Significance
The mechanisms and location of polymerization and disassembly direct the function of cytoskeletal proteins. Septins are far less understood than other cytoskeletal elements such as actin and microtubules, yet they have a conserved function acting as scaffolds at cell membranes and are implicated in cancers, neurodegenerative diseases, and microbial pathogenesis. We have defined a key role of the membrane in directing septin filament formation in live cells and reconstituted dynamic septin polymerization, using purified components. We find that septins grow into filaments and form higher-order structures by diffusing, colliding, and annealing on the plasma membrane. This work is important because it defines previously unidentified basic steps of polymerization and construction of higher-order assemblies of septin proteins.
Keywords: cytoskeleton, biophysics
Abstract
Septins assemble into filaments and higher-order structures that act as scaffolds for diverse cell functions including cytokinesis, cell polarity, and membrane remodeling. Despite their conserved role in cell organization, little is known about how septin filaments elongate and are knitted together into higher-order assemblies. Using fluorescence correlation spectroscopy, we determined that cytosolic septins are in small complexes, suggesting that septin filaments are not formed in the cytosol. When the plasma membrane of live cells is monitored by total internal reflection fluorescence microscopy, we see that septin complexes of variable size diffuse in two dimensions. Diffusing septin complexes collide and make end-on associations to form elongated filaments and higher-order structures, an assembly process we call annealing. Septin assembly by annealing can be reconstituted in vitro on supported lipid bilayers with purified septin complexes. Using the reconstitution assay, we show that septin filaments are highly flexible, grow only from free filament ends, and do not exchange subunits in the middle of filaments. This work shows that annealing is a previously unidentified intrinsic property of septins in the presence of membranes and demonstrates that cells exploit this mechanism to build large septin assemblies.
Septin filaments form rings, bars, and gauzes that serve as a scaffold at cell division sites; act to retract blebbed regions of membrane; and restrict diffusion between cell compartments (1–4). Septin function is required for cell division and viability in many eukaryotes whereas misregulation is associated with cancers and neurodegenerative disorders (5–8). Furthermore, septins mediate entry of both bacterial and fungal pathogens into host cells (9–11). In vivo, septin assembly is restricted both in time and in space through local activation of small GTPases such as Cdc42. Localized signaling leads to higher-order septin structures forming closely apposed to the plasma membrane at the plane of division, sites of polarity, and curved membranes (10, 12–14). Notably, eukaryotic cells of different geometries build higher-order septin assemblies of various shapes, sizes, and functions (4, 15, 16). Although septins are critical for spatial organization of cell plasma membranes, their assembly and disassembly dynamics are not understood (15).
Electron microscopy (EM) studies of recombinant and immunoprecipitated Saccharomyces cerevisiae septins have shown that septins form nonpolar hetero-octameric rod-shaped complexes in high-salt buffers (>300 mM) and elongated filaments when dialyzed into low-salt buffers (<100 mM) (17, 18). Structural analyses of worm and mammalian septins have revealed that the heteromeric, rod-shaped complex is conserved (19–21). Thus, septin rods characterized to date contain two copies of each septin subunit assembled into a nonpolar, heteromeric complex (Fig. S1). Association of purified septin proteins with phosphoinositide-containing membrane monolayers placed on EM grids can promote the assembly of septin filaments in otherwise nonpermissive conditions such as high salt or the presence of mutant septins in the complex (22). A polybasic region in the N terminus of septin proteins as well as other surfaces of the septin protein have been proposed to be the basis for septin association with phosphoinositides; however, the functional role of membranes in filament formation is not yet known (22–24).
Previous work has defined a possible starting state of assembly (the rod) and endpoint (filaments and gauzes); however, there is nothing known about how filaments elongate either in vivo or in vitro. Do septin filaments extend by stepwise addition of rods? Does addition occur from both ends of the filament? Can subunits be added in the middle and/or sides of a filament? Do septin filaments grow in the cytosol or on plasma membranes? Thus, it is still not known where filaments form in vivo, how filaments elongate, and how filaments are brought together to construct higher-order assemblies.
The goal of this study was to identify the locations and mechanism of septin filament polymerization. Using fluorescence correlation spectroscopy (FCS), we observed that cytosolic septins are likely rods, not monomers or filaments. Using total internal reflection fluorescence (TIRF) microscopy, we found septins form short filaments on the plasma membrane and then long filaments and higher-order assemblies grow when filaments merge. We have called the process of short filaments coming together “annealing” as this has previously been described for F-actin in vitro (25). We reconstituted septin assembly, using purified proteins and supported lipid bilayers, and found that annealing is an intrinsic property of septins that occurs at very low protein concentrations in the presence of a supported phospholipid bilayer. Our results suggest that the plasma membrane concentrates septins and provides a platform for 2D diffusion that promotes polymerization.
Results
Septins Are in Small, Oligomeric Complexes in Cytosol.
To determine how and where septins transition from monomers to filaments, we observed septins in the cytosol of three different fungal organisms: Schizosaccharomyces pombe, Saccharomyces cerevisiae, and Ashbya gossypii. These three systems have distinct cell morphologies and cell cycle controls influencing septin assembly and function. In all three systems, a septin was tagged with GFP at the native locus and expressed as the sole copy of the gene to monitor endogenous levels and enable quantitative analysis of fluorescence intensity (Tables S1–S3). To examine composition and properties of septin complexes in the cytoplasm, fluctuations in fluorescence intensity were monitored and autocorrelated using FCS. The cytoplasmic concentration of fluorescently labeled septins was measured to be 100–200 nM, which is on the order of previous estimates in S. pombe, using different approaches (Fig. S2A) (26).
We expected to observe cytosolic septins behaving as if they were in one of three states: as individual proteins, as rod complexes with two copies of each septin, or as filaments (Fig. S1). Molecular brightness measurements by FCS indicated that septins in the cytosol of all three model systems are in complexes containing one to two fluorescently labeled molecules (Fig. 1A and Fig. S2 B–D). Importantly, similar molecular brightness values were measured regardless of which septin family member was tagged with GFP (Spn1/Cdc3 in S. pombe, Cdc11 in A. gossypii, and Cdc11 or Cdc3 in S. cerevisiae; Fig. 1A and Fig. S2 B–D). We suspect that in some cases a single molecule rather than two fluorescent molecules is detected because a subset of rods is in an intermediate state of assembly or a state of disassembly or both chromophores in the complex have not fully matured. Furthermore, cytoplasmic septins diffused at 0.67 ± 0.29 µm2/s (S. pombe), which is much slower than diffusion of cytoplasmic mEGFP alone, which was 25.80 ± 7.97 µm2/s. This diffusion behavior is consistent with a complex that is likely larger in size than a single septin protein (Fig. 1A and Fig. S2 B–D). The reproducible detection of septin complexes with one to two fluorescent molecules and the slow diffusion of these complexes relative to mEGFP suggest septins are predominantly in heteromeric rods in the cytosol rather than in monomers or filaments.
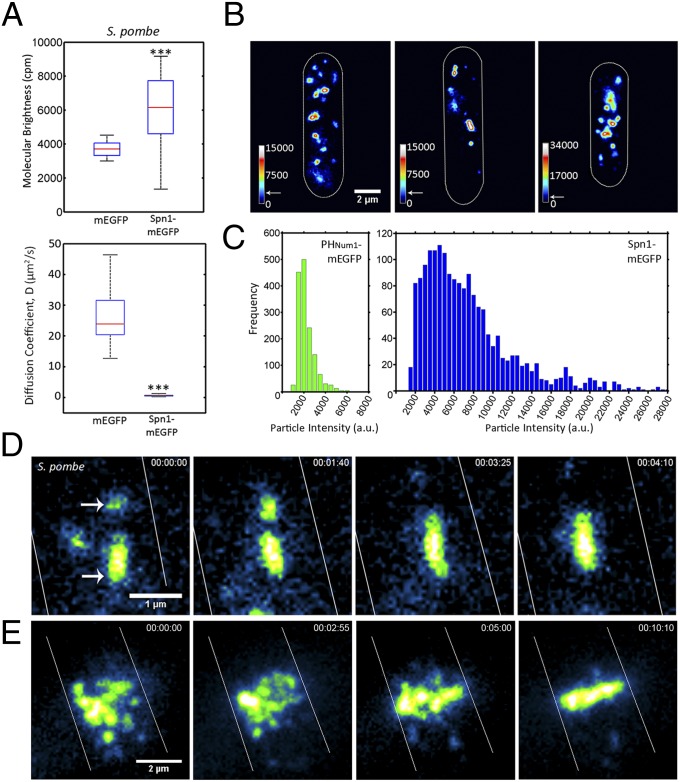
Fig. 1.
Septins grow by annealing on the plasma membrane of S. pombe. (A) Molecular brightness and diffusion coefficient of cytoplasmic Spn1-mEGFP (AGY093) determined by FCS compared with those of cells expressing cytoplasmic mEGFP (AGY109) alone in S. pombe. Box plots display median, lower, and upper quartiles and whiskers show range n > 20 cells, ***P < 0.001 by Kruskal–Wallace one-way analysis of variance. (B) Spn1-mEGFP particles and short filaments localized on the cortex of S. pombe monitored using TIRF microscopy. Heat maps represent relative particle intensities. Arrows on the intensity scale indicate the estimated intensity of a complex containing two Cdc11-mEGFP molecules, ∼4,000 a.u., based on calibration. Cell outlines are shown in white. (C) Distribution of PHNum1-GFP and Spn1-mEGFP particle intensities on the plasma membrane measured from images collected using the same laser intensity and exposure time. (D) Annealing event between two diffusing short septin filaments on the cell cortex of S. pombe. Arrows indicate particles involved in annealing. Cell outines are shown in white. (E) Formation of Spn1-mEGFP septin ring by annealing of intermediate-size particles at the plane of division in S. pombe. Cell outines are shown in white. With the exception of FCS (A), all images were acquired using TIRF microscopy.
Septins Elongate by Annealing on Plasma Membranes.
We next assessed the properties of septins at the plasma membrane of living cells, using TIRF microscopy. Septins were found to diffuse in two dimensions as discrete spots and filaments (defined as linear-shaped signals that in fact may be more complex than a single filament) at the plasma membrane in both S. pombe and A. gossypii cells (Fig. 1B, Fig. S3, and Movies S1 and S2). These signals could be seen moving all over the cell cortex and presumably have not been detected in previous studies due to the substantial cytosolic background fluorescence that obscures them in widefield or spinning-disk confocal imaging.
To determine the composition of the signals, we calculated the number of fluorescent septin-mEGFP molecules in individual spots in S. pombe cells. To do this, signal intensities of septins were detected and measured using custom MATLAB-based software. To estimate the number of fluorescent molecules in each spot, the measured intensity values of septin signals were compared with the intensity of mEGFP fused to a PH domain, previously shown to localize to the plasma membrane in spots containing one to two molecules of mEGFP (Fig. 1C) (27). Based on this calibration, septin spots contained highly variable numbers of fluorescent molecules with the vast majority greater in intensity than a predicted octameric complex, which we predict should be twice the intensity of a single mEGFP (population mean PHNum1-mEGFP = 1,974 ± 820 a.u., n = 1,510 particles; Spn1-mEGFP = 7,196 ± 4,896 a.u., n = 1,774 particles; Fig. 1C). Thus, septins are found in complexes of heterogeneous size on the cortex of fission yeast cells.
Fluorescent septin spots and short filaments collide and fuse, a process that we term annealing, and a number of these events occurring sequentially leads to the eventual formation of elongated filaments (Fig. 1D, Movies S1 and S2, and Fig. S3). Once formed, filaments can be observed to fragment (Movies S1 and S2). As the cell cycle progressed in S. pombe, short filaments localized to the center of the cell, annealed there, and subsequently formed continuous bright ring structures at the site of cell division (Fig. 1E and Movie S3). Our results suggest that septins arrive at the plasma membrane from the cytosol as rod complexes, which then elongate through diffusion-mediated collisions and annealing to construct large assemblies.
Reconstitution of Septin Filament Elongation.
To determine whether elongation by annealing is a fundamental behavior of septin complexes or is mediated by other factors in the cell, we reconstituted septin assembly, using recombinant proteins on supported lipid bilayers (Fig. S4A). S. cerevisiae septin complexes containing Cdc11-mEGFP, Cdc12-(His6), Cdc3, and Cdc10 were coexpressed in Escherichia coli and purified (Fig. S4B). Complexes were diluted to a final concentration of 0.5–3 nM of the octameric complex in low-salt buffer (50 mM NaCl). We first determined the soluble state of purified septin complexes in a low-salt solution. Using FCS, we found septin-containing particles fluoresced to a median molecular brightness of slightly less than twice the value of purified mEGFP-(His6). This intensity of signal is consistent with the interpretation that the majority of the population of septins are in complexes with two labeled septins (Fig. 2A). Purified septin complexes diffused at a mean of 31 ± 5 µm2/s in solution compared with 101 ± 6 µm2/s for purified mEGFP-(His6) (Fig. 2B). This measured diffusion constant for septin complexes is consistent with a theoretical estimate for a rod with a diameter of 4 nm, a length of 32 nm, and a molecular weight of eight septin proteins (Fig. 2C). Thus, purified septins at low concentration (3 nM) in low-salt solution likely exist as an octameric rod rather than as elongated filaments.
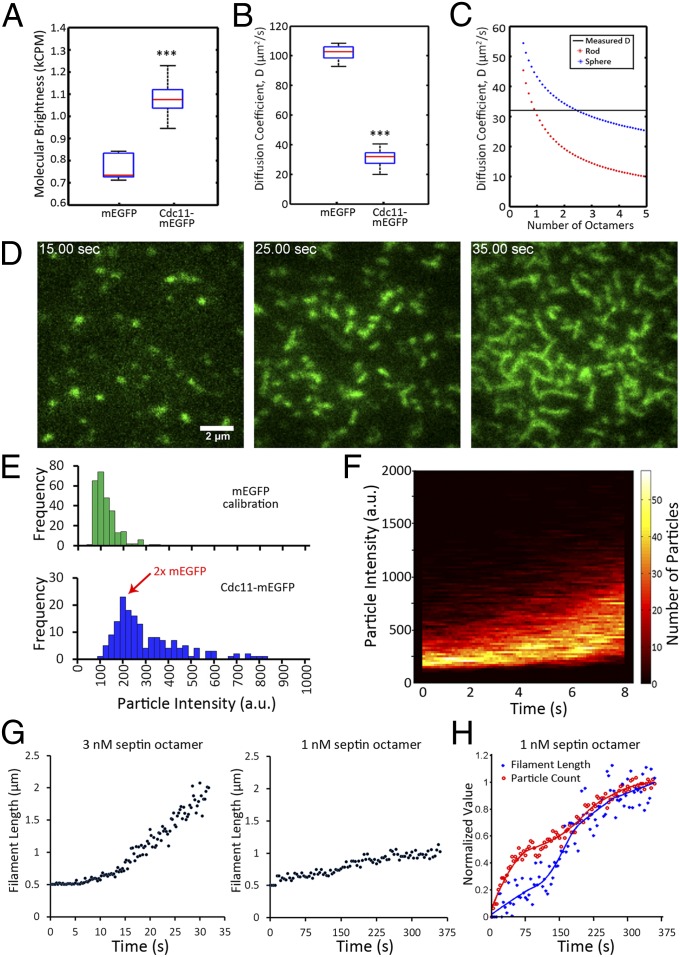
Fig. 2.
Reconstituted septin assembly on supported lipid bilayers. (A) Molecular brightness of pure mEGFP compared with that of copurified, recombinant septins containing Cdc11-mEGFP in solution, as measured by FCS. Box plots display median, lower, and upper quartiles and whiskers show range. ***P < 0.001 by Kruskal–Wallace one-way analysis of variance. (B) Diffusion coefficients of pure mEGFP compared with Cdc11-mEGFP containing septin complexes, estimated by fitting FCS results to a single-component anamolous diffusion model. (C) Diffusion coefficients were predicted for complexes of different numbers of septin octamers, using previously measured dimensions (i.e., length = 32 nm, diameter = 4 nm, molecular mass = 443 kDa) modeled as either rod-like or spherical in organization for a given molecular mass. Horizontal black line is measured diffusion constant, red asterisks are predicted diffusion constants for septin complexes in a rod-like shape, and blue asterisks are diffusion constants of complexes arranged in a sphere. (D) Cdc11-mEGFP signal accumulates and forms filaments at the lipid bilayer over time (3 nM octamer concentration), monitored by TIRF microscopy. (E) Quantification of fluorescence intensity (background subtracted) of single molecules of mEGFP (green histogram) and Cdc11-mEGFP complexes in the first moment of arrival at the bilayer (3 nM, blue histograms). (F) TIRF microscopy was used to measure septin particle brightness in each frame during the first 8 s of assembly (3 nM) in conditions where single molecules of mEGFP fluoresce to 100 a.u. The number of particles of a given intensity is represented by the color code beside the plot (n > 10,000 particles). (G) Filament length was measured over time and the mean length of elongated particles is plotted for each time point in a 3-nM and a 1-nM mixture of septin complex. (H) Particle count and filament length over time in a 1-nM assembly (same data as in G). Values were normalized to maximum values of length or particle count and minimum values were set to zero. These values were fitted and plotted normalized to the best-fit curve.
We next added purified septin complex to supported lipid bilayers in an attempt to observe septin filament formation. Bilayers were prepared with 75:22:3 mol% phosphatidylcholine:phosphatidylinositol:phosphatidylinositol-4,5-bisphosphate and were judged to be fluid based on observation of 2D diffusion of individual dye molecules when a trace amount of rhodamine-labeled phosphatidylethanolamine was included in the mixture (D = 1.46 ± 1.05 μm2/s, n = 25 particles; Fig. S4C and Movie S4). Septins were then monitored using TIRF microscopy to determine their behavior in the plane of the model lipid bilayer.
At 3 nM septin octamer, short filaments formed on the bilayer within 30 s, enabling an analysis of the filament assembly process in real time (Fig. 2D and Movie S5). To determine the smallest unit of assembly, we measured the number of septin-GFP molecules in each diffraction-limited spot at the first moments of detection. To do this, we used custom MATLAB-based software to detect the spots and calibrated the measured intensities, using intensity measurements of single molecules of pure mEGFP. Septin spots were initially found to fluoresce at an intensity comparable to ∼2× mEGFP, supporting the conclusion that the rod complex containing two copies of Cdc11-mEGFP is the initial unit of assembly (Fig. 2E). Thereafter, fluorescent spots were continually recruited to the bilayer and an increase in the fluorescence intensity of each spot was noted with time (Fig. 2F). When combined, these data suggest Cdc11 is present in an octamer when it arrives at the membrane (with two molecules of Cdc11) and then rapidly forms short filaments (composed of multiple octamers) in the initial steps of assembly.
We then asked how filament formation was influenced by the concentration of septins in solution added to the bilayer. At 3 nM, puncta were clearly elongated in shape after 5–10 s and within 30 s, filaments of 1.5–2 μm were readily detected (Fig. 2G). Late in the assembly process at 3 nM, the network of septin filaments became too dense to accurately measure individual filament lengths; however, continued recruitment of septins to the bilayer was indicated by increasing fluorescence intensity (Fig. S4D). In contrast, at 1 nM, elongation occurred much more slowly and was found to reach equilibrium at shorter filament lengths (Fig. 2G). The number of particles at the bilayer was found to increase from the initial addition of septins through elongation and this count represents a balance between recruitment of new particles from solution and merging of particles into filaments over time (Fig. 2H). Notably, elongated puncta and very short filaments could be seen at concentrations as low as 0.5 nM. These data support that the length of filaments and kinetics of assembly are impacted by the concentration of septins used in the assay.
Septins Elongate by Annealing on Supported Lipid Bilayers.
We next examined the mechanism of filament elongation on supported lipid bilayers. Filaments were defined as elongated structures that moved as a single unit. The elongated structures were produced by fusion of rod complexes to produce short filaments and subsequently short filaments merged together to form longer filaments (Fig. 3A and Movie S6). Thus, pure septin complexes on diffusive lipid bilayers recapitulated the annealing behavior observed in vivo.
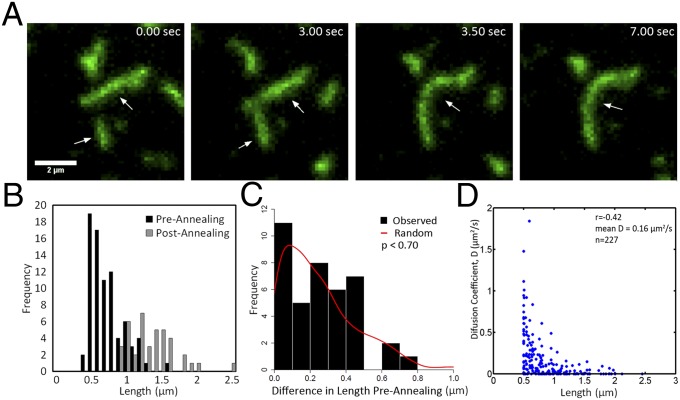
Fig. 3.
Septin filaments grow by annealing on supported lipid bilayers. (A) Representative TIRF image series of filament growth by annealing. Arrows indicate short filaments that merge. (B) Measured filament lengths before annealing and after annealing are plotted (n = 40 annealing events). (C) Histogram shows the length difference between pairs of filaments before annealing. These filaments that participated in annealing events were then paired at random, and differences were calculated from the random pairs and plotted as a red line. The simulated random distribution of differences is not distinguishable from the actual data (insignificant two-sample Kolmogorov–Smirnov test). (D) Filament length and diffusion coefficients were measured and plotted for a population of septin particles at the bilayer.
We then evaluated whether filament length impacts annealing events or diffusive behavior of complexes on the membrane. We wondered whether there was any bias in the size of filaments that anneal; however, we could not detect a systematic pattern in the sizes of filaments that participate in an annealing event (Fig. 3 B and C). This means that filaments can merge regardless of their relative sizes. This property can be seen when we calculate the difference in length between two filaments that anneal and compare this measured distribution to a distribution made from a simulation in which the same collection of filaments is paired at random (Fig. 3C, no significant difference between observed and simulated random distribution). Length does, however, have a modest impact on filament diffusion rates in that we detect a moderate negative correlation between filament length and diffusion rate as measured by mean square displacement (r = −0.42, mean = 0.16 µm2/s, Fig. 3D). Thus, filament length does not restrict septin annealing events directly; however, filament length does modestly impact rates of diffusion.
Next, we evaluated whether filaments can elongate at both free ends and whether subunits can be incorporated in the middle/sides of a preexisting filament. For these experiments, septin complexes containing Cdc11-mCherry were purified and mixed with Cdc11-mEGFP complexes. Specifically, Cdc11-mEGFP complexes were first assembled on a supported lipid bilayer and excess, non-membrane–bound complexes were removed. Cdc11-mCherry septin complexes were then added and the locations of new additions were monitored. We found addition occurred at filament ends and in many cases preformed filaments elongated at both ends (Fig. 4A and Movie S7). The majority of newly incorporated septin arrived at preformed filament ends after 2D diffusion on the membrane rather than arriving directly from the bulk solution (81%, n = 21). No Cdc11m-Cherry–labeled complexes were found to add to the middle of preexisting green filaments. Over time, however, mCherry subunits could be seen in the middle of filaments due to repeated fragmentation and reannealing of the original GFP-labeled filaments. These results support the conclusion that elongation occurs exclusively at the ends of preexisting filaments and suggest that septins do not exchange subunits internally or bind to sides of preexisting filaments in these conditions.
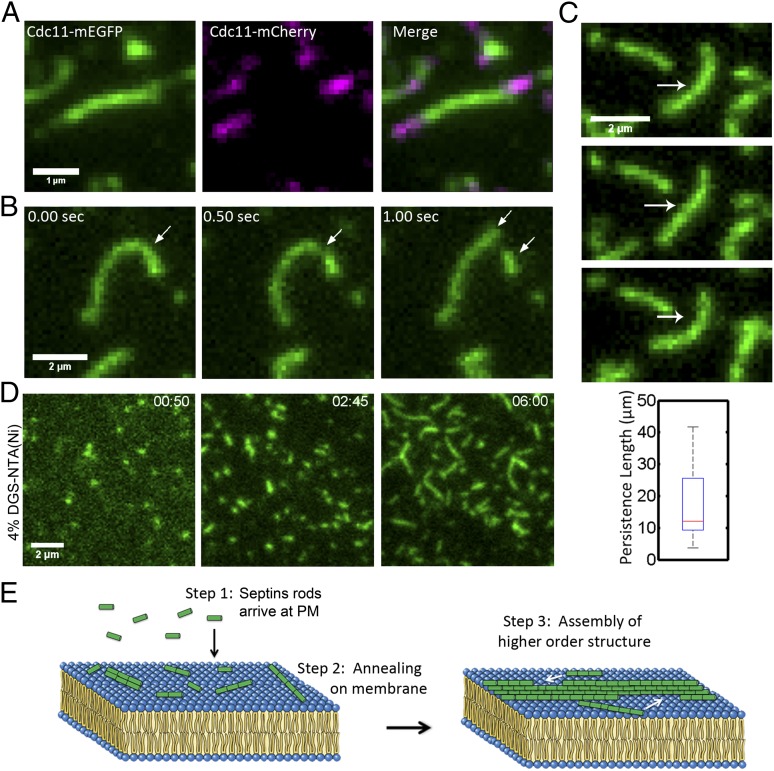
Fig. 4.
Septin filaments elongate at ends, are flexible, and can be formed on a membrane without phosphatidylinositide. (A) Cdc11-mEGFP septin filaments were assembled on a bilayer, unbound complexes were washed out, and Cdc11-mCherry complexes were added and monitored in TIRF. Addition of new complexes occurred at both filament ends. (B) Cdc11-mEGFP filaments fragment on a supported lipid bilayer. Arrows indicate site of fragmentation event and new short filaments arising from the break. (C) Example of filament-bending image series used for determining persistence length of septin filaments. Arrow indicates bending filament. Box plot shows a median persistence length of 12 μm. Filaments used ranged between 1.2 µm and 2.1 µm, with a median of 1.8 µm (n = 10). Box plots display median, lower, and upper quartiles and whiskers show range. (D) Cdc11-mEGFP containing septin filaments assembled on supported lipid bilayers containing 96% (mol%) phosphatidylcholine and 4% (mol%) DGS-NTA(Ni). (E) Model. Step 1: Septins arrive at plasma membranes from cytoplasm as short rods. Step 2: Rods then assemble into short filaments on the plasma membrane through annealing. Step 3: Short filaments then build higher-order structures.
In addition to annealing, fragmentation of filaments was also readily observed in vitro as well as in vivo (Fig. 4B and Movie S8). Filaments can fragment at a variety of places and breakage is not limited to the ends or the middle. Fragmentation occurred after filaments bent, indicating that the flexibility is integral to setting the filament length and stability on the bilayer.
To measure the flexibility of septin filaments, bending over time was used to determine persistence length. Persistence length is a measure of the stiffness of a polymer, and we calculated this property of septins as described by Gittes et al. for actin and microtubules (28). Septins were found to have a median persistence length of 12 µm on bilayers and there was no correlation between filament length and persistence length (Fig. 4C). These estimates are in contrast to 18 µm for actin and 5,200 µm for microtubules on glass. These measurements support that septins are highly flexible filaments and that in this reconstituted setting these are unlikely to be bundled filaments.
In vitro, two possible mechanisms could favor septin filament elongation on supported lipid bilayers rather than in solution. It is possible that an effective concentration increase and restriction of diffusion to two dimensions control the formation of septin filaments. Alternatively, a phosphatidylinositol-induced conformational change that occurs within septins upon association with the membrane could promote filament formation. To differentiate between these two scenarios, an alternative approach to bilayer recruitment was used. Because septin complexes were purified using Cdc12-(His6), bilayers containing 4% (mol%) DGS-NTA(Ni) lipids and 96% (mol%) phosphatidylcholine were prepared and analyzed for their ability to promote septin filament assembly (Fig. 4D). The same property of elongation by annealing was observed on membranes of this composition of lipids although the assembly of filaments was slower than in phosphatidylinositol-containing bilayers (Movie S9). Notably, no filaments formed when purified septin complexes were added to cleaned or casein- or BSA-coated glass, indicating that diffusion provided by a fluid membrane is a critical component of filament assembly at low septin concentrations (Fig. S4E). Thus, in a reduced system containing septin heteromeric complexes and a supported lipid bilayer, septins assemble filaments and higher-order structures through 2D diffusion and annealing. These data suggest that annealing is a fundamental property of the septin polymer that drives the assembly process.
Discussion
Dynamic transitions between soluble and assembled states lend cytoskeletal polymers many of their functional properties, from force generation and motility to contraction and scaffolding (29, 30). Based on the work reported here, we are able to propose a three-step process for the formation of septin assemblies (Fig. 4E). First, as measured by FCS, septins oligomerize into small complexes that are not filaments in the cytoplasm. These complexes are likely to be rods that are heterooctamers or decamers in fungi based on previous work with purified and recombinant proteins in high-salt conditions (17, 18, 31, 32). Subsequently, when rods contact the plasma membrane, they are concentrated, diffuse, and merge to grow into short filaments through annealing. These short filaments then diffuse in the plane of the membrane, merging into each other and nascent higher-order structures such as the rings and collars that demarcate sites of cytokinesis in fungal cells (Fig. 4E). In summary, this study shows septin membrane associations effectively concentrate subunits and limit the dimensionality of diffusion to promote filament formation.
In cells, it is unknown what controls the ratio of the amount of septins on the plasma membrane to the amount in the cytoplasm. It is interesting to note that septins assemble in vitro at concentrations that are orders of magnitude lower than the measured cytoplasmic concentration of septins. This could indicate that even at high concentrations, three-dimensional diffusion is insufficient to support annealing of rods in the cytosol. Alternatively there could be a conformational change induced by membrane association or a capping protein that dissociates at the membrane to restrict the location of elongation. This possible function of the membrane need not necessarily be phosphoinositide dependent based on the in vitro experiments where filaments form with Ni-NTA–based recruitment (Fig. 4). It will be critical in the future to determine whether septin rods are capped to limit cytosolic polymerization and whether the membrane provides a distinct role in assembly beyond restricting diffusion to two dimensions.
With the reconstitution assay, we can make predictions about the mechanisms driving septin polymerization. We speculate filaments form through what is likely an isodesmic rather than a cooperative, nucleation–elongation process. An isodesmic assembly is consistent with a relatively low concentration for polymerization in the presence of a membrane and a broad filament size distribution (33). It is possible that in early stages of assembly, in diffraction-limited spots, septins do cooperatively assemble via a process not detectable by this assay. Furthermore, it is conceivable that the assembly of heteromeric rods in the cytosol is a cooperative process that cannot be monitored using the present approaches in vivo. Mixed (i.e., isodesmic and cooperative) modes of assembly have been speculated to exist for FtsZ and even actin displays isodesmic or nucleation–elongation assembly kinetics depending on the species of origin (33, 34). For example, animal F-actin assembly clearly displays a nucleation–elongation mechanism of assembly, whereas parasite F-actin forms by an isodesmic process reminiscent of what we see with septins on membranes (35).
The behavior of septins in the vitro assay supports the conclusion that filament assembly begins with the recruitment of individual septin rods from solution to membranes and that filament growth progresses in a bidirectional, end-dependent manner. It is clear new rods are not added in the middle of premade filaments; nor do they associate with the sides of preexisting filaments. This indicates both that exchange of subunits within a filament does not occur under these conditions and that rods are not joining filaments by lateral associations but rather by only end-on associations. Furthermore, in vitro, we do not see filaments elongate due to addition of subunits arriving from the bulk solution but rather primarily through collisions between particles diffusing in two dimensions. Importantly, in vivo it cannot be determined whether soluble septins are also able to join larger assemblies directly from the cytosol in addition to diffusion-driven annealing. Analyzing this process in vivo is complicated by the fact that it is unclear whether all septin filaments are membrane associated or whether there are layers of septin filaments stacked on membranes with some filaments directly binding membranes and some filaments associating with other filaments. Future work in vivo should consider how to address the issue of exchange of subunits within filaments and bundling in assembled higher-order structures. These facets of septin assembly likely require additional cytosolic factors, posttranslational modifications, and/or lateral associations between filaments.
How do the properties of septins we have found here in terms of flexibility, annealing, and fragmentation relate to the properties of higher-order structures of septins in cells? We speculate that flexibility of filaments and their capacity to anneal may accelerate assembly, rearrangement, and disassembly of higher-order structures. Previous work using polarization microscopy has demonstrated that the septin hour-glass structure in yeast is anisotropic, suggesting it is highly ordered both before and after cytokinesis (13, 36, 37). We imagine that the order emerges from a septin binding or bundling partner that constrains and contains the flexible filaments. Alternatively, tight lateral associations between adjacent filaments may be sufficient to bundle filaments and create the highly anisotropic assemblies observed in cells. If present, these lateral associations either form in a concentration-dependent manner or require a cytosolic factor because we did not obtain any evidence for lateral associations on membranes in vitro. Despite appearing highly anisotropic through most of the cell cycle, the septin hourglass displays a notably more “fluid” or isotropic transition at cytokinesis (1, 13, 36, 37). The flexibility, annealing, and fragmentation properties are likely important for transit through the isotropic phase. The loss, addition, or modification of a binding partner or septins themselves could initiate this transition seen by polarization microscopy at cytokinesis. Future work adding in putative septin regulators to the in vitro system will be illuminating to determine whether any cell-cycle–regulated, septin-binding proteins can bring order to flexible filaments.
This report presents reconstitution of septin polymerization in vitro that captures and quantitatively measures the dynamics of septin assembly on a bilayer. This will be a powerful model system on which to test the role of nucleotides, posttranslational modifications, and accessory factors in the organization of septins. By combining a reconstitution assay with live cell imaging, this work reveals a role for a plasma membrane in driving septin filament elongation and the construction of higher-order assemblies.
Materials and Methods
Supported Lipid Bilayer Preparation.
Supported lipid bilayers were prepared on clean glass as detailed in SI Materials and Methods. Briefly, after sonication and plasma cleaning, small unilamellar vesicles (SUVs) of desired lipid mixture were prepared by rehydration and bath sonication until the solution clarified and SUVs were allowed to fuse with glass. Bilayers were washed with 50 mM Tris, pH 8.0, 300 mM NaCl, and 1 mg/mL fatty acid-free BSA (Sigma A6003) before the addition of septins. Detailed methods can be found in SI Materials and Methods.
Image Analysis.
Images were processed and analyzed using ImageJ, MATLAB, and Nikon Elements software (38). Detailed descriptions of particle detection, tracking, and quantification can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the laboratories of T. Mitchison, C. Field, R. Oldenbourg, and the cytoskeletal community at Dartmouth College for thoughtful discussion; R. Sloboda for critically reading the manuscript; M. Loose for training in the preparation of supported lipid bilayers; A. Lavanway for support with microscopes; J. Q. Wu, J. Moseley, and H. Ewers for sharing strains; C. Anderson for statistical advice; J. Mayor and D. Köster for lipids and protocols; and J. Thorner for sharing plasmids. This project was supported by funding from the National Science Foundation (MCB-507511, to A.S.G.) and the National Institutes of Health (GM100160, to T.T. and A.S.G.) and by Colwin, Lemann, and Spiegel summer fellowships and The Nikon Award for summer investigation at the Marine Biological Laboratory in Woods Hole, MA (A.S.G.) and instrument support from Micro Video Instruments.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314138111/-/DCSupplemental.
References
- 1.Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305(5682):393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- 2.Gilden JK, Peck S, Chen YC, Krummel MF. The septin cytoskeleton facilitates membrane retraction during motility and blebbing. J Cell Biol. 2012;196(1):103–114. doi: 10.1083/jcb.201105127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69(3):717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh Y, Bi E. Septin structure and function in yeast and beyond. Trends Cell Biol. 2011;21(3):141–148. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culotti J, Hartwell LH. Genetic control of the cell division cycle in yeast. 3. Seven genes controlling nuclear division. Exp Cell Res. 1971;67(2):389–401. doi: 10.1016/0014-4827(71)90424-1. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita A, et al. Identification of septins in neurofibrillary tangles in Alzheimer’s disease. Am J Pathol. 1998;153(5):1551–1560. doi: 10.1016/S0002-9440(10)65743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall PA, Russell SE. The pathobiology of the septin gene family. J Pathol. 2004;204(4):489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- 8.Russell SE, Hall PA. Septin genomics: A road less travelled. Biol Chem. 2011;392(8-9):763–767. doi: 10.1515/BC.2011.079. [DOI] [PubMed] [Google Scholar]
- 9.Dagdas YF, et al. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science. 2012;336(6088):1590–1595. doi: 10.1126/science.1222934. [DOI] [PubMed] [Google Scholar]
- 10.Mostowy S, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8(5):433–444. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Ryder LS, et al. NADPH oxidases regulate septin-mediated cytoskeletal remodeling during plant infection by the rice blast fungus. Proc Natl Acad Sci USA. 2013;110(8):3179–3184. doi: 10.1073/pnas.1217470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tooley AJ, et al. Amoeboid T lymphocytes require the septin cytoskeleton for cortical integrity and persistent motility. Nat Cell Biol. 2009;11(1):17–26. doi: 10.1038/ncb1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeMay BS, et al. Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J Cell Biol. 2011;193(6):1065–1081. doi: 10.1083/jcb.201012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gladfelter AS, Bose I, Zyla TR, Bardes ES, Lew DJ. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J Cell Biol. 2002;156(2):315–326. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beise N, Trimble W. Septins at a glance. J Cell Sci. 2011;124(Pt 24):4141–4146. doi: 10.1242/jcs.087007. [DOI] [PubMed] [Google Scholar]
- 16.Spiliotis ET, Gladfelter AS. Spatial guidance of cell asymmetry: Septin GTPases show the way. Traffic. 2012;13(2):195–203. doi: 10.1111/j.1600-0854.2011.01268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertin A, et al. Saccharomyces cerevisiae septins: Supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105(24):8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frazier JA, et al. Polymerization of purified yeast septins: Evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143(3):737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barral Y, Kinoshita M. Structural insights shed light onto septin assemblies and function. Curr Opin Cell Biol. 2008;20(1):12–18. doi: 10.1016/j.ceb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 20.John CM, et al. The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 2007;26(14):3296–3307. doi: 10.1038/sj.emboj.7601775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirajuddin M, et al. Structural insight into filament formation by mammalian septins. Nature. 2007;449(7160):311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- 22.Bertin A, et al. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J Mol Biol. 2010;404(4):711–731. doi: 10.1016/j.jmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol. 1999;9(24):1458–1467. doi: 10.1016/s0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
- 24.Casamayor A, Snyder M. Molecular dissection of a yeast septin: Distinct domains are required for septin interaction, localization, and function. Mol Cell Biol. 2003;23(8):2762–2777. doi: 10.1128/MCB.23.8.2762-2777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy DB, Gray RO, Grasser WA, Pollard TD. Direct demonstration of actin filament annealing in vitro. J Cell Biol. 1988;106(6):1947–1954. doi: 10.1083/jcb.106.6.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310(5746):310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 27.Tang X, Punch JJ, Lee WL. A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle. 2009;8(19):3182–3190. doi: 10.4161/cc.8.19.9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120(4):923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312(5991):237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 30.Estensen RD, et al. Cytochalasin B: Microfilaments and “contractile” processes. Science. 1971;173(3994):356–359. doi: 10.1126/science.173.3994.356. [DOI] [PubMed] [Google Scholar]
- 31.Field CM, et al. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol. 1996;133(3):605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meseroll RA, Howard L, Gladfelter AS. Septin ring size scaling and dynamics require the coiled-coil region of Shs1p. Mol Biol Cell. 2012;23(17):3391–3406. doi: 10.1091/mbc.E12-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romberg L, Simon M, Erickson HP. Polymerization of Ftsz, a bacterial homolog of tubulin. Is assembly cooperative? J Biol Chem. 2001;276(15):11743–11753. doi: 10.1074/jbc.M009033200. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Bjornson K, Redick SD, Erickson HP. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys J. 2005;88(1):505–514. doi: 10.1529/biophysj.104.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skillman KM, et al. The unusual dynamics of parasite actin result from isodesmic polymerization. Nat Commun. 2013;4:2285. doi: 10.1038/ncomms3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vrabioiu AM, Mitchison TJ. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 2006;443(7110):466–469. doi: 10.1038/nature05109. [DOI] [PubMed] [Google Scholar]
- 37.DeMay BS, Noda N, Gladfelter AS, Oldenbourg R. Rapid and quantitative imaging of excitation polarized fluorescence reveals ordered septin dynamics in live yeast. Biophys J. 2011;101(4):985–994. doi: 10.1016/j.bpj.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.