, Wu HC, Perrin M, Liao Y, Kappil MA, Ferris JS, et al. Global DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Epigenetics : official journal of the DNA Methylation Society. 2012;7:868–74. doi: 10.4161/epi.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic acids research. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrich CM, Toriola AT, Koepl LM, Sandifer T, Poole EM, Duggan C, et al. Metabolic, hormonal and immunological associations with global DNA methylation among postmenopausal women. Epigenetics : official journal of the DNA Methylation Society. 2012;7:1020–8. doi: 10.4161/epi.21464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, Baccarelli A, Shu XO, Ji BT, Yu K, Tarantini L, et al. Blood leukocyte Alu and LINE-1 methylation and gastric cancer risk in the Shanghai Women’s Health Study. British journal of cancer. 2012;106:585–91. doi: 10.1038/bjc.2011.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu HC, Delgado-Cruzata L, Flom JD, Perrin M, Liao Y, Ferris JS, et al. Repetitive element DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Carcinogenesis. 2012;33:1946–52. doi: 10.1093/carcin/bgs201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirabello L, Schiffman M, Ghosh A, Rodriguez AC, Vasiljevic N, Wentzensen N, et al. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. International journal of cancer Journal international du cancer. 2013;132:1412–22. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wentzensen N, Sun C, Ghosh A, Kinney W, Mirabello L, Wacholder S, et al. Methylation of HPV18, HPV31, and HPV45 genomes and cervical intraepithelial neoplasia grade 3. Journal of the National Cancer Institute. 2012;104:1738–49. doi: 10.1093/jnci/djs425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirabello L, Kratz CP, Savage SA, Greene MH. Promoter methylation of candidate genes associated with familial testicular cancer. International journal of molecular epidemiology and genetics. 2012;3:213–27. [PMC free article] [PubMed] [Google Scholar]
- 29.Wong EM, Southey MC, Fox SB, Brown MA, Dowty JG, Jenkins MA, et al. Constitutional methylation of the BRCA1 promoter is specifically associated with BRCA1 mutation-associated pathology in early-onset breast cancer. Cancer prevention research (Philadelphia, Pa) 2011;4:23–33. doi: 10.1158/1940-6207.CAPR-10-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian B, Katsaros D, Lu L, Canuto EM, Benedetto C, Beeghly-Fadiel A, et al. IGF-II promoter specific methylation and expression in epithelial ovarian cancer and their associations with disease characteristics. Oncology reports. 2011;25:203–13. [PMC free article] [PubMed] [Google Scholar]
- 31.Li WQ, Pan KF, Zhang Y, Dong CX, Zhang L, Ma JL, et al. RUNX3 methylation and expression associated with advanced precancerous gastric lesions in a Chinese population. Carcinogenesis. 2011;32:406–10. doi: 10.1093/carcin/bgq259. [DOI] [PubMed] [Google Scholar]
- 32.Rizvi MM, Alam MS, Ali A, Mehdi SJ, Batra S, Mandal AK. Aberrant promoter methylation and inactivation of PTEN gene in cervical carcinoma from Indian population. Journal of cancer research and clinical oncology. 2011;137:1255–62. doi: 10.1007/s00432-011-0994-0. [DOI] [PubMed] [Google Scholar]
- 33.Levine AJ, Win AK, Buchanan DD, Jenkins MA, Baron JA, Young JP, et al. Cancer risks for the relatives of colorectal cancer cases with a methylated MLH1 promoter region: data from the Colorectal Cancer Family Registry. Cancer prevention research (Philadelphia, Pa) 2012;5:328–35. doi: 10.1158/1940-6207.CAPR-11-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturgeon SR, Balasubramanian R, Schairer C, Muss HB, Ziegler RG, Arcaro KF. Detection of promoter methylation of tumor suppressor genes in serum DNA of breast cancer cases and benign breast disease controls. Epigenetics : official journal of the DNA Methylation Society. 2012;7:1258–67. doi: 10.4161/epi.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. The Journal of pathology. 2013;231:63–76. doi: 10.1002/path.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes LA, Melotte V, de Schrijver J, de Maat M, Smit VT, Bovee JV, et al. The CpG island methylator phenotype: what’s in a name? Cancer research. 2013;73:5858–68. doi: 10.1158/0008-5472.CAN-12-4306. [DOI] [PubMed] [Google Scholar]
- 37.Nishihara R, Morikawa T, Kuchiba A, Lochhead P, Yamauchi M, Liao X, et al. A prospective study of duration of smoking cessation and colorectal cancer risk by epigenetics-related tumor classification. American journal of epidemiology. 2013;178:84–100. doi: 10.1093/aje/kws431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes LA, Simons CC, van den Brandt PA, Goldbohm RA, de Goeij AF, de Bruine AP, et al. Body size, physical activity and risk of colorectal cancer with or without the CpG island methylator phenotype (CIMP) PloS one. 2011;6(4):e18571. doi: 10.1371/journal.pone.0018571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schernhammer ES, Giovannucci E, Baba Y, Fuchs CS, Ogino S. B vitamins, methionine and alcohol intake and risk of colon cancer in relation to BRAF mutation and CpG island methylator phenotype (CIMP) PloS one. 2011;6:e21102. doi: 10.1371/journal.pone.0021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Razzak AA, Oxentenko AS, Vierkant RA, Tillmans LS, Wang AH, Weisenberger DJ, et al. Alcohol intake and colorectal cancer risk by molecularly defined subtypes in a prospective study of older women. Cancer Prev Res (Phila) 2011;4:2035–43. doi: 10.1158/1940-6207.CAPR-11-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nature reviews Gastroenterology & hepatology. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Relton CL, Davey Smith G. Is epidemiology ready for epigenetics? International journal of epidemiology. 2012;41:5–9. doi: 10.1093/ije/dys006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cash HL, Tao L, Yuan JM, Marsit CJ, Houseman EA, Xiang YB, et al. LINE-1 hypomethylation is associated with bladder cancer risk among nonsmoking Chinese. International journal of cancer Journal international du cancer. 2012;130:1151–9. doi: 10.1002/ijc.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alshatwi AA. Breast cancer risk, dietary intake, and methylenetetrahydrofolate reductase (MTHFR)single nucleotide polymorphisms. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association. 2010;48:1881–5. doi: 10.1016/j.fct.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 45.Cebrian A, Pharoah PD, Ahmed S, Ropero S, Fraga MF, Smith PL, et al. Genetic variants in epigenetic genes and breast cancer risk. Carcinogenesis. 2006;27:1661–9. doi: 10.1093/carcin/bgi375. [DOI] [PubMed] [Google Scholar]
- 46.Chung MT, Sytwu HK, Yan MD, Shih YL, Chang CC, Yu MH, et al. Promoter methylation of SFRPs gene family in cervical cancer. Gynecologic oncology. 2009;112:301–6. doi: 10.1016/j.ygyno.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Kaaks R, Stattin P, Villar S, Poetsch AR, Dossus L, Nieters A, et al. Insulin-like growth factor-II methylation status in lymphocyte DNA and colon cancer risk in the Northern Sweden Health and Disease cohort. Cancer research. 2009;69:5400–5. doi: 10.1158/0008-5472.CAN-08-3020. [DOI] [PubMed] [Google Scholar]
- 48.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clinical chemistry. 2010;56:1871–9. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 49.Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. European journal of cancer (Oxford, England : 1990) 2011;47:784–91. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 50.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:108–14. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 51.Claus R, Hackanson B, Poetsch AR, Zucknick M, Sonnet M, Blagitko-Dorfs N, et al. Quantitative analyses of DAPK1 methylation in AML and MDS. International journal of cancer Journal international du cancer. 2012;131:E138–42. doi: 10.1002/ijc.26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akkiz H, Bayram S, Bekar A, Akgollu E, Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. Journal of viral hepatitis. 2011;18:e399–407. doi: 10.1111/j.1365-2893.2010.01414.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:4781–8. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 54.Cao Y, Miao XP, Huang MY, Deng L, Liang XM, Lin DX, et al. Polymorphisms of methylenetetrahydrofolate reductase are associated with a high risk of nasopharyngeal carcinoma in a smoking population from Southern China. Molecular carcinogenesis. 2010;49:928–34. doi: 10.1002/mc.20669. [DOI] [PubMed] [Google Scholar]
- 55.Bastian PJ, Palapattu GS, Yegnasubramanian S, Rogers CG, Lin X, Mangold LA, et al. CpG island hypermethylation profile in the serum of men with clinically localized and hormone refractory metastatic prostate cancer. The Journal of urology. 2008;179:529–34. doi: 10.1016/j.juro.2007.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao LM, Brennan P, van Bemmel DM, Zaridze D, Matveev V, Janout V, et al. LINE-1 methylation levels in leukocyte DNA and risk of renal cell cancer. PloS one. 2011;6:e27361. doi: 10.1371/journal.pone.0027361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsubayashi H, Sato N, Brune K, Blackford AL, Hruban RH, Canto M, et al. Age- and disease-related methylation of multiple genes in nonneoplastic duodenum and in duodenal juice. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:573–83. [PubMed] [Google Scholar]
- 58.Franzmann EJ, Reategui EP, Pedroso F, Pernas FG, Karakullukcu BM, Carraway KL, et al. Soluble CD44 is a potential marker for the early detection of head and neck cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:1348–55. doi: 10.1158/1055-9965.EPI-06-0011. [DOI] [PubMed] [Google Scholar]
- 59.Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, et al. Global levels of histone modifications predict prognosis in different cancers. The American journal of pathology. 2009;174:1619–28. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss G, Cottrell S, Distler J, Schatz P, Kristiansen G, Ittmann M, et al. DNA methylation of the PITX2 gene promoter region is a strong independent prognostic marker of biochemical recurrence in patients with prostate cancer after radical prostatectomy. The Journal of urology. 2009;181:1678–85. doi: 10.1016/j.juro.2008.11.120. [DOI] [PubMed] [Google Scholar]
- 61.Curtin K, Samowitz WS, Ulrich CM, Wolff RK, Herrick JS, Caan BJ, et al. Nutrients in folate-mediated, one-carbon metabolism and the risk of rectal tumors in men and women. Nutrition and cancer. 2011;63:357–66. doi: 10.1080/01635581.2011.535965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsubayashi H, Canto M, Sato N, Klein A, Abe T, Yamashita K, et al. DNA methylation alterations in the pancreatic juice of patients with suspected pancreatic disease. Cancer research. 2006;66:1208–17. doi: 10.1158/0008-5472.CAN-05-2664. [DOI] [PubMed] [Google Scholar]
- 63.Sun W, Zaboli D, Wang H, Liu Y, Arnaoutakis D, Khan T, et al. Detection of TIMP3 promoter hypermethylation in salivary rinse as an independent predictor of local recurrence-free survival in head and neck cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1082–91. doi: 10.1158/1078-0432.CCR-11-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tessema M, Belinsky SA. Mining the epigenome for methylated genes in lung cancer. Proceedings of the American Thoracic Society. 2008;5:806–10. doi: 10.1513/pats.200805-045TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baryshnikova E, Destro A, Infante MV, Cavuto S, Cariboni U, Alloisio M, et al. Molecular alterations in spontaneous sputum of cancer-free heavy smokers: results from a large screening program. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:1913–9. doi: 10.1158/1078-0432.CCR-07-1741. [DOI] [PubMed] [Google Scholar]
- 66.Dudziec E, Miah S, Choudhry HM, Owen HC, Blizard S, Glover M, et al. Hypermethylation of CpG islands and shores around specific microRNAs and mirtrons is associated with the phenotype and presence of bladder cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:1287–96. doi: 10.1158/1078-0432.CCR-10-2017. [DOI] [PubMed] [Google Scholar]
- 67.Mueller WC, Spector Y, Edmonston TB, St Cyr B, Jaeger D, Lass U, et al. Accurate classification of metastatic brain tumors using a novel microRNA-based test. The oncologist. 2011;16:165–74. doi: 10.1634/theoncologist.2010-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dietrich D, Krispin M, Dietrich J, Fassbender A, Lewin J, Harbeck N, et al. CDO1 promoter methylation is a biomarker for outcome prediction of anthracycline treated, estrogen receptor-positive, lymph node-positive breast cancer patients. BMC cancer. 2010;10:247. doi: 10.1186/1471-2407-10-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rawson JB, Mrkonjic M, Daftary D, Dicks E, Buchanan DD, Younghusband HB, et al. Promoter methylation of Wnt5a is associated with microsatellite instability and BRAF V600E mutation in two large populations of colorectal cancer patients. British journal of cancer. 2011;104:1906–12. doi: 10.1038/bjc.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Worthley DL, Whitehall VL, Buttenshaw RL, Irahara N, Greco SA, Ramsnes I, et al. DNA methylation within the normal colorectal mucosa is associated with pathway-specific predisposition to cancer. Oncogene. 2010;29:1653–62. doi: 10.1038/onc.2009.449. [DOI] [PubMed] [Google Scholar]
- 71.Jin Z, Cheng Y, Gu W, Zheng Y, Sato F, Mori Y, et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer research. 2009;69:4112–5. doi: 10.1158/0008-5472.CAN-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang MS, Uozaki H, Chong JM, Ushiku T, Sakuma K, Ishikawa S, et al. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:2995–3002. doi: 10.1158/1078-0432.CCR-05-1601. [DOI] [PubMed] [Google Scholar]
- 73.Delfino KR, Serao NV, Southey BR, Rodriguez-Zas SL. Therapy-, gender- and race-specific microRNA markers, target genes and networks related to glioblastoma recurrence and survival. Cancer genomics & proteomics. 2011;8:173–83. [PMC free article] [PubMed] [Google Scholar]
- 74.Avissar M, McClean MD, Kelsey KT, Marsit CJ. MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis. 2009;30:2059–63. doi: 10.1093/carcin/bgp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dikshit RP, Gillio-Tos A, Brennan P, De Marco L, Fiano V, Martinez-Penuela JM, et al. Hypermethylation, risk factors, clinical characteristics, and survival in 235 patients with laryngeal and hypopharyngeal cancers. Cancer. 2007;110:1745–51. doi: 10.1002/cncr.22975. [DOI] [PubMed] [Google Scholar]
- 76.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. The New England journal of medicine. 2009;361:1437–47. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Limsui D, Vierkant RA, Tillmans LS, Wang AH, Weisenberger DJ, Laird PW, et al. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. Journal of the National Cancer Institute. 2010;102:1012–22. doi: 10.1093/jnci/djq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, et al. DNA methylation markers and early recurrence in stage I lung cancer. The New England journal of medicine. 2008;358:1118–28. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 79.Bray I, Bryan K, Prenter S, Buckley PG, Foley NH, Murphy DM, et al. Widespread dysregulation of MiRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: association of miRNA expression with survival. PloS one. 2009;4:e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao J, Zhou J, Gao Y, Gu L, Meng H, Liu H, et al. Methylation of p16 CpG island associated with malignant progression of oral epithelial dysplasia: a prospective cohort study. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:5178–83. doi: 10.1158/1078-0432.CCR-09-0580. [DOI] [PubMed] [Google Scholar]
- 81.An J, Wei Q, Liu Z, Lu KH, Cheng X, Mills GB, et al. Messenger RNA expression and methylation of candidate tumor-suppressor genes and risk of ovarian cancer-a case-control analysis. International journal of molecular epidemiology and genetics. 2010;1:1–10. [PMC free article] [PubMed] [Google Scholar]
- 82.Dai W, Teodoridis JM, Zeller C, Graham J, Hersey J, Flanagan JM, et al. Systematic CpG islands methylation profiling of genes in the wnt pathway in epithelial ovarian cancer identifies biomarkers of progression-free survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:4052–62. doi: 10.1158/1078-0432.CCR-10-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Donahue TR, Tran LM, Hill R, Li Y, Kovochich A, Calvopina JH, et al. Integrative survival-based molecular profiling of human pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1352–63. doi: 10.1158/1078-0432.CCR-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim SJ, Kelly WK, Fu A, Haines K, Hoffman A, Zheng T, et al. Genome-wide methylation analysis identifies involvement of TNF-alpha mediated cancer pathways in prostate cancer. Cancer letters. 2011;302:47–53. doi: 10.1016/j.canlet.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 85.Slattery ML, Curtin K, Wolff RK, Herrick JS, Caan BJ, Samowitz W. Diet, physical activity, and body size associations with rectal tumor mutations and epigenetic changes. Cancer causes & control : CCC. 2010;21(8):1237–45. doi: 10.1007/s10552-010-9551-4. Epub 2010/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(15):4726–34. doi: 10.1158/1078-0432.ccr-07-4921. Epub 2008/08/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Costa VL, Henrique R, Danielsen SA, Duarte-Pereira S, Eknaes M, Skotheim RI, et al. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(23):5842–51. doi: 10.1158/1078-0432.ccr-10-1312. Epub 2010/10/27. [DOI] [PubMed] [Google Scholar]
- 88.Baden J, Green G, Painter J, Curtin K, Markiewicz J, Jones J, et al. Multicenter evaluation of an investigational prostate cancer methylation assay. The Journal of urology. 2009;182(3):1186–93. doi: 10.1016/j.juro.2009.05.003. Epub 2009/07/25. [DOI] [PubMed] [Google Scholar]
- 89.Vener T, Derecho C, Baden J, Wang H, Rajpurohit Y, Skelton J, et al. Development of a multiplexed urine assay for prostate cancer diagnosis. Clinical chemistry. 2008;54(5):874–82. doi: 10.1373/clinchem.2007.094912. Epub 2008/03/15. [DOI] [PubMed] [Google Scholar]