Significance
Strigolactones are a new class of plant hormones regulating plant shoot and root architecture in response to the environment. Also present in root exudates, strigolactones stimulate the germination of parasitic plant seeds. This report describes a genomic polymorphism—associated with the Indica/Japonica subspecies divide in rice that has a major impact on the biosynthesis of strigolactones, plant tillering, and germination of the parasitic plant Striga hermonthica—consisting of the deletion of two strigolactone biosynthetic genes orthologous to Arabidopsis MAX1. Both of these genes rescued the Arabidopsis max1-1 highly branched mutant phenotype and increased the strigolactone level when overexpressed in the Indica rice variety Bala. This finding is of great interest for plant physiologists, plant evolutionary biologists, and breeders.
Keywords: QTL, plant hormone, CYP450
Abstract
Rice (Oryza sativa) cultivar Azucena—belonging to the Japonica subspecies—exudes high strigolactone (SL) levels and induces high germination of the root parasitic plant Striga hermonthica. Consistent with the fact that SLs also inhibit shoot branching, Azucena is a low-tillering variety. In contrast, Bala, an Indica cultivar, is a low-SL producer, stimulates less Striga germination, and is highly tillered. Using a Bala × Azucena F6 population, a major quantitative trait loci—qSLB1.1—for the exudation of SL, tillering, and induction of Striga germination was detected on chromosome 1. Sequence analysis of the corresponding locus revealed a rearrangement of a 51- to 59-kbp stretch between 28.9 and 29 Mbp in the Bala genome, resulting in the deletion of two cytochrome P450 genes—SLB1 and SLB2—with high homology to the Arabidopsis SL biosynthesis gene, MAX1. Both rice genes rescue the Arabidopsis max1-1 highly branched mutant phenotype and increase the production of the SL, ent-2′-epi-5-deoxystrigol, when overexpressed in Bala. Furthermore, analysis of this region in 367 cultivars of the publicly available Rice Diversity Panel population shows that the rearrangement at this locus is a recurrent natural trait associated with the Indica/Japonica divide in rice.
The root parasitic Striga spp. parasitize on roots of crops in tropical and subtropical areas. The species typically parasitize cereals, including economically important crops such as maize, sorghum, millet, and rice (1). The parasitic relationship is dependent on the ability of the parasite to detect the host, which is mediated by the perception of strigolactones (SLs), molecules exuded by the host into the rhizosphere, by the seeds of the parasite (2). SLs are also signaling molecules for the establishment of the symbiosis with arbuscular mycorrhizal (AM) fungi (3) that help the plant to improve nutrient uptake. Under low phosphate availability, SL exudation into the rhizosphere is strongly enhanced, hence promoting AM symbiosis (4). As a negative consequence, however, agricultural areas with poor soils and low fertilizer input are strongly affected by Striga (1, 4). In addition to their rhizosphere role, SLs also function as plant hormones inhibiting shoot branching and modulating root architecture (5–7), also in response to phosphate deficiency (8, 9). SL biosynthesis or signaling mutants have increased axillary bud outgrowth, resulting in a bushy and dwarf phenotype (10). Biosynthesis of SLs proceeds through isomerization of β-carotene by β-CAROTENE ISOMERASE (D27), followed by cleavage by CAROTENOID CLEAVAGE DIOXYGENASE 7 (CCD7) and CAROTENOID CLEAVAGE DIOXIGENASE 8 (CCD8), which results in the formation of carlactone (11–16). The gene(s) responsible for the conversion of carlactone to a SL has/have not been identified, although MORE AXILLARY GROWTH 1 (MAX1), encoding a cytochrome P450 (CYP) in Arabidopsis, has been suggested to be a candidate (8, 11, 17, 18). SL signaling is mediated by an F-Box protein (MAX2 in Arabidopsis; D3 in rice) and an α/β-hydrolase protein (D14) (5, 6, 19, 20).
In the present study, molecular genetics was used to further elucidate the SL biosynthetic pathway. We had observed that the rice cultivars Bala and Azucena differ greatly in SL biosynthesis and susceptibility to Striga infection. The Bala × Azucena F6 recombinant inbred line (RIL) population was used to map quantitative trait loci (QTL) related to SLs. A major QTL was detected explaining most of the variation in the concentrations of all five SLs detected in rice exudates. This locus was also detected as a QTL for rice–Striga interaction in a previous study that used the same population (21). Here we show that the QTL is due to a rearrangement of a 51- to 59-kbp stretch between 28.9 and 29.0 Mbp of chromosome 1 in the Bala genome. This rearrangement results in the deletion of two CYP genes, which we show are orthologs of the Arabidopsis MAX1. The rearrangement of this locus is a recurrent natural trait, observed in several rice cultivars.
Results
Rice Varieties Bala and Azucena Show Differential Susceptibility to Striga hermonthica Infection.
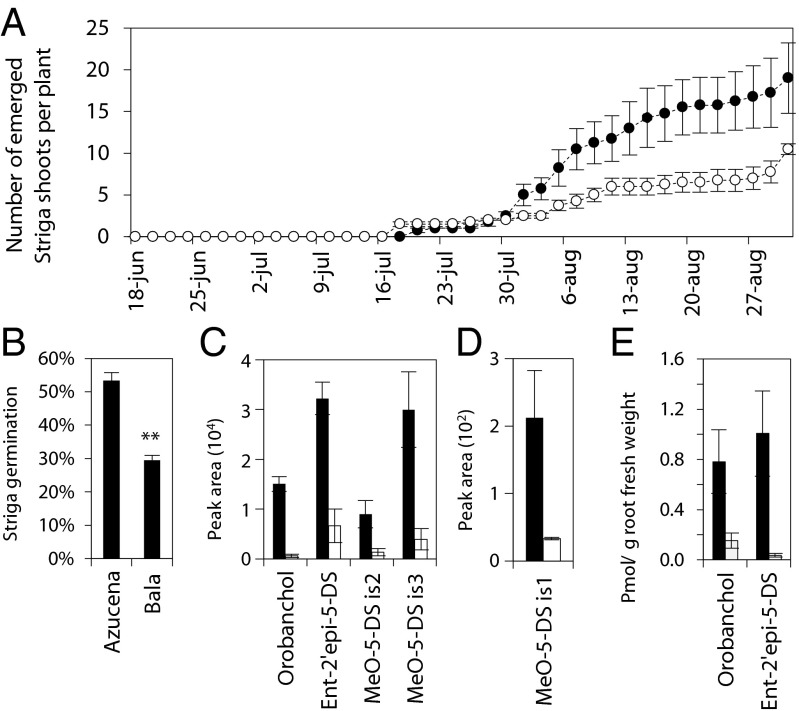
When the two parental lines were grown in soil infected with Striga seeds, emergence of Striga occurred fastest on Bala, and during the 2 wk after first emergence, there was on average one Striga shoot in Azucena and two in Bala (Fig. 1A). After 2 wk, however, the number of Striga shoots increased more rapidly in Azucena, and after 5 wk Azucena had on average 19 Striga shoots, whereas Bala had only 10.
Fig. 1.
Analysis of the parental lines of the RIL population Bala × Azucena. (A) Emergence of Striga shoots per pot during a period of 78 d (●, Azucena; ○, Bala). (B) Germination percentage of Striga seeds exposed to crude exudates of the parental lines. (C and D) LC-MS peak areas of orobanchol, ent-2′-epi-5-deoxystrigol (ent-2′-epi-5-DS), and methoxy-5-deoxystrigol isomers 1–3 (MeO-5-DS is) in root exudates. (E) Orobanchol and ent-2′-epi-5-deoxystrigol in root extract. Error bars represent SEM [n = 4 (A); n = 3 (B–E)]. (B–E) Plants were grown under low-P nutrition for 1 wk, before exudate collection. (C–E) Filled bars correspond to Azucena and open bars to Bala.
Low Striga Infection Rate in Bala Correlates with Reduced SL Exudation.
To investigate whether the difference in Striga emergence and infection between Bala and Azucena is the result of differences in SL exudation, root exudates and extracts of the parental lines were analyzed. Azucena root exudates induced a higher percentage of Striga seed germination than Bala exudates (Fig. 1B). Liquid chromatography (LC)-MS analysis of root exudates and extracts of Bala and Azucena showed that the higher germination in Azucena root exudate correlates with higher amounts of the SLs orobanchol, ent-2′-epi-5-deoxystrigol, and three methoxy-5-deoxystrigol isomers in Azucena (22, 23) (Fig. 1 C–E).
QTL Mapping of SL Levels and Related Phenotypes.
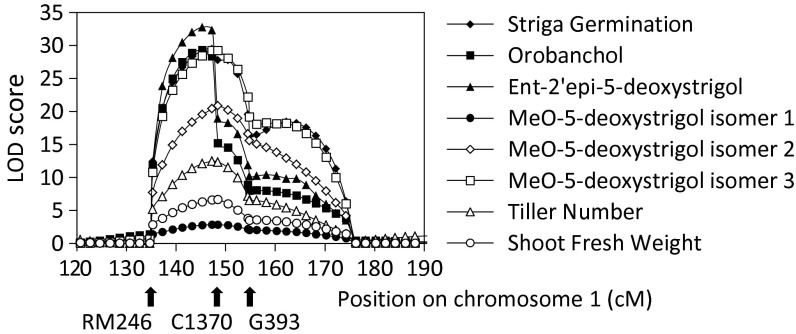
Given the different amounts of SLs found in the parental lines, we used the Bala × Azucena mapping population (21) to map Striga germination, the level of SLs in root exudates, shoot and root fresh weight (fwt), and tillering. The broad sense heritabilities of all traits were high (78–97%) with the exception of methoxy-5-deoxystrigol isomer 1 (44%). A major QTL for Striga germination, qSTRIGOLACTONE BIOSYNTHESIS 1.1 or qSLB1.1 [logarithm of the odds ratio (LOD) = 29.42, R2 = 66%], was identified on chromosome 1 at 143,8 cM near marker C1370 (Fig. 2, Table S1, and Fig. S1). At the same position, QTLs were detected for the levels of orobanchol (LOD = 29.25, R2 = 70.2%), ent-2′-epi-5-deoxystrigol (LOD = 32.7, R2 = 71.2%), and methoxy-5-deoxystrigol isomers 1, 2, and 3 (LOD = 2.83, R2 = 6.7%; LOD = 20.83, R2 = 49.2%; and LOD = 29.17, R2 = 52.8%, respectively). For all these traits, the positive allele was from Azucena. In addition, tiller number (LOD = 12.43, R2 = 28.8%) and shoot fwt (LOD = 6.64, R2 = 14.6%) mapped to this region, however, with the positive effect from Bala. Minor QTLs for SL levels and tillering mapped to chromosome 6 and 10 (Table S1 and Fig. S1). Although these QTLs have lower LOD scores (2.5–4.6), the colocalization of QTLs for SL level and a SL-related phenotypic trait makes them interesting candidate loci for additional SL regulatory, biosynthetic, and/or signaling genes.
Fig. 2.
LOD score distribution of the QTLs for SL production (MeO-5-DS, methoxy-5-deoxystrigol), Striga germination, tiller number, and plant shoot fwt that colocate on rice chromosome 1. The 5% genome-wide threshold for QTL detection was determined after 1,000 permutations and ranged from LOD 3.3 to 4.2. Arrows mark the position of the three closest markers.
Molecular Analysis of the Major QTL on Chromosome 1 Reveals a Genome Rearrangement in Bala.
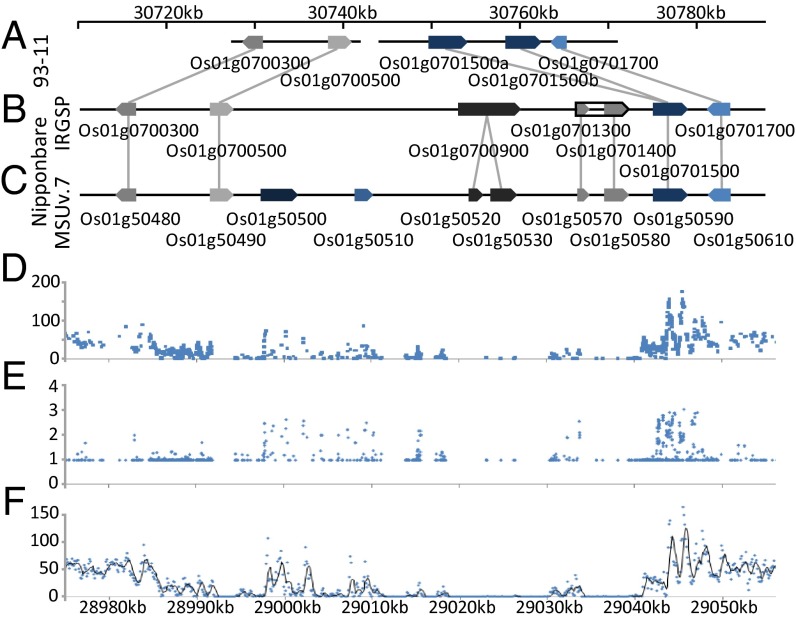
The genomic region for the major QTL contained a genomic rearrangement (Fig. 3). Alignment of the genomic sequence and predicted genes of Indica cultivar 93-11 from the Beijing Institute of Genomics (http://rise2.genomics.org.cn/page/rice/download.jsp) with the International Rice Genome Sequencing Project (IRGSP-1.0: http://rapdb.dna.affrc.go.jp/viewer/gbrowse/irgsp1/) (24, 25) Nipponbare reference sequence showed the following: (i) three predicted Nipponbare genes between 30,750 and 30,771 kbp are missing in the Indica sequence; (ii) there is a gap in the Indica sequence within that region; and (iii) the Indica cultivar appears to have two genes with homology to a single gene, Os01g0701500, in Nipponbare (Fig. 3 A–C). Alignment of Bala genomic sequence reads (88- to 120-bp lengths at 55× genome coverage) to the IRGSP Nipponbare reference sequence revealed a stretch of between 51 and 59 kbp with a Bala read depth close to zero (Fig. 3F). This alignment also confirms that Os01g0701500 is duplicated (Fig. 3 D–F) and that the Bala genes in this region share 100% homology with the 93-11 Indica sequence. Two of the genes in the rearranged region that appear to be missing in 93-11 and Bala, have conserved domains that classify them as CYP (Os01g0700900 and Os01g0701400). At the border of the deleted region, Os01g0701500, which is present in Nipponbare and is duplicated in 93-11 and Bala, also contains this conserved domain. When annotation of the recent Build 5 of the IRGSP-1.0 was compared with the MSU v.7 annotation [Rice Genome Annotation Project (RGAP)], it was found that Os01g0700900 is split into two genes, LOC_Os01g50520 and LOC_Os01g50530 (Fig. 3 B and C). Os01g0701300 and Os01g0701400 correspond to LOC_Os01g50570 and LOC_Os01g50580, respectively, whereas Os01g0701500 is the same as LOC_Os01g50590 (Fig. 3 B and C).
Fig. 3.
Alignment of genomic sequences of cultivar 93-11, Nipponbare, and Bala in the region where the major QTL for SL production was identified. (A) Gene models for cultivar 93-11 were adapted from the genome browser of the Beijing Institute of Genomics (http://rice.genomics.org.cn/rice/index2.jsp). (B) Gene models for cultivar Nipponbare as predicted in the genome browser of the International Rice Genome Sequencing Project (IRGSP-1.0; http://rapdb.dna.affrc.go.jp/viewer/gbrowse/irgsp1/). Os01g0700900 was confirmed by RACE PCR; Os01g0701300 and Os01g0701400 turned out to be one single gene indicated with black outline. (C) Annotation by the genome browser of the Rice Genome Annotation Project (RGAP) (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/). Genes that are homologous between the different annotations are linked by a line. (D–F) Alignment of the 55x coverage of Bala Illumina sequence reads to the MSU Nipponbare reference. Shown are SNP frequency (D), ratio of Bala sequence read depth to SNP frequency (E), and Bala sequence read depth (F). Note the evidence of a deletion between 28,985 and 29,040 kb because read depth and SNP frequency are close to zero (except around the transposon Os01g50500 and the expressed protein Os01g50510), and also note the evidence of a duplication for Os01g50590 because both read depth and SNP frequency are high and a ratio of read depth to SNP frequency of 2 indicates that two alleles align to the same place. Note the coordinates at the bottom of the image that refer to the RGAP (MSU; Version 7) annotation to which Bala sequences were aligned and do not match the IRGSP build5 coordinates that are indicated at the top of the image.
The Rearranged Region on Bala Chromosome 1 Contains MAX1 Homologs.
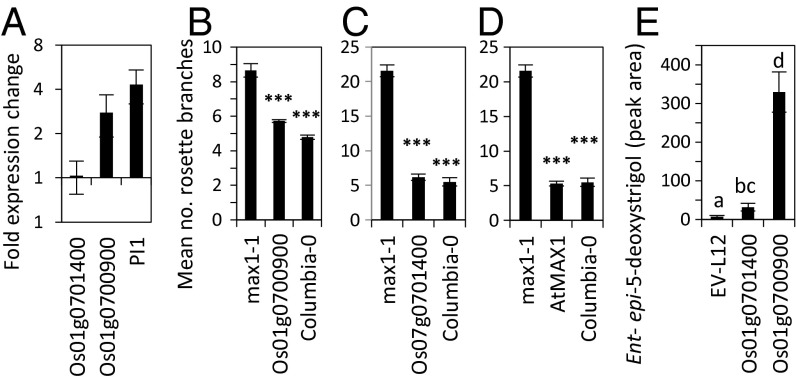
A BLASTX search, using the predicted ORFs of the deleted genes Os010700900 and Os01g0701400, revealed high similarity to the SL biosynthetic gene MORE AXILLARY BRANCHES 1 (At2g26170) from Arabidopsis, which belongs to the CYP711A1 family. We could confirm experimentally by RACE PCR that the IRGSP annotation for Os01g0700900 is correct, although with different splicing form and start codon (Genbank accession no.: JX235697, Fig. S2A), whereas the RGAP annotation for Os01g50520/30 is incorrect. We could also confirm that Os01g0701300 and Os01g0701400 and the corresponding RGAP-annotated Os01g50570/80 are one single gene, which from here on will be referred to as Os01g0701400 (Genbank accession no.: JX235696; Fig. S2B). The protein sequences of Os01g0700900 and Os01g0701400 show, respectively, 57.6% and 60.3% identity to AtMAX1 (Fig. S3). Quantitative RT-PCR (qRT-PCR) showed that expression of Os01g0700900 in Nipponbare was induced by low-P treatment—just as expression of the P starvation marker OsPI1 (26)—but not of Os01g0701400 (Fig. 4A).
Fig. 4.
Expression analysis (A) and functionality of rice CYP genes Os01g0700900 and Os01g0701400 in Arabidopsis (B–D) and rice (E). (A) Regulation of gene expression of Os01g0700900, Os01g0701400, and the low-P marker gene PI by low P nutrition measured by qRT-PCR (n = 4–5). (B–D) Complementation of axillary bud outgrowth of max1-1 by overexpression of Os01g0700900 (B), Os01g07001400 (C), and AtMAX1 (D) under control of the 35S promoter (n = 2–9). Significance values (Student t test) are shown as follows: *P < 0.05; **P < 0.01); ***P < 0.001. (E) Levels of ent-2′-epi-5-deoxystrigol measured in root exudates of Bala cultivar transformed with Os01g0701400 or Os01g0700900 under control of the 35S promoter and empty vector control pHm43GW (n = 2–4). Different letters indicate significantly different means at P < 0.05, using ANOVA followed by Student t test on log-transformed data. Values shown in the graph are back transformed. Bars represent mean values ± SE.
Os01g0700900 and Os01g0701400 Rescue the Branched Phenotype of Arabidopsis max1.
Arabidopsis max1-1 was transformed with the Os01g0700900 and Os01g07001400 cDNA under the control of the CaMV35S promoter (p35S). p35S:AtMAX1 was used as a positive control. Os01g0700900, Os01g07001400, and AtMAX1 all fully restored the branching phenotype of max1-1 (P < 0.001) (Fig. 4 B–D and Fig. S4), showing that these two rice CYP450 genes are AtMAX1 orthologs.
Os01g0700900 and Os01g0701400 Increase SL Levels in Root Exudates of Bala.
Bala was independently transformed with Os01g0700900 and Os01g0701400, driven by the p35S promoter. The levels of ent-2′-epi-5-deoxystrigol in root exudates of P-starved p35S:Os01g0700900- and p35S:Os01g0701400-transformed T1 Bala plants (for which transgene expression was confirmed) both significantly increased compared with the empty vector control, but the effect of p35S:Os01g0700900 was much stronger than that of p35S:Os01g0701400 (Fig. 4E).
Rearrangement in Rice Chromosome 1 Is Associated with Low SL Levels in a Collection of Accessions.
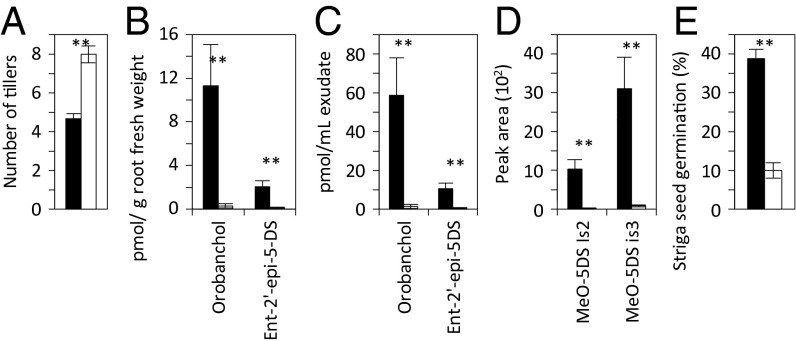
To establish the presence of the Bala rearrangement on chromosome 1 across rice germplasm, a PCR assay was developed using multiplexed primers for three genes (Os01g0700900, Os01g0701400, and Os01g0701500), which all give a product in Azucena, but for which only Os01g0701500 gives a product in Bala. The multiplex test was applied to 367 cultivars of the publicly available Rice Diversity Panel. In the indica and aus subpopulations within the Indica subspecies (27) the Nipponbare/Azucena allele frequency was 3/74 (4.1%) and 4/59 (6.8%), respectively, whereas in the temperate and tropical japonicas within the Japonica subspecies (27) it was 93/96 (96.9%) and 63/94 (67%), respectively, showing that the genome rearrangement is associated with the Indica/Japonica divide in rice. To evaluate how the deletion affects SL biosynthesis in different genetic backgrounds, the SL content of root exudates and root extracts was analyzed in pairs of cultivars that differed in the allele under study but were otherwise from the same subspecies and the same country of origin (Table 1). The lines containing the Bala alleles (carrying the rearrangement) had more tillers, exuded lower amounts of SL, had lower SL root content, and induced lower Striga germination (Fig. 5 and Table S2) than the genotypes containing the Azucena alleles in both the Indica and the Japonica genetic backgrounds.
Table 1.
Lines from the Diversity Panel selected to test the impact of the genomic rearrangement on SL levels, tillering, and Striga germination
| Line | Subgroup* | Country of origin |
| Azucena allele | ||
| Azucena | TRJ | Philippines |
| DZ-78 | AUS | Bangladesh |
| Aswina-330 | AUS | Bangladesh |
| Kun-Min-TH | IND | China |
| Ta-Mao-T | TEJ | China |
| Sinaguing | TRJ | Philippines |
| Bala allele | ||
| Bala | IND | India |
| Dhala-S | AUS | Bangladesh |
| Kachilon | AUS | Bangladesh |
| Guan-Yin-T | IND | China |
| Sung-Liao-2 | TEJ | China |
| Asse Y Pung | TRJ | Philippines |
| Kinastano | TRJ | Philippines |
According to Zhao et al. (27).
Fig. 5.
Impact of the rearrangement on the number of tillers (A), the SL content in root tissue (B) and root exudates (C and D), and the stimulation of Striga germination by root exudates (E). (B and C) Orobanchol and Ent-2′-epi-5-deoxystrigol (ent-2′-epi-5-DS) are determined in picomoles per gram fwt (B) or picomoles per milliliter exudate (C). (D) Levels of methoxy-5-deoxystrigol isomers 1–2 (MeO-5-DS is) in root exudates are determined in peak area. The lines used for this comparison are listed in Table 1. Bars represent the mean values ± SE obtained from lines containing the Azucena (filled bars) or the Bala (open bars) allele with the rearrangement. Significance values (Wilcoxon rank sum test) are shown as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
A strong QTL for SL production in rice—qSTRIGOLACTONE BIOSYNTHESIS (qSLB1.1)—was mapped to chromosome 1. In this locus the Bala genome contains a 51- to 59-kbp rearrangement compared with Nipponbare. The rearrangement spans two CYP genes (Os01g0700900, SLB1; and Os01g0701400, SLB2) that are present in Azucena but absent in Bala. Both genes share high similarity with AtMAX1, the CYP that is required for the biosynthesis of SLs (8, 17). Overexpression of either of the two genes in Arabidopsis max1-1 rescued the branched mutant phenotype, and overexpression in Bala—particularly of Os01g0700900—increased the level of ent-2′-epi-5-deoxystrigol in the root exudate. Although it is possible that there are other elements or genomic features present in the rearranged region that may also contribute to the differential expression of SLs, the results show that Os01g0700900 and Os01g0701400 contribute to SL biosynthesis in rice and that the deletion of these genes in the Bala genome causes the low SL levels that are observed.
Our SL analyses show an overall reduction of all SL in Bala exudates compared with Azucena but no differences in the composition (Fig. 1C). This result suggests that the MAX1 orthologs in rice discovered in the present work contribute to the synthesis of all SLs present in rice, possibly at an earlier step of the biosynthetic pathway, rather than to the biosynthesis of specific structural SL variants. The big difference in SL levels driven by the Azucena allele also suggest that the SL biosynthetic CYP genes characterized in this study make an important contribution to SL biosynthesis across different genetic backgrounds. Nevertheless, SLs are still produced in Bala, showing that there must be redundancy for this biosynthetic step. In Arabidopsis, only a single MAX1 ortholog is present, but indeed in rice, besides the two CYP genes described in the present study, three other CYP genes homologous to MAX1 are present in the rice genome: Os01g0701500, Os06g0565100, and Os02g0221900. Two of these, Os02g0221900 and Os06g0565100, also rescued the branched phenotype of Arabidopsis max1-1 (28), suggesting that they have MAX1-like activity as well. In fact, several other monocotyledonous species such as maize and sorghum have two to five MAX1 orthologs, whereas in dicotyledonous species such as petunia and Medicago, generally only one and sometimes two are present (28, 29).
Gene duplication allows for diversification in metabolic regulation. This diversification was also observed for Os01g0700900 and Os01g0701400 in the present study. The expression of Os01g0700900 was increased by P starvation but not that of Os01g0701400 (Fig. 4A). In line with this finding, the expression of Os01g0700900 and Os02g0221900, as well as the SL biosynthetic genes D10, D17, and D27, was repressed by P replenishment, whereas expression of Os01g07001400, Os01g07001500, and Os06g0565100 were not (9). Finally, the levels of ent-2′-epi-5-deoxystrigol obtained in root exudates of Bala overexpressing Os01g0701400 were considerably lower than those of Bala overexpressing Os01g0700900 (Fig. 4E). This difference may be caused by diversification in gene function after duplication, resulting in differences in enzymatic efficiency or specificity. Combined, these observations suggest that MAX1 duplication has led to diversification in the regulation of SL biosynthesis in rice and other grass species, but not in dicots, for as-yet-unknown reasons. The existence of multiple MAX1 orthologs in the rice genome does explain why mutant screens have been unsuccessful in detecting these genes, whereas in Arabidopsis this approach was successful (10). It also demonstrates the power of QTL mapping to address more fundamental questions, such as plant hormone biosynthesis, because it allowed us to obtain more insight into the SL biosynthetic pathway in rice. Two rice MAX1 orthologs were revealed, and additional QTLs with minor influence on SL levels and tillering were also detected on chromosomes 6 and 10.
Population genetic analysis provides insights into the evolutionary history of genomes and traits, and our analysis of a Rice Diversity Panel revealed a striking difference in the frequency of the rearrangement found in this study between the two major rice varietal groups. The prevalence of the high-SL Azucena allele in the Japonica group and its virtual absence in the Indica group suggests that it is likely to have originated in a Japonica ancestor. By extension, this finding also supports the hypothesis that Indica and Japonica were domesticated from divergent populations of their wild ancestor, Oryza rufipogon (30). The fact that the Indica low-SL allele is present in ∼33% of the tropical japonica varieties in the Diversity Panel is consistent with introgression from indica and aus varieties into tropical japonica varieties growing sympatrically in tropical environments. This pattern of allele divergence and introgression in Oryza sativa has been documented in other studies (31–34).
SLs regulate overall plant architecture. In shoots, SLs inhibit tillering; in roots, SLs influence root and root hair elongation and lateral root development (5, 7, 9, 35, 36). Indeed, tillering mapped to the same SL locus on chromosome 1. Interestingly, in previous QTL studies using an IR64–Azucena and the Bala–Azucena mapping populations, several root architectural traits were mapped in the same region, suggesting that they may also be controlled by this locus (37, 38). These findings emphasize the potential of qSLB1.1 for the improvement of important agronomic traits in rice. Our study demonstrates once more the positive correlation between SL production and Striga germination (22, 39). However, although decreased SL biosynthesis results in less Striga germination, it is unclear whether SL levels also affect Striga attachment. In earlier work, Striga tolerance mapped to the same region as qSLB1.1, with the Azucena allele increasing Striga tolerance (21), even though in the present study we showed there were more Striga emerging on Azucena in controlled environments (Fig. 1A). The later establishment of infection in Azucena likely explains the higher tolerance to Striga, similar to what was observed in sorghum, where tolerant varieties generally exhibit later Striga emergence (40). Intriguingly, Striga postattachment resistance mapped to the same position on chromosome 1 as the QTL discussed in the present work (near marker C1370) in a cross between Nipponbare and Kasalath (21), with the Nipponbare allele conferring greater resistance (41). Nipponbare contains the same allele as Azucena, and we confirmed that Kasalath carries the same allele as Bala for the locus under study. This observation offers the intriguing hypothesis that higher SL levels increase Striga germination but reduce the subsequent efficiency of parasitization.
Materials and Methods
Mapping Population Plant Material.
A mapping population of 115 F6 RILs derived from Bala × Azucena described in Price et al. (42) was used. The experiments were conducted under controlled conditions (28 °C/25 °C; 450 µM⋅m−2⋅s−1; 10-h light/14-h dark; and 70% relative humidity) in randomized design with three replicates, each consisting of one pot with five plants. After root exudate collection, the number of tillers per pot was counted, plants were removed from the pots, and root and shoot fwt were determined. Allelic frequencies of the rearrangement in the different subpopulations were assayed by using 367 diverse accessions from the publicly available Rice Diversity Panel (27, 34). Eleven of these lines (Table 1) were multiplied at the University of Aberdeen and sent to Wageningen University for physiological characterization. Arabidopsis growing conditions are described in SI Materials and Methods, Complementation of Arabidopsis max1-1 Mutants.
Complementation of Bala.
Transformation of Bala with constructs p35S:Os01g0700900, p35S:Os01g0701400, and empty vector pHm43GW is described in SI Materials and Methods, Rice Bala Variety Complementation. Transgenic T1 plants were selected for exudate collection after confirmation on selection medium with hygromycin and verification of transgene expression by qRT-PCR. Equivalent average expression levels of the transgenes were ensured when plants were pooled and transferred to pots (three plants per pot) for root exudate collection (see below). Statistical tests were performed by using Genstat (Genstat for Windows 15th Edition; VSN International).
SL Collection from Root Exudates and Root Extracts.
For the mapping population, SLs were collected from 5-wk-old rice plants in three replicates with each replicate consisting of one pot with five plants as described (39). For the transgenic rice, root exudates were collected from pots containing three 4-wk-old plants. The exudates were collected at 3, 6, and 9 d after the start of P starvation, and the three samples were pooled for SL analysis. The root exudates were passed through an SPE C18-Fast column (500 mg per 3 mL), and the SL was eluted with 6 mL of 100% acetone. For root extracts, 1 g fwt of ground root tissue was extracted following the method described (39), but the resulting extracts were evaporated to dryness, taken up in hexane, loaded on preequilibrated Silica gel Grace Pure SPE (200 mg/3 mL) columns, and eluted with 2 mL of hexane:ethyle acetate (1:9) for further purification. The solvent was evaporated, and the residue was redissolved in 200 µL of 25% (vol/vol) acetonitrile in water and filtered through Minisart SRP4 0.45-µm filters (Sartorius) before LC–tandem MS (LC-MS/MS) analysis.
SL Analysis Using LC-MS/MS.
SLs were analyzed by comparing retention times and mass transitions with those of SL standards using a Waters Xevo TQ mass spectrometer equipped with an electrospray-ionization source and coupled to a Waters Acquity ultraperformance LC system using the settings described (39) with some modifications specified in SI Materials and Methods, Detection and Quantification of Strigolactones by Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS). The analyses were performed in three biological replicates.
Striga Germination Bioassay.
Root exudate germination stimulatory activity was assessed by using a germination bioassay with S. hermonthica as described (39, 43). Approximately 50–100 preconditioned Striga seeds on a 9-mm diameter glass fiber filter paper disk (Sartorius) were exposed to the column-purified root exudates (50 μL per disk) after acetone evaporation. Germination was scored after 48-h incubation in darkness at 30 °C. The synthetic SL GR24 (3.3 µM) was used as positive and water as negative control. Three biological replicates and three discs per replicate were used.
Striga Emergence.
Striga emergence was studied as described (39). Approximately 25 mg of Striga seeds were mixed thoroughly with 1 L of the 50:50 sand and soil mixture, which was then used to fill 1.5-L pots. Pregerminated rice seeds were transferred to the pots (one seed per pot) and grown at 28 °C day (14 h) and 25 °C night (10 h) with relative humidity at 70% and 400 µM⋅m−2⋅s−1 of light. Striga emergence was assessed at 2-d intervals in four replicates.
RNA Extraction.
RNA was extracted from roots of Nipponbare rice grown with full nutrition or P deprived for 5 d before tissue collection of roots. The RNA was purified from 70 mg of homogenized ground roots using 500 ml Trizol (Invitrogen) and further purified with chloroform. After precipitation with 70% (vol/vol) ethanol, the RNA was recovered with an RNAeasy Mini Kit column (Qiagen) and DNA was removed using the DNAase I Kit (Qiagen), according to manufacturer's instructions.
Gene Expression Analysis.
cDNA was synthetized by using the iScript cDNA Synthesis Kit (BioRad) using 1 µg of total RNA per sample following the manufacturer’s instructions. The qRT-PCR reactions were prepared by using iQ SYBR Green Supermix (BioRad). Per reaction, each primer at a concentration of 0.3 µM and 1 µL of 10-fold diluted template cDNA was used. The amplification was detected with a BioRad qRT-PCR detection system and thermocycler. The primers are listed in Table S3. The expression data are the average of five (control) or four biological replicates (P deprived).
Characterization of Os01g0700900 and Os01g0701400 Transcripts.
RNA from Nipponbare rice roots was used for cDNA synthesis according to instructions for the SMART RACE cDNA Amplification Kit (Clontech). The primers and nested primers of RACE PCR were designed based on the predicted mRNA sequences in the National Center for Biotechnology Information (NCBI) (GI: 115439412; accession no. NM_001050521) and are listed in Table S4. The 5′ and 3′ RACE PCR products were cloned into pJET1.2 (Fermentas) and sequenced.
Complementation of Arabidopsis max1-1.
Constructs p35S:Os01g0700900 and p35S:Os01g0701400 were transformed into Arabidopsis max1-1 plants (Columbia-0 background) as described in SI Materials and Methods, Complementation of Arabidopsis max1-1 Mutants. Rosette branching was measured on independent single-insertion homozygous lines: nine lines (20 plants per line) for p35S:Os01g0700900, five lines (three to five plants per line) for p35S:AtMAX1, and two lines (five to seven plants per line) for p35S:Os01g0701400. Rosette branching was measured by using the decapitation method (44).
Phylogenetic Studies of Rice MAX1 Homologs.
The confirmed amino acid sequences of Os01g0700900 and Os01g0701400 and the predicted sequences of Os01g0701500, Os06g0565100, and Os02g0221900 in IRGSP were aligned with Arabidopsis MAX1, and a phylogenetic tree made with ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2).
QTL Mapping.
Before QTL analysis, germination percentage and SL production were log-transformed. QTL analysis was conducted as described in Price et al. (45). The molecular map (46) contains 164 markers covering 1,833 cM on 12 linkage groups. The identification of QTLs was performed by composite interval mapping using QTLCartographer (47). Background markers (maximum of 10) for composite interval mapping were selected by forward stepwise regression with backward elimination using the default setting. The 5% genome-wide threshold for QTL detection was determined after 1,000 permutations.
Bala and Azucena Genome Sequencing.
The Bala and Azucena genomic DNA was extracted, made into pair-end libraries, and sequenced on an Illumina Genome Analyzer II at Cornell University providing reads of 88-, 100-, and 120-bp lengths for Bala and 100 bp lengths for Azucena. To report SNP calls, reads were aligned to the Nipponbare reference by using Panati (48) and ref. 49. Fastq data have been deposited in the NCBI Short Read Archive (Acc_ID SRA050654.1).
Assessing Allelic Diversity.
Allelic diversity was accessed by multiplex PCR in a single reaction (25-µL mix) of 5-min denaturation at 95 °C, 35 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min, and 5 min of final extension at 72 °C. Primers are listed in Tables S5.
Supplementary Material
Acknowledgments
We thank Koichi Yoneyama (Utsunomiya University, Japan), Yukihiro Sugimoto (Kobe University, Japan), and Tadao Asami (University of Tokyo, Japan) for the kind provision of ent-2′-epi-orobanchol, orobanchol, 5-deoxystrigol, 2′-epi-5-deoxystrigol, and [2H]6-2′-epi-5-deoxystrigol; and Binne Zwanenburg (University of Nijmegen, The Netherlands) for providing GR24. A.H.P. and H.J.B. thank Pieter Ouwerkerk for bringing them together. This work was supported by the Higher Education Commission Pakistan (to M.J.); Netherlands Organization for Scientific Research Vici Grant 865.06.002 (to H.J.B.) and Equipment Grant 834.08.001 (to H.J.B.); the Centre for BioSystems Genomics, part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (to H.J.B.); National Science Foundation Plant Genome Research Grant 1026555 (to S.R.M.); US Department of Agriculture National Institute for Food and Agriculture Grant 2009-65300-05661 (to S.R.M.); United Kingdom Biotechnology and Biological Science Research Council (to J.H.); Government of the Rivers State of Nigeria Postgraduate Scholarship (to S.O.N.D.); and National Natural Science Foundation of China Grant 31161130535 (to Y.W.). Bala sequencing was supported by European Union FP6 Project 015468 (“CEDROME”) and a Monitoring Agricultural Resources grant; Azucena sequencing was supported by Biotechnology and Biological Sciences Research Council–Department for International Development Grant BBF0041841.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The rearranged region of chromosome 1 has been deposited in the NCBI Short Read Archive (Acc_ID SRA050654.1). The sequences reported in this paper have been deposited in the GenBank database (accession nos. JX235697 and JX235696 [confirmed coding sequences of SLB1 (Os01g0700900) and SLB2 (Os01g0701400), respectively]).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317360111/-/DCSupplemental.
References
- 1.Parker C. Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci. 2009;65(5):453–459. doi: 10.1002/ps.1713. [DOI] [PubMed] [Google Scholar]
- 2.Cardoso C, Ruyter-Spira C, Bouwmeester HJ. Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp. Plant Sci. 2011;180(3):414–420. doi: 10.1016/j.plantsci.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435(7043):824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 4.Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 2007;12(5):224–230. doi: 10.1016/j.tplants.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Roldan V, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455(7210):189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 6.Umehara M, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455(7210):195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 7.Ruyter-Spira C, et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2011;155(2):721–734. doi: 10.1104/pp.110.166645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohlen W, et al. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 2011;155(2):974–987. doi: 10.1104/pp.110.164640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 2010;51(7):1118–1126. doi: 10.1093/pcp/pcq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domagalska MA, Leyser O. Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol. 2011;12(4):211–221. doi: 10.1038/nrm3088. [DOI] [PubMed] [Google Scholar]
- 11.Alder A, et al. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335(6074):1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- 12.Lin H, et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell. 2009;21(5):1512–1525. doi: 10.1105/tpc.109.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arite T, et al. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007;51(6):1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x. [DOI] [PubMed] [Google Scholar]
- 14.Zou J, et al. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006;48(5):687–698. doi: 10.1111/j.1365-313X.2006.02916.x. [DOI] [PubMed] [Google Scholar]
- 15.Booker J, et al. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol. 2004;14(14):1232–1238. doi: 10.1016/j.cub.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 16.Sorefan K, et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003;17(12):1469–1474. doi: 10.1101/gad.256603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Booker J, et al. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell. 2005;8(3):443–449. doi: 10.1016/j.devcel.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Ruyter-Spira C, Bouwmeester H. Strigolactones affect development in primitive plants. The missing link between plants and arbuscular mycorrhizal fungi? New Phytol. 2012;195(4):730–733. doi: 10.1111/j.1469-8137.2012.04261.x. [DOI] [PubMed] [Google Scholar]
- 19.Arite T, et al. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009;50(8):1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- 20.Hamiaux C, et al. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol. 2012;22(21):2032–2036. doi: 10.1016/j.cub.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Kaewchumnong K, Price AH. A study on the susceptibility of rice cultivars to Striga hermonthica and mapping of Striga tolerance quantitative trait loci in rice. New Phytol. 2008;180(1):206–216. doi: 10.1111/j.1469-8137.2008.02568.x. [DOI] [PubMed] [Google Scholar]
- 22.Jamil M, Rodenburg J, Charnikhova T, Bouwmeester HJ. Pre-attachment Striga hermonthica resistance of New Rice for Africa (NERICA) cultivars based on low strigolactone production. New Phytol. 2011;192(4):964–975. doi: 10.1111/j.1469-8137.2011.03850.x. [DOI] [PubMed] [Google Scholar]
- 23.Xie X, et al. Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol Plant. 2013;6(1):153–163. doi: 10.1093/mp/sss139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawahara Y, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 2013;6(1):4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai H, et al. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54(2):e6. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wasaki J, Yonetani R, Shinano T, Kai M, Osaki M. Expression of the OsPI1 gene, cloned from rice roots using cDNA microarray, rapidly responds to phosphorus status. New Phytol. 2003;158:239–248. [Google Scholar]
- 27.Zhao K, et al. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS ONE. 2010;5(5):e10780. doi: 10.1371/journal.pone.0010780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Challis RJ, Hepworth J, Mouchel C, Waites R, Leyser O. A role for more axillary growth1 (MAX1) in evolutionary diversity in strigolactone signaling upstream of MAX2. Plant Physiol. 2013;161(4):1885–1902. doi: 10.1104/pp.112.211383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond RSM, et al. The expression of petunia strigolactone pathway genes is altered as part of the endogenous developmental program. Front Plant Sci. 2011;2:115. doi: 10.3389/fpls.2011.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23(11):578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 31.Famoso AN, et al. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. 2011;7(8):e1002221. doi: 10.1371/journal.pgen.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweeney MT, et al. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3(8):e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takano-Kai N, et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics. 2009;182(4):1323–1334. doi: 10.1534/genetics.109.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao K, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koltai H, et al. Strigolactones’ effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. J Plant Growth Regul. 2010;29:129–136. [Google Scholar]
- 36.Kapulnik Y, et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta. 2011;233(1):209–216. doi: 10.1007/s00425-010-1310-y. [DOI] [PubMed] [Google Scholar]
- 37.Topp CN, et al. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci USA. 2013;110(18):E1695–E1704. doi: 10.1073/pnas.1304354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Champoux MC, et al. Locating genes associated with root morphology and drought avoidance in rice via linkage to molecular markers. Theor Appl Genet. 1995;90(7-8):969–981. doi: 10.1007/BF00222910. [DOI] [PubMed] [Google Scholar]
- 39.Jamil M, Charnikhova T, Houshyani B, van Ast A, Bouwmeester HJ. Genetic variation in strigolactone production and tillering in rice and its effect on Striga hermonthica infection. Planta. 2012;235(3):473–484. doi: 10.1007/s00425-011-1520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurney AL, Press MC, Scholes JD. Infection time and density influence the response of sorghum to the parasitic angiosperm Striga hermonthica. New Phytol. 1999;143:573–580. doi: 10.1046/j.1469-8137.1999.00467.x. [DOI] [PubMed] [Google Scholar]
- 41.Gurney AL, Slate J, Press MC, Scholes JD. A novel form of resistance in rice to the angiosperm parasite Striga hermonthica. New Phytol. 2006;169(1):199–208. doi: 10.1111/j.1469-8137.2005.01560.x. [DOI] [PubMed] [Google Scholar]
- 42.Price AH, Steele KA, Moore BJ, Barraclough PP, Clark LJ. A combined RFLP and AFLP linkage map of upland rice (Oryza sativa L.) used to identify QTLs for root-penetration ability. Theor Appl Genet. 2000;100:49–56. [Google Scholar]
- 43.Matusova R, et al. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 2005;139(2):920–934. doi: 10.1104/pp.105.061382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greb T, et al. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003;17(9):1175–1187. doi: 10.1101/gad.260703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price AH, Steele KA, Moore BJ, Jones RGW. Upland rice grown in soil-filled chambers and exposed to contrasting water-deficit regimes: II. Mapping quantitative trait loci for root morphology and distribution. Field Crops Res. 2002;76:25–43. [Google Scholar]
- 46.Norton GJ, Price AH. Mapping of quantitative trait loci for seminal root morphology and gravitropic response in rice. Euphytica. 2008;166:229–237. [Google Scholar]
- 47. Basten CJ, Weir BS, Zeng ZB (2001) QTLCartographer (Department of Statistics, North Carolina State University, Raleigh, NC), Version 1.15.
- 48. Wright MH (2009–2012) Panati. Available at http://panati.sourceforge.net/. Accessed June 2012.
- 49.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.